Abstract
Although previous studies on N2-fixing legumes have demonstrated the contribution of acid phosphatases to their phosphorus (P) use efficiency under P-deficient growth conditions, localization of these enzymes in bean nodules has not been demonstrated. In this study, phosphoenol pyruvate phosphatase (PEPase) gene transcripts were localized within the nodule tissues of two recombinant inbred lines, RIL115 (P-deficiency tolerant) and RIL147 (P-deficiency sensitive), of Phaseolus vulgaris. Nodules were induced by Rhizobium tropici CIAT899 under hydroaeroponic conditions with a sufficient versus a deficient P supply. The results indicated that PEPase transcripts were particularly abundant in the nodule infected zone and cortex of both RILs. Analysis of fluorescence intensity indicated that nodule PEPase was induced under conditions of P deficiency to a significantly higher extent in RIL147 than in RIL115, and more in the inner cortex (91%) than in the outer cortex (71%) or the infected zone (79%). In addition, a significant increase (39%) in PEPase enzyme activity in the P-deficient RIL147 correlated with an increase (58%) in the efficiency of use in rhizobial symbiosis. It was concluded that nodule PEPase is upregulated under conditions of P deficiency in the P-deficiency-sensitive RIL147, and that this gene may contribute to adaptation of rhizobial symbiosis to low-P environments.
Key words: nitrogen; nodule; Phaseolus vulgaris; phosphoenol pyruvate phosphatase, phosphorus; transcript; nitrogen
Introduction
Phosphorus (P) is an essential, but limiting macronutrient that plays critical roles in plant metabolism and development. It is the least accessible macronutrient in many soils because it readily forms insoluble calcium salts in alkaline soils, or complexes with iron and aluminum oxides in acidic soils, rendering it inaccessible for root uptake (Richardson et al., 2009). The available P in many soils usually ranges from 1 to 10 µM (Hinsinger, 2001). This is far lower than the intracellular Pi concentrations of 5–20mM that are required for optimal plant growth (Vance et al., 2003; Fang et al., 2009) and legume nodule development (Bargaz et al., 2011a). The legume nodules where symbiotic nitrogen fixation (SNF) takes place are particularly sensitive to low P availability, and N2-fixing legumes usually require more P than plants dependent on mineral nitrogen (N) fertilizers (Serraj and Adu-Gyamfi, 2004).
Two major mechanisms are involved in P nutrition of legumes under P-deficient conditions: (i) increasing P acquisition using such mechanisms as root morphology, root exudation, and P uptake systems; and (ii) enhancing P utilization by internal mechanisms associated with efficient use of P at the cellular level (Raghothama, 1999; Vance et al., 2003). Secretion of acid phosphatases (APases) that hydrolyse esterified P is among the intricate array of adaptations used to enhance P acquisition and utilization from their environment (Vance et al., 2003; Richardson et al., 2009). Induction of intracellular and secreted APases appears to be a universal plant response to nutritional Pi limitation that participates in systemic Pi mobilization from soil organic matter-localized P, including nucleic acids (Tran et al., 2010). Likewise, several studies have suggested that high levels of APases in the rhizosphere may hydrolyse Pi from external organophosphates, which can comprise up to 80% of total soil P (Radersma and Grierson, 2004).
Generally, most APases are non-specific and may hydrolyse Pi from a broad spectrum of Pi mono-esters over a wide pH range. These enzymes may constitute an adaptive mechanism for N2-fixing legumes to tolerate P deficiency, as attested by the increase in the activities of APases and phytase in nodules of P-deficient Phaseolus vulgaris (Araújo et al., 2008). Moreover, a report on APases purified from soybean (Glycine max) nodules suggests that these enzymes are involved in the conversion of purines into ureides during SNF (Penheiter et al., 1997). Moreover, an APase recently identified as PvPAP3 in the roots of P-deficient P. vulgaris plants was most active with ATP as a substrate, suggesting that it might function in acclimation of these plants to Pi starvation through the use of extracellular ATP as a Pi source from the environment (Liang et al., 2010). Likewise, results on the examination of the vegetative vacuole proteome of Arabidopsis thaliana rosette leaf tissue have confirmed that AtPAP26 is the predominant vacuolar APase exhibiting high intracellular APase activities with phosphoenol pyruvate (PEP) as a substrate (Veljanovski et al., 2006, Tran et al., 2010).
The results of the above studies, the marked activation of PEP phosphatase (PEPase) in P. vulgaris roots under conditions of P deficiency (Juszczuk and Rychter, 2002), and the characterization in embryonic axes of P. vulgaris of an APase encoding a dimer enzyme that presents its highest activity against PEP (Yoneyama et al., 2004) led us to assume that PEPase may play a key role in internal nodule P metabolism and thus may contribute to P use efficiency for SNF. Thus, the overall aims of this study were to utilize biochemical and molecular approaches to analyse the effect of P deficiency on both PEPase activity and transcript abundance in nodules of P. vulgaris–rhizobia symbiosis.
Materials and methods
Plant material and growth conditions
Experiments were conducted in a glasshouse under natural light with day/night temperatures of 28/20 °C and a 16h photoperiod with additional illumination of 400 µmol photons m–2 s–1 and 70% relative humidity during the day. This study was carried out using two recombinant inbred lines (RILs) originating from the International Centre of Tropical Agriculture (CIAT), RIL115 and RIL147. The RIL115 and RIL147 lines were characterized previously under conditions of P deficiency as tolerant and sensitive genotypes, respectively, based on plant growth and seed yield in relation to P availability (Drevon et al., 2011). Seeds were surface sterilized with 3% calcium hypochlorite and then washed thoroughly in ten successive baths of sterile distilled water. Thereafter, seeds were germinated for 4 d at 28 ºC in rolls of germination paper soaked in sterile distilled water.
Seedling roots were inoculated with the reference strain Rhizobium tropici CIAT899 grown in liquid yeast extract mannitol medium at 28 °C for 3 d to an approximate cell density of 109 ml–1. Thereafter, the seedlings were transferred into hydroaeroponic culture consisting of vats filled with 40 l of nutrient solution, which were aerated intensely and arranged in a fully randomized block design. The roots of selected uniform seedlings were passed through the hole of a rubber stopper on the vat cover. Cotton wool was fitted at the hypocotyl level to maintain the root system suspended in the nutrient solution (Vadez et al., 1999) with either 75 or 250 µmol P per plant per week, defined as P-deficient or P-sufficient supplies, respectively. Urea was supplied at 2mM per plant into the nutrient solution during the initial 2 weeks of growth to avoid N deficiency during nodule development. Thereafter, the plants were grown in N-free nutrient solution. Each week, the nutrient solution pH was adjusted to around pH 7 by the addition of 0.2g l–1 of CaCO3 and the medium was aerated by an air flow of ambient air at 400ml min–1 during the experiment.
