Abstract
The abscisic acid (ABA) signalling core in plants include the cytosolic ABA receptors (PYR/PYL/RCARs), the clade-A type 2C protein phosphatases (PP2CAs), and the subclass III SNF1-related protein kinases 2 (SnRK2s). The aim of this work was to identify these ABA perception system components in sweet orange and to determine the influence of endogenous ABA on their transcriptional regulation during fruit development and ripening, taking advantage of the comparative analysis between a wild-type and a fruit-specific ABA-deficient mutant. Transcriptional changes in the ABA signalosome during leaf dehydration were also studied. Six PYR/PYL/RCAR, five PP2CA, and two subclass III SnRK2 genes, homologous to those of Arabidopsis, were identified in the Citrus genome. The high degree of homology and conserved motifs for protein folding and for functional activity suggested that these Citrus proteins are bona fide core elements of ABA perception in orange. Opposite expression patterns of CsPYL4 and CsPYL5 and ABA accumulation were found during ripening, although there were few differences between varieties. In contrast, changes in expression of CsPP2CA genes during ripening paralleled those of ABA content and agreeed with the relevant differences between wild-type and mutant fruit transcript accumulation. CsSnRK2 gene expression continuously decreased with ripening and no remarkable differences were found between cultivars. Overall, dehydration had a minor effect on CsPYR/PYL/RCAR and CsSnRK2 expression in vegetative tissue, whereas CsABI1, CsAHG1, and CsAHG3 were highly induced by water stress. The global results suggest that responsiveness to ABA changes during citrus fruit ripening, and leaf dehydration was higher in the CsPP2CA gene negative regulators than in the other ABA signalosome components.
Key words: Abscisic acid (ABA), Citrus, fruit ripening, gene expression, leaf dehydration, PP2CA, PYR/PYL/RCAR, receptor, signalling, SnRK2
Introduction
A number of studies have attempted to obtain a deeper insight into the cellular and molecular responses to ABA in plants, comprising the perception, signalling, metabolism, and transport of this phytohormone (Nambara and Marion-Poll, 2005; Verslues and Zhu, 2007; Kang et al., 2010; Kuromori et al., 2010; Antoni et al., 2011). Natural or induced plant mutants of ABA biosynthetic and signalling genes have been extensively used to elucidate the involvement of ABA in several physiological processes (Karssen et al., 1983; Peña-Cortés et al., 1989; Groot and Karssen, 1992; Armstrong et al., 1995; Schwartz et al., 1997; Galpaz et al., 2008; Sun et al., 2012). However, less information is available about the effects of this plant regulator on non-climacteric fruit performance and physiology (Rodrigo et al., 2006; Zhang et al., 2009a; Giribaldi et al., 2010; Chai et al., 2011; Jia et al., 2011).
In woody plants, artificially generated mutants are less affordable, but spontaneous mutants are more widely found (Koornneef et al., 2004). A spontaneous fruit-specific ABA-deficient mutant from the ‘Navelate’ orange (Citrus sinensis L. Osbeck), named ‘Pinalate’, has been biochemically characterized (Rodrigo et al., 2003). ‘Pinalate’ orange presents distinctive yellow-coloured fruit because of a partial blockage of the carotenoid biosynthetic pathway, causing, consequently, a fruit-specific ABA deficiency. During natural ripening, the onset of fruit degreening is delayed in ‘Pinalate’ as compared with its wild-type cultivar (Rodrigo et al., 2003). Moreover, the sensitivity to ABA and the molecular responses to fruit dehydration during post-harvest storage have been shown to be impaired in this mutant, which suggested that the ABA perception system may fail in sensing the phytohormone (Romero et al., 2012). Therefore, the fruit-specific ABA-deficient ‘Pinalate’ orange offers an exceptional experimental system to investigate the role of endogenous ABA in the regulation of the hormone perception system components during citrus fruit ripening.
Several pieces of evidence support that multiple ABA receptors perceive the ABA signal outside and inside the cells, this perception being tissue specific (Finkelstein et al., 2002). The PYR/PYL/RCAR soluble proteins (Ma et al., 2009; Park et al., 2009), belonging to the START protein superfamily (Klingler et al., 2010), and the downstream complex composed of protein phosphatase type 2C (PP2CA) and SNF1-related kinases family 2 (SnRK2) proteins (Umezawa et al., 2009; Vlad et al., 2009; Hirayama and Umezawa, 2010), have been shown to regulate the well-known ABA responses in the model plant Arabidopsis thaliana. Thus, the ABA signalling core is composed of the cytoplasmic ABA receptors (PYR/PYL/RCAR) and the clade A PP2CAs as negative regulators (Gosti et al., 1999; Merlot et al., 2001), and a number of protein kinases, including the subclass III of the SnRK2s, as positive regulators of the pathway (Yoshida et al., 2002). The PYR/PYL/RCAR proteins contain a ligand-binding pocket in a cavity that closes after ABA binding through conformational changes of two conserved β-loops that serve as a gate and a latch. ABA binding to the receptors is enhanced when PYR/PYL/RCAR proteins are bound to their negative regulator PP2CAs (Ma et al., 2009; Melcher et al., 2009; Park et al., 2009). This new conformation locks the receptor in a closed structure and inhibits the PP2CA active site (Melcher et al., 2009; Santiago et al., 2009a). Consequently, SnRK2 is released and can phosphorylate downstream proteins or transcription factors that trigger the expression of ABA-responsive genes (Umezawa et al., 2010). Some investigations have been conducted on ABA signalling core components at the transcriptional and functional levels. In general, concomitant with increases in ABA, positive effectors (PYR/PYL/RCAR and SnRKs) were transcriptionally repressed whereas negative regulators (PP2CAs) increased, together modulating downstream signalling and, consequently, physiological ABA responses in model and crop plants (Huai et al., 2008; Park et al., 2009; Santiago et al., 2009b; Umezawa et al., 2009; Nishimura et al., 2010; Szostkiewicz et al., 2010; Sun et al., 2011). Currently, limited information is available in non-climacteric fruit (Jia et al., 2011; Chai et al., 2011; Li et al., 2012) and there is no report analysing the expression of this set of genes as a whole.
In this study, 13 genes belonging to the PYR/PYL/RCAR, PP2CA, and SnRK2 families have been identified in sweet orange. In order to obtain a deeper insight into the modulation of the ABA signalling components during fruit development and ripening of this non-climacteric fruit, as well as the relationship existing between these components and the changes in the endogenous ABA accumulation during these processes, the expression of the ABA signalosome components has been investigated in fruits of ‘Navelate’ orange and its ABA-deficient mutant ‘Pinalate’ during different developmental stages. Moreover, expression analysis of the ABA signalling core elements was performed in detached leaves from both cultivars subjected to dehydration. This has allowed a comparative analysis between fruit and vegetative tissue, providing further insights into the role of the different ABA signalosome genes, and has helped to decipher whether the key genes in this system are common or tissue specific.
Materials and methods
Plant material and colour measurement