Design of PEPase gene primers with nodule DNA and RNA
Nodules of approximately 3mm diameter for each RIL corresponding to 50±5mg of nodule fresh weight were carefully detached at 42 d after transplantation (DAT) and immediately frozen in liquid N2 and stored at –80 °C until use. Prior to nucleic acid extraction, all solutions and glassware were rendered RNase free by diethyl pyrocarbonate (DEPC) treatment, and only certified RNase- and DNase-free plastic ware was used. Total DNA and RNA extraction was performed using a FastDNA Spin kit for soil, an RNase kit, and a Fast Prep Instrument (MP Biomedicals, Santa Ane, CA, USA). Extraction of DNA and RNA was confirmed and quantified using a Quan-iTTM Pico Green DNA Assay kit and a Quan-iTTM Ribo Green RNA Assay kit (Molecular Probes, Carlsbad, NM, USA) and visualized in an RNase-free agarose gel (1.5%). Yields of approximately 25 µg of DNA and 4 µg of total RNA g–1 of fresh weight of nodule were observed in the total extracts.
Design of the PEPase gene primers was performed online with the only PEPase found in GenBank, an enzyme characterized in Allium cepa (Shinano et al., 2001; GenBank accession no. AB052619.1). The protein sequence was analysed using BLAST against enzymes of plants in the family Fabaceae in the National Center for Biotechnology Information protein database. Subsequently, several primer pairs were designed for use in an in situ RT-PCR approach.
Total nodule RNA was reverse transcribed using Moloney murine leukemia virus (MMLV) reverse transcriptase H– (Promega, Madison, WI, USA) following the manufacturer’s recommendations. The different primer pairs were used to amplify gene products using 30 cycles of 95 °C for 45 s, 60 °C for 30 s, and 72 °C for 45 °C, with final extension at 72 °C for 2min, using cDNA and genomic DNA as templates. The amplified bands were ligated into a pGEM®-T Easy vector (Promega) using bacteriophage T4 DNA ligase, and recombinant plasmids were transformed into Escherichia coli grown on Luria–Bertani/ampicillin/IPTG/X-Gal plates at 37 °C for 24h. The PCR products were sequenced (Beckman Coulter Genomics, UK) to verify the amplification of the desired gene.
Finally, we used the primers PEPdir (5'-ATGTCCATGCTTATGAAAGATC-3') and PEPrev (5'-CTTGCCATCATCATTGCGGT-3') to quantify and localize the P. vulgaris gene transcript of PEPase. These primers show 100% nucleotide identity to a P. vulgaris mRNA for APase (GenBank accession no. AB116720.1; Yoneyama et al., 2004).
In situ RT-PCR of PEPase transcripts
Sample preparation and fixation for in situ RT-PCR were carried out according to the protocol described by Molina et al. (2011). The method involves in situ amplification of specific nucleic acid sequences on nodule sections, followed by fluorescence detection (Van Aarle et al., 2007) of the localized PCR product using epifluorescence microscopy. Briefly, three nodules of 3mm diameter for each RIL and P treatment were harvested from plants at 42 DAT, coinciding with the flowering stage and optimum nodule functioning activities. The nodules were washed with DEPC-treated water and fixed in 4% (v/v) paraformaldehyde, 45% (v/v) ethanol and 5% (v/v) acetic acid, kept for 2h under vacuum, and stored overnight at 4 °C. Fixed nodules were included in 9% (w/v) low-melting-point agarose and cut using a microtome (Micro-cut H1200 Vibrating Microtome; Bio-Rad, Marnes la Coquette, France) into 50 µm thick sections, which were collected into small tubes containing 0.5ml of DEPC-treated water, and freed from residual agarose by three washes with DEPC-treated water heated at 60 °C.
For reverse transcription, the first cDNA strand was synthesized from total RNA of the 50 µm nodules slices using MMLV reverse transcriptase following the manufacturer’s recommendations. Nodules slices were incubated in 40 µl reverse transcriptase mix containing the PEPase gene-specific reverse primer PEPrev. Negative controls were prepared by omitting the reverse transcriptase. The reverse transcriptase mix was removed and 40 µl of PCR mix was added including 0.25 µM each of the gene-specific PEPdir and PEPrev primers. Thermocycling was performed using 30 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 45 s, with extension at 72 °C for 2min.
The amplified cDNA in the fixed tissue was then detected after removing the PCR mix. Samples were washed and incubated in 100 µl of blocking solution under gentle agitation in the dark at 37 °C. The blocking solution was replaced by 100 µl of alkaline phosphatase-conjugated anti-dioxygenin Fab fragment (Roche Diagnostics, Basel, Switzerland) diluted 1:1000 in 2% BSA. The samples were incubated at room temperature for 90min and then washed three times to remove excess antibody. Detection of alkaline phosphatase was carried out using an ELF-97® endogenous phosphatase detection kit (Molecular Probes, Leiden, The Netherlands). Observations were made with an Olympus BX61® microscope (Olympus, Hamburg, Germany) equipped with an epifluorescence condenser, a Hoechst/DAPI longpass filter set configured at an excitation filter of 360–370nm, a dichroic mirror of 400nm and an emission longpass of 420nm. Images were photographed with a grey View II® camera (ORCA AG; Hamamastu) using Analysis® software (Soft Imaging System, Munster, Germany). Image analysis and signal quantification were performed using ImageJ software as an image analysis program. Overall, 16 in situ RT-PCR experiments were performed to visualize the PEPase transcript and ten images per RIL and P treatment were used for statistical analyses of the signal quantification.
PEPase and APase assays
Samples of nodules of 3mm diameter corresponding to 100±5mg of nodule fresh weight were carefully detached from roots at 42 DAT and immediately frozen at –80 °C. Each nodule sample was ground with an extraction mixture consisting of 500 µl of 0.1M sodium acetate buffer (pH 5.6) containing 1mM dithiothreitol. Homogenates were centrifuged at 13 000 g at 4 °C for 30min and aliquots of 50 µl of supernatant were used for PEPase and APase assays, and for Pi determination.
For measurements of PEPase activity, hydrolysis of PEP to pyruvate was coupled to a lactate dehydrogenase reaction and assayed at 25 °C by continuously monitoring the oxidation of reduced NADH at 340nm for 5min (Bozzo et al., 2004). The assay conditions were 50mM sodium acetate (pH 5.6), 2mM PEP, 10mM MgCl2, 0.2mM NADH, and 3U ml−1 of desalted rabbit muscle lactate dehydrogenase. All assays were initiated by the addition of enzyme preparation and corrected for any NADH oxidase activity by omitting PEP from the reaction mixture.
For measurements of total APase activity, p-nitrophenyl phosphate (pNPP) was used as a substrate. A total reaction volume of 1ml was prepared for each sample and incubated at 37 °C for 30min. The reaction was stopped with 1ml of 0.5M NaOH and the absorbance was measured at 405nm.
The activities of APases and PEPase were defined as the amount of protein hydrolysing 1 µmol of pNPP or reducing NADH min–1 mg–1 of protein, respectively. Protein concentrations were determined using a Bradford assay with BSA as a standard.
Determination of P and N in nodules and shoots, and statistical analysis
Plants were harvested at 42 DAT, and the dry weight of shoots and nodules was used to determine the efficiency of use in rhizobial symbiosis (EURS). This parameter was estimated as the slope of the regression model (i.e. y=ax+b) of plant biomass as a function of nodule biomass (Bargaz et al., 2011b). Dry samples were ground to a fine powder in a vibrating mill for plant material to determine the amount of P and N in nodules. Subsamples of shoot and nodule tissue (50mg) were digested in concentrated HNO3 in a microwave oven (ETHOS; Milestone) at 40 bars for 15min. Total P and Pi content was determined using the vanadomolybdate method (AFNOR, 1969) and the absorbance was determined at 460nm. For N determination, samples of 1.5±0.5mg (nodules) and 2.0±0.5mg (shoots) were analysed for N using a CHN elemental analyser (Fison EA 1108; Thermo Scientific, Waltham, USA).
The experimental design was a randomized complete block. Data were analysed using analysis of variance, and subsequent comparison of means was performed using Fisher’s least significant difference test at 1% probability. The relationship between nodule and shoot dry weights was tested by regression analysis.
Results
Localization of the PEPase transcript
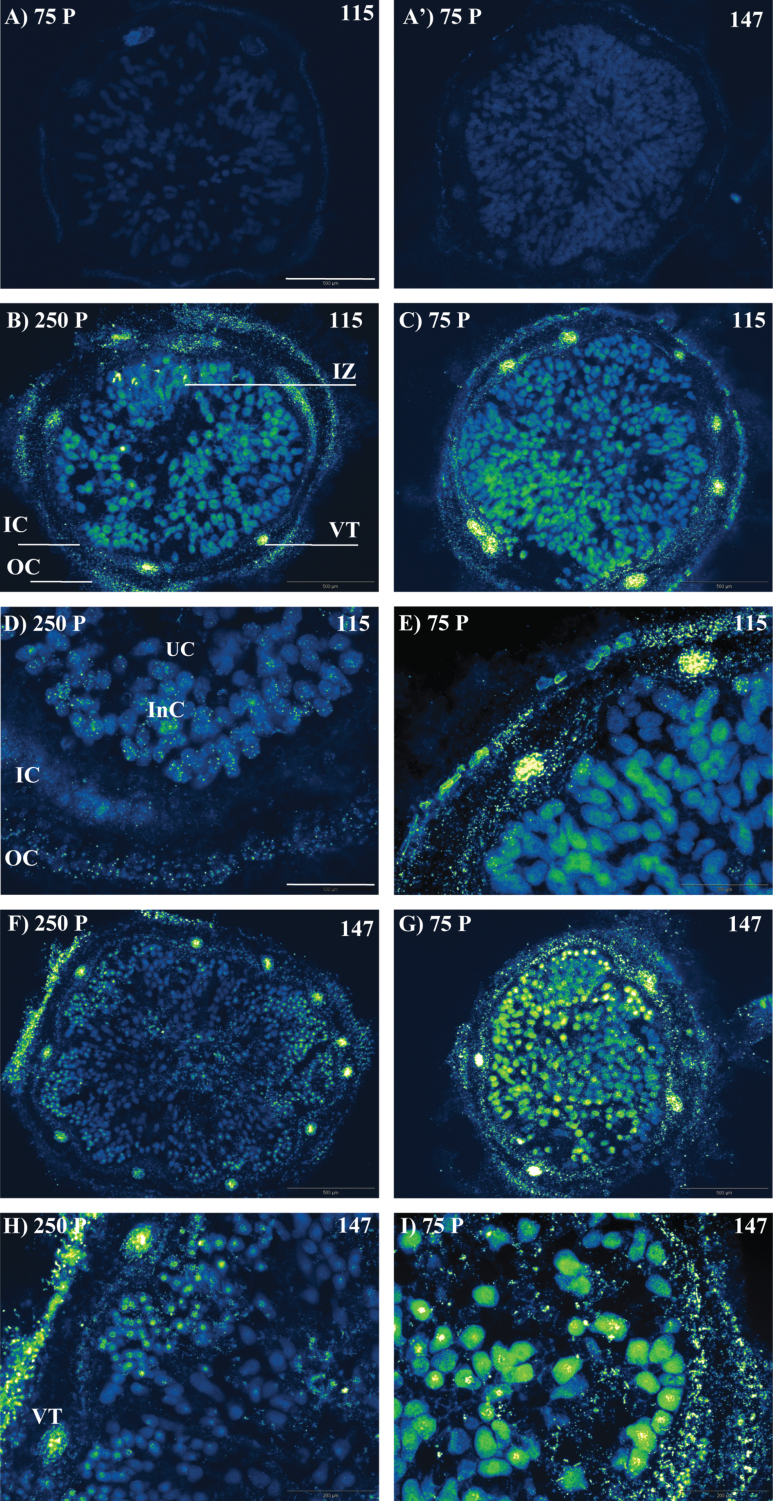
The PEPase transcript was detected in the infected zone, nodule cortex, and vascular traces for both RIL115 and RIL147 and under both P treatments (Fig. 1). Under conditions of P deficiency, the PEPase transcript was more abundant in nodules of the sensitive RIL147 (Fig. 1G) than in the tolerant RIL115 (Fig. 1C), most clearly in the infected zone. More precisely, at a higher magnification, the PEPase transcript was detected mainly in infected cells (Fig. 1I), particularly those localized next to the inner cortex (Fig. 1D, E and H), while there was no transcript in uninfected cells. Moreover, it could clearly be seen that more PEPase transcript was localized in the outer cortex than in the inner cortex (Fig. 1D, E and H).
Fig. 1.
In situ localization of PEPase transcripts (green spots) in nodules of common bean RIL115 and RIL147 (as indicated) inoculated with R. tropici CIAT899 and grown under a sufficient (250 P) versus a deficient (75 P) P supply. (A, A') Negative controls without reverse transcriptase; (B, D) P-sufficient RIL115; (C, E) P-deficient RIL115; (F, H) P-sufficient RIL147; (G, I) P-deficient RIL147. InC, infected cell; IC, inner cortex; IZ, infected zone; OC, outer cortex; UC, uninfected cell; VT, vascular trace parenchyma. Bars, 500 µm (A, A', B, C, F, G); 200 µm (D, E, H, I). (This figure is available in colour at JXB online.)
Quantification of PEPase transcripts and phosphatase activities
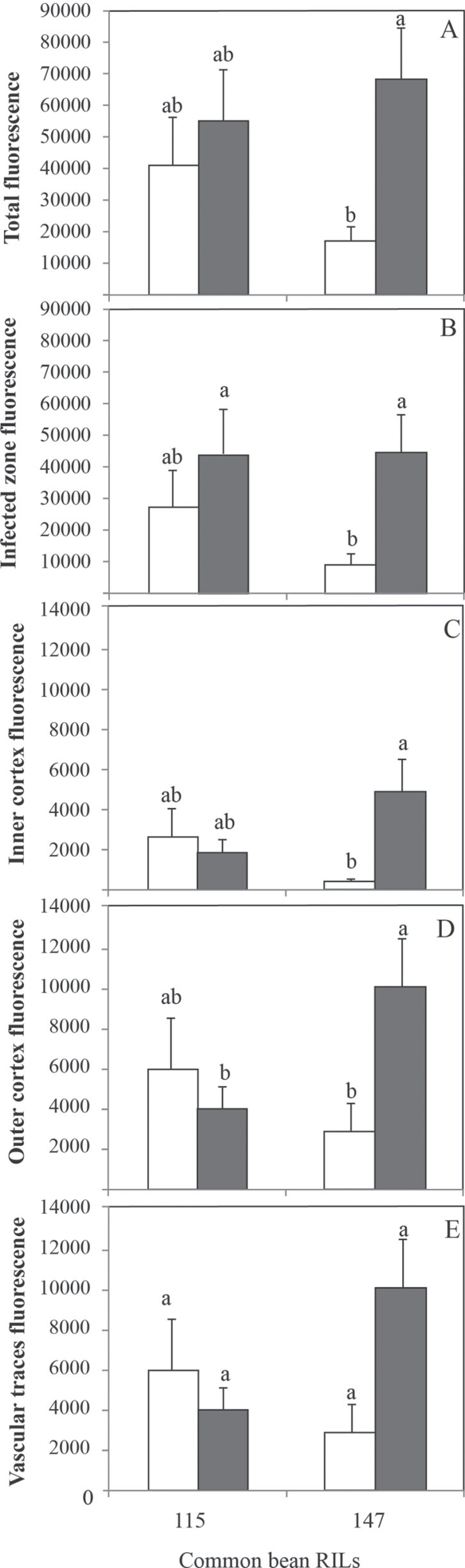
The results indicated that the PEPase transcript signal intensity increased significantly in the sensitive RIL147 under conditions of P deficiency (Fig. 2A). This sensitive RIL147 markedly increased the PEPase transcript abundance by 79, 91 and 71% in the infected zone, inner cortex, and outer cortex, respectively (Fig. 2B–D). Whatever the RIL and the P supply, the PEPase transcript signal intensity was higher in the outer cortex than in the inner cortex. By contrast, the signal intensity did not vary significantly in vascular traces (Fig. 2E).
Fig. 2.
PEPase transcript signal (pixel number of the green spots) in (A) total nodule sections, (B) infected zone, (C) inner cortex, (D) outer cortex, and (E) vascular traces of common bean RIL115 and RIL147 inoculated with R. tropici CIAT899 and grown under a sufficient (open columns) versus a deficient (filled columns) P supply. Data are means ±SD of ten images of nodules harvested at 42 DAT. Mean values labelled with the same letter were not significantly different at P <0.01.
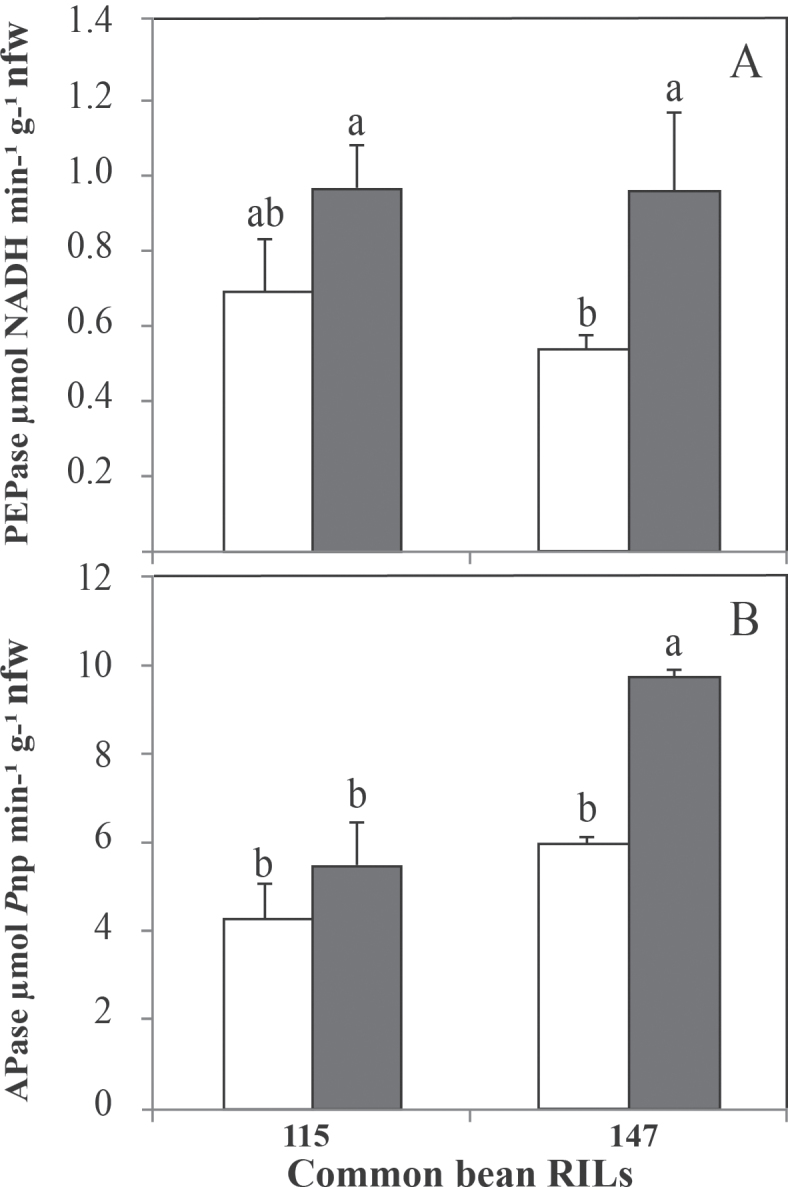
Although P deficiency increased the PEPase activity for both RIL115 (29%) and RIL147 (44%), the increase was only significant in RIL147 (Fig. 3A). Likewise, pNPP APase activity showed the same trend as PEPase (Fig. 3B), with a significant increase under P deficiency in the sensitive RIL147 (39%) compared with RIL115 (23%).
Fig. 3.
PEPase (A) and APase (B) activities in nodules of common bean RIL115 and RIL147 inoculated with R. tropici CIAT899 and grown under a sufficient (open columns) versus deficient (filled columns) P supply. Data are means ±SD of six replicates harvested at 42 DAT. nfw, Nodule fresh weight. Mean values labelled with the same letter were not significantly different at P <0.01.
Overall, considering both P treatments and the two common bean RILs, the proportion of PEPase was approximately 15 % of the total APases measured in this study.
Nodule and shoot P and N content, and efficiency of use in rhizobial symbiosis
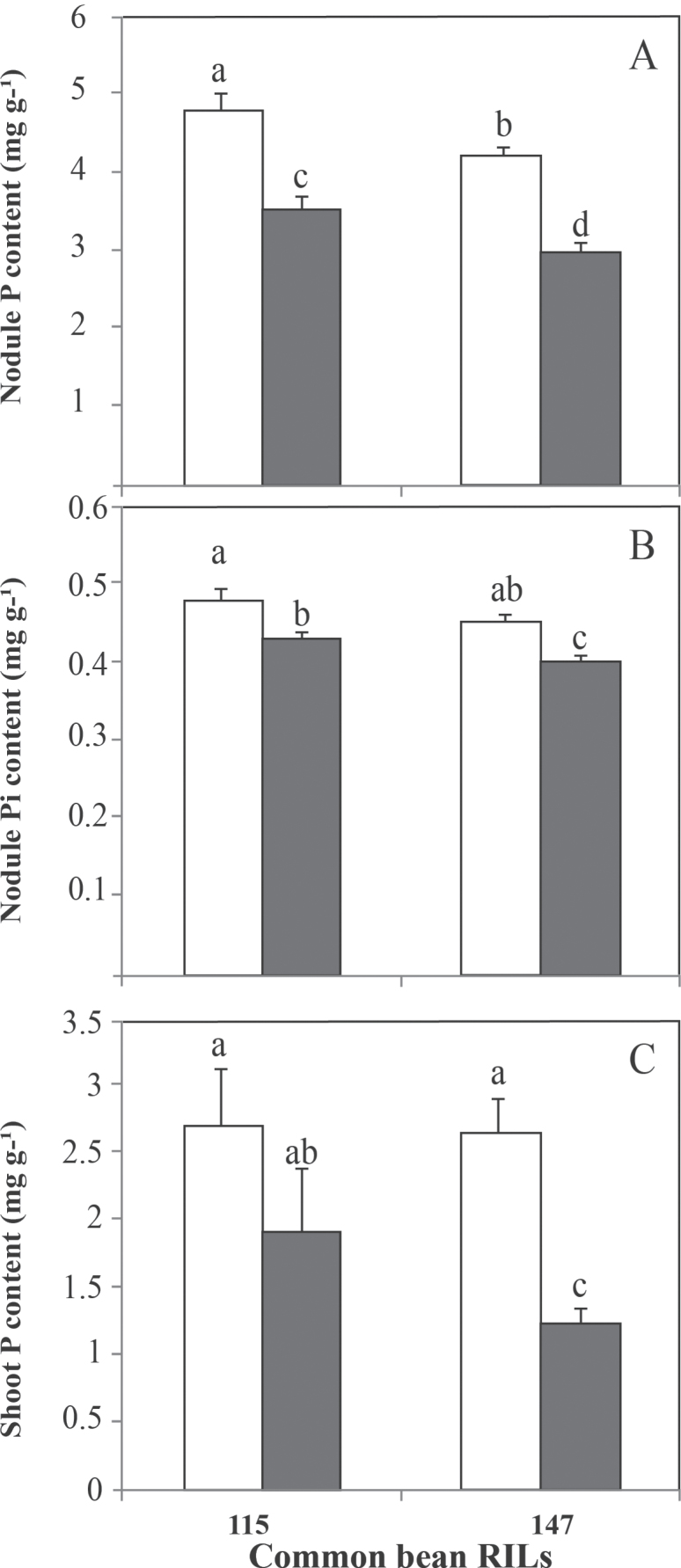
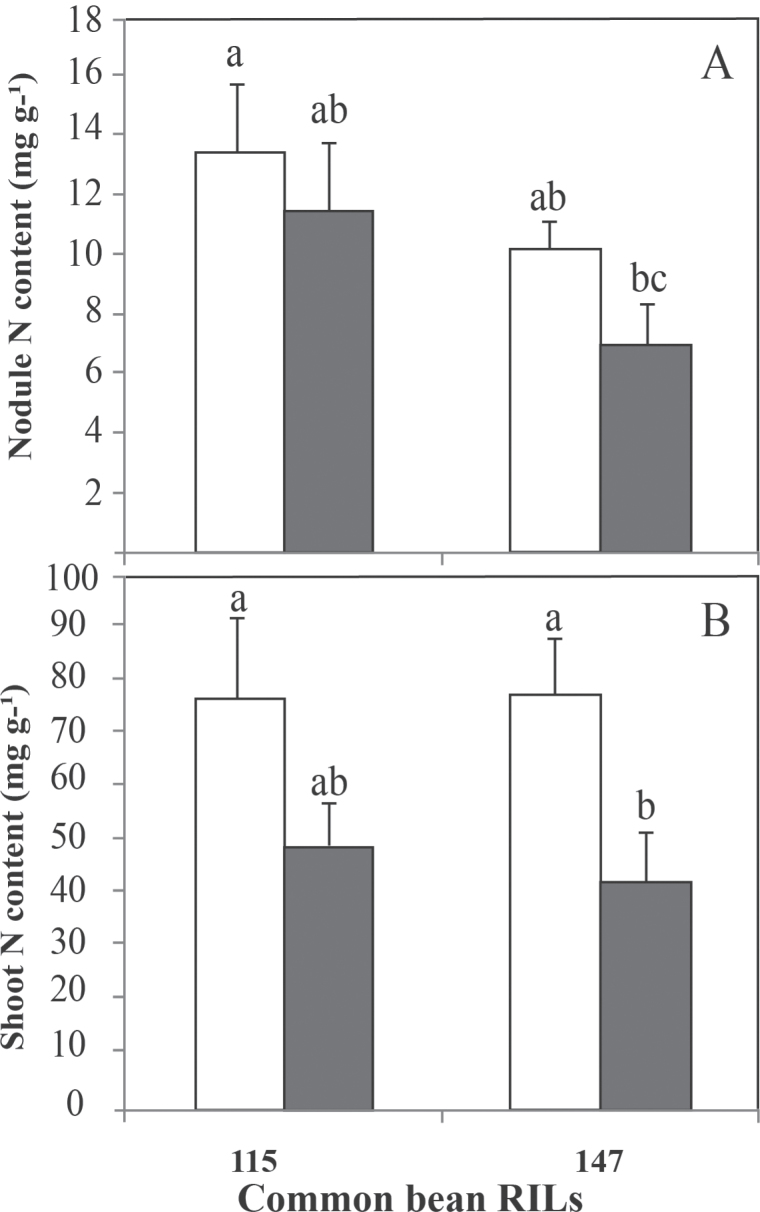
The results indicated that P deficiency significantly decreased the total nodule P content by 26 and 30% for RIL115 and RIL147, respectively (Fig. 4A), while the inorganic nodule P was significantly decreased by about 10% for both RILs (Fig. 4B).
Fig. 4.
Nodule (A, B) and shoot (C) P content of common bean RIL115 and RIL147 inoculated with R. tropici CIAT899 and grown under a sufficient (open columns) versus deficient (filled columns) P supply. Data are means ±SD of six replicates harvested at 42 DAT. Mean values labelled with the same letter were not significantly different at P <0.01.
Although shoot P content was decreased in both RILs, this parameter was not statistically different for the tolerant RIL115, whereas, for the sensitive RIL147, shoot P content was almost twice as high under P-sufficient compared with P-deficient conditions (Fig. 4C).
P deficiency decreased the nodule and shoot N contents for both RILs tested (Fig. 5). However, the decrease in nodule N content was not significant for RIL147 (31%) or RIL115 (14%). By contrast, shoot N content decreased significantly for RIL147 (46%). Overall, the decrease in N content was more pronounced in shoots than in nodules for plants grown under conditions of P deficiency.
Fig. 5.
Nodule (A) and shoot (B) N contents of common bean RIL115 and RIL147 inoculated with R. tropici CIAT899 and grown under a sufficient (open columns) versus a deficient (filled columns) P supply. Data are means ±SD of six replicates harvested at 42 DAT. Mean values labelled with the same letter were not significantly different at P <0.01.
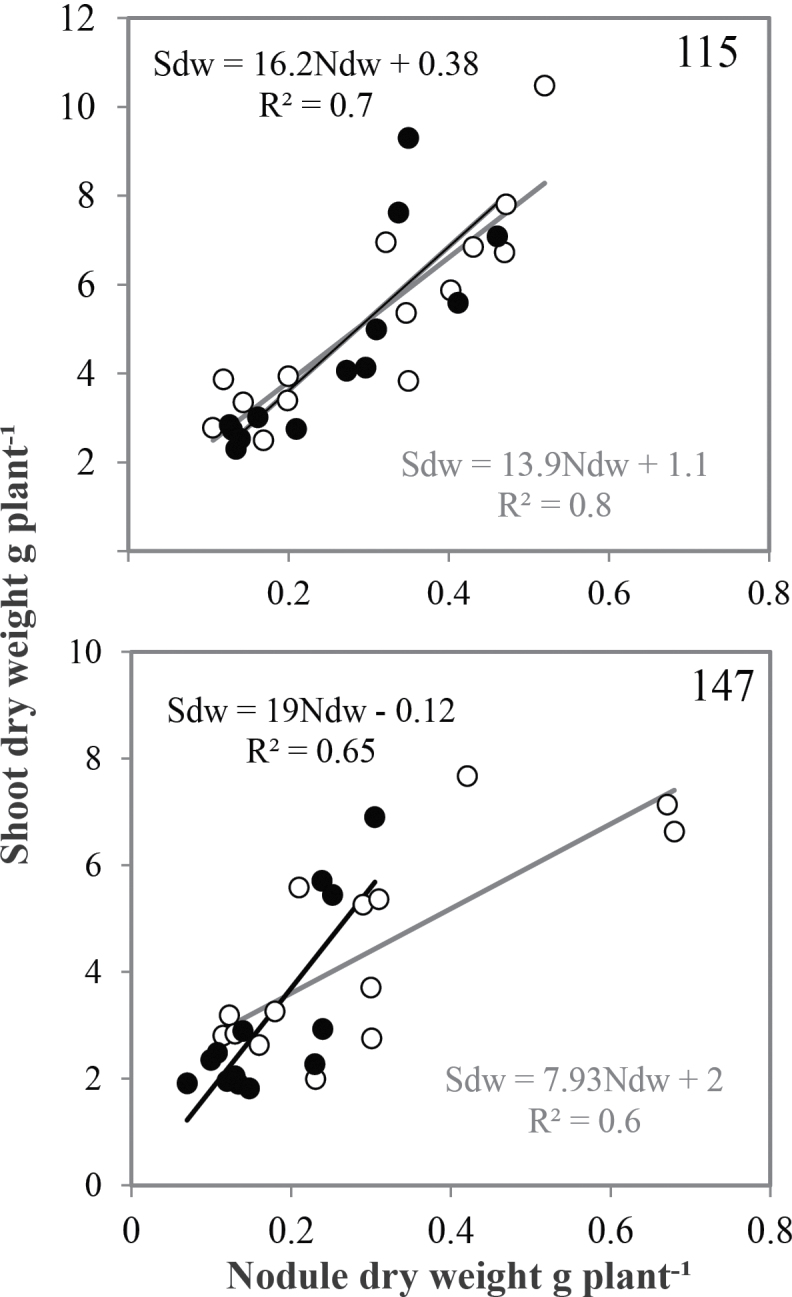
The decrease in both P and N content was associated with a significant reduction in growth and nodulation of the tested RILs under conditions of P deficiency (Fig. 6). However, the decrease in both nodule and shoot biomass was higher for the sensitive RIL147 (41 and 29%, respectively) than for the tolerant RIL115 (14 and 15%, respectively). The biomass of shoots and nodules of the RIL115 and RIL147 were positively correlated (up to r 2 = 0.6) under both P treatments (Fig. 6). The slope value of the regression curve, namely the EURS, was higher under conditions of P deficiency than under P sufficiency, by 58 and 14 % for RIL147 and RIL115, respectively.
Fig. 6.
Efficiency of use of rhizobial symbiosis of common bean RIL115 and RIL147 inoculated with R. tropici CIAT899 and grown under a sufficient (open circles) versus a deficient (filled circles) P supply. Data represent individual values of 14 replicates harvested at 42 DAT. Ndw, Nodule dry weight.
Discussion
The development of a successful in situ RT-PCR methodology provides an interesting opportunity to link localization and distribution of candidate genes within organs such as nodules of N2-fixing legumes. In addition, the technique allows quantification of the effects of environmental constraints on gene expression. Our work holds significant promise in addressing the effect of P deficiency on the nodule localization of a PEPase transcript that very probably correlates with the activity of PEP hydrolysis and presents the highest sequence homology with an APase gene expressed specifically in the embryonic axes of P. vulgaris (Yoneyama et al., 2004).
The detection of a PEPase transcript in common bean nodules is, to our knowledge, the first observation of this gene expression among APase genes that are known to be overexpressed in legume nodules in response to P deficiency (Figs 1 and 2). Increased transcript levels for PEPase under conditions of P deficiency in nodules of RIL147 (Figs 1G and I and A), in contrast to that in nodules of the tolerant RIL115, which exhibited a similar level under both P treatments, suggest that PEPase may be involved in the tolerance of rhizobial symbiosis to P deficiency. This is consistent with a previous conclusion that P deficiency specifically affects SNF by limiting growth and survival of rhizobia (O’Hara et al., 1988), nodule function (Tang et al., 2001), and host plant growth (Tsvetkova and Georgiev, 2003).
Although the high level of PEPase transcripts under conditions of P deficiency is linked to a rise in nodule PEPase and total APase activities (Fig. 3), transcript abundance does not always predict intra- or extracellular proteome remodelling orin the gene products that result from nutrient limitations (Li et al., 2008; Tran and Plaxton, 2008). Nevertheless, the increases in these phosphatase activities were linked to a decrease in nodule P content, which may subsequently serve for ATP generation via O2 respiration, with at least 16 ATPs consumed per N2 reduced (Salsac et al., 1984). The link of PEPase expression to EURS, particularly for the sensitive RIL147 under P deficiency (Fig. 6), suggests tight regulation between EURS and the nodule P requirement (Fig. 4), probably in relation to the high energy requirement of the SNF process. In agreement with this, Li et al. (2011) reported that some members of the purple acid phosphatase gene family in soybean are possibly involved in the response of the host plant to symbiosis with rhizobia or arbuscular mycorrhizal fungi under P-deficient conditions.
Regarding the differential expression of the PEPase gene among nodule tissues (Figs 1 and 2), the high level of transcripts in the infected zone (Figs 1C and G and B) is consistent with a large P requirement for optimum metabolism of bacteroids, as well as for multiplication and survival of bacteroids. All of this is consistent with the high P content of nodules compared with other plant organs (Schulze and Drevon, 2005; Bargaz et al., 2011a). The increase in PEPase transcripts in the nodule cortex might be an adaptive response to P deficiency. Furthermore, PEPase expression in the inner cortex may play a role in the relation of SNF to O2 flux (Hunt and Layzell, 1993), which is postulated to be a regulator of osmotic conditions (Schulze and Drevon, 2005). The higher level of transcripts in the outer cortex compared with the inner cortex (Figs 1D, E and H and D) may be a response to the high P demand of the infected zone and the Pi mobilization from senescing tissues in the nodule external cortex. The production of pyruvate in P-deficient nodules may have an essential role in limiting the production of active oxygen species, as pyruvate has been proposed to be an antioxidant undergoing a non-enzymatic oxidative decarboxylation by hydrogen peroxide (Nath et al., 1995).
We conclude that PEPase can be classified as a P-starvation-inducible gene involved in the tolerance of nodulated legumes to P deficiency. The marked increase in PEPase transcripts in the nodule cortex of P-deficient nodules not only suggests an increase in intracellular Pi scavenging but also opens up a challenge to understand whether such sublocalization is involved in the scavenging of Pi from extracellular organophosphates. Although PEPase appears to be transcriptionally induced during P deficiency, additional studies are needed to understand whether this phosphatase plays a secondary role in response to constraints other than P deficiency, such as oxidative stresses, which may occasionally occur.
Acknowledgments
This work was supported by a Great Federative FABATROPIMED Project (2011–2014) and AVERROES scholarship programme provided by the EU for the stay of Adnane Bargaz in Montpellier. The authors thank Claude Plassard, Catherine Pernot, and Estelle Tournier (INRA Montpellier, France) for their technical assistance.
Glossary
Abbreviations:
- APase
acid phosphatase;
- CIAT
International Centre of Tropical Agriculture;
- DAT
days after transplantation;
- EURS
efficiency of use in rhizobial symbiosis;
- P
phosphorus;
- PEP
phosphoenol pyruvate;
- PEPase
PEP phosphatase;
- pNPP
p-nitrophenyl phosphate;
- RIL
recombinant inbred line;
- SNF
symbiotic nitrogen fixation.
References
- AFNOR (1969). Dosage spectrophotométrique de l’anhydride phosphorique: méthode vanadomolybdique Paris: : AFNOR; 224 246 [Google Scholar]
- Araújo AP, Plassard C, Drevon JJ. Phosphatase and phytase activities in nodules of common bean genotypes at different levels of phosphorus supply. Plant and Soil. 2008;312:129–138. [Google Scholar]
- Bargaz A, Drevon JJ, Oufdou K, Mandri B, Faghire M, Ghoulam C. Nodule phosphorus requirement and O2 uptake in common bean genotypes under phosphorus deficiency. Acta Agriculturae Scandinavica B Soil and Plant Sciences. 2011a;61:602–611. [Google Scholar]
- Bargaz A, Ghoulam C, Faghire M, Aslan Attar H, Drevon JJ. The nodule conductance to the O2 diffusion increases with high phosphorus content in the Phaseolus vulgaris–rhizobia symbiosis. Symbiosis. 2011b;53:157–164. [Google Scholar]
- Bozzo GG, Raghothama KG, Plaxton WC. Structural and kinetic properties of a novel purple acid phosphatase from phosphate-starved tomato (Lycopersicon esculentum) cell cultures. Biochemical Journal. 2004;377:419–428. doi: 10.1042/BJ20030947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drevon JJ, Alkama N, Araujo A, et al. Nodular diagnosis for ecological engineering of the symbiotic nitrogen fixation with legumes. Procedia Environmental Sciences. 2011;9:40–46. [Google Scholar]
- Fang ZY, Shao C, Meng YJ, Wu P, Chen M. Phosphate signaling in Arabidopsis and Oryza sativa. Journal of Plant Science. 2009;176:170–180. [Google Scholar]
- Hinsinger P. 2001. Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: a review Plant and Soil 237 173 195 [Google Scholar]
- Hunt S, Layzell DB. Gas exchange of legume nodules and the regulation of nitrogenase activity. Annual Review of Plant Physiology and Plant Molecular Biology. 1993;44:483–511. [Google Scholar]
- Juszczuk IM, Rychter AM. Pyruvate accumulation during phosphate deficiency stress of bean roots. Plant Physiology and Biochemistry. 2002;40:783–788. [Google Scholar]
- Li C, Gui S, Yang T, Walk T, Wang X, Liao H. Identification of soybean purple acid phosphatase genes and their expression responses to phosphorus availability and symbiosis. Annals of Botany. 2011;109:275–285. doi: 10.1093/aob/mcr246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WY, Shao G, Lam HM. Ectopic expression of GmPAP3 alleviates oxidative damage caused by salinity and osmotic stresses. New Phytologist. 2008;178:80–91. doi: 10.1111/j.1469-8137.2007.02356.x. [DOI] [PubMed] [Google Scholar]
- Liang C, Tian J, Lam HM, Lim BL, Yan X, Liao H. Biochemical and molecular characterization of PvPAP3, a novel purple acid phosphatase isolated from common bean enhancing extracellular ATP utilization. Plant Physiology. 2010;152:854–865. doi: 10.1104/pp.109.147918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina C, Zaman-Allah M, Khan F, et al. The salt-responsive transcriptome of chickpea roots and nodules via deepSuperSAGE. BMC Plant Biology. 2011;11:31. doi: 10.1186/1471-2229-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath KA, Ngo EO, Hebbel RP, Croatt AJ, Zhou B, Nutter LM. α-Ketoacids acids scavenge H2O2 in vitro and in vivo and reduce menadione-induced injury and cytotoxicity. American Journal of Physiology – Cell Physiology. 1995;268:227–236. doi: 10.1152/ajpcell.1995.268.1.C227. [DOI] [PubMed] [Google Scholar]
- O’Hara GW, Boonkered N, Dilworth MJ. Mineral constraints to nitrogen fixation. Plant and Soil. 1988;108:93–110. [Google Scholar]
- Penheiter AR, Duff SMG, Sarath G. Soybean root nodule acid phosphatase. Plant Physiology. 1997;114:597–604. doi: 10.1104/pp.114.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radersma S, Grierson PF. Phosphorus mobilization in agroforestry: organic anions, phosphatase activity and phosphorus fractions in the rhizosphere. Plant and Soil. 2004;259:209–219. [Google Scholar]
- Raghothama KG. Phosphate acquisition. Annual Review of Plant Physiology and Plant Molecular Biology. 1999;50:665–693. doi: 10.1146/annurev.arplant.50.1.665. [DOI] [PubMed] [Google Scholar]
- Richardson AE, Hocking PJ, Simpson RJ, George TS. Plant mechanisms to optimize access to soil phosphorus. Crop Pasture Science. 2009;60:124–143. [Google Scholar]
- Salsac L, Drevon JJ, Zengbe M, Cleyet-Marel JC, Obaton M. Energy requirement of symbiotic nitrogen fixation. Physiologie Végétale. 1984;22:509–521. [Google Scholar]
- Schulze J, Drevon JJ. P-deficiency increases the O2 uptake per N2 reduced in alfalfa. Journal of Experimental Botany. 2005;56:1779–1784. doi: 10.1093/jxb/eri166. [DOI] [PubMed] [Google Scholar]
- Serraj R, Adu-Gyamfi J. Role of symbiotic nitrogen fixation in the improvement of legume productivity under stressed environments. West Africa Journal of Applied Ecology. 2004;6:95–109. [Google Scholar]
- Shinano T, Yonetani R, Ushihara N, Adachi H, Wasaki J, Matsui H, Osaki M. Characteristics of phosphoenolpyruvate phosphatase purified from Allium cepa. Plant Science. 2001;161:861–869. [Google Scholar]
- Tang C, Hinsinger P, Jaillard B, Rengelz Z, Drevon JJ. Effect of phosphorus deficiency on the growth, symbiotic N2 fixation and proton release by two bean (Phaseolus vulgaris) genotypes. Agronomie. 2001;21:683–689. [Google Scholar]
- Tran HT, Plaxton WC. Proteomic analysis of alterations in the secretome of Arabidopsis thaliana suspension cells subjected to nutritional phosphate deficiency. Proteomics. 2008;8:4317–4326. doi: 10.1002/pmic.200800292. [DOI] [PubMed] [Google Scholar]
- Tran HT, Qian W, Hurley BA, She Y, Wang D, Plaxton WC. Biochemical and molecular characterization of AtPAP12 and AtPAP26: the predominant purple acid phosphatase isozymes secreted by phosphate-starved Arabidopsis thaliana. Plant, Cell and Environment. 2010;33:1789–1803. doi: 10.1111/j.1365-3040.2010.02184.x. [DOI] [PubMed] [Google Scholar]
- Tsvetkova GE, Georgiev GI.2003Effect of phosphorus nutrition on the nodulation, nitrogen fixation and nutrient-use efficiency of Bradyrhizobium japonicun soybean (Glycine max L. Merr.) symbiosis Bulgarian Journal of Plant Physiology Special issue,331–335. [Google Scholar]
- Vadez V, Lasso JH, Beck DP, Drevon JJ. Variability of N2-fixation in common bean (Phaseolus vulgaris L.) under P-deficiency is related to P use efficiency. Euphytica. 1999;106:231–242. [Google Scholar]
- Van Aarle IM, Viennois G, Amenc LK, Tatry MV, Luu DT, Plassard C. Fluorescent in situ RT-PCR to visualise the expression of a phosphate transporter gene from an ectomycorrhizal fungus. Mycorrhiza. 2007;17:487–494. doi: 10.1007/s00572-007-0127-4. [DOI] [PubMed] [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL. Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytologist. 2003;157:427–447. doi: 10.1046/j.1469-8137.2003.00695.x. [DOI] [PubMed] [Google Scholar]
- Veljanovski V, Vanderbeld B, Knowles VL, Snedden WA, Plaxton WC. Biochemical and molecular characterization of AtPAP26, a vacuolar purple acid phosphatase up-regulated in phosphate-deprived Arabidopsis suspension cells and seedlings. Plant Physiology. 2006;142:1282–1293. doi: 10.1104/pp.106.087171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama T, Shiozawa M, Nakamura M, Suzuki T, Sagane Y, Katoh Y, Watanabe T, Ohyama T. Characterization of a novel acid phosphatase from embryonic axes of kidney bean exhibiting vanadate-dependent chloroperoxidase activity. Journal of Biological Chemistry. 2004;279:37477–37484. doi: 10.1074/jbc.M405305200. [DOI] [PubMed] [Google Scholar]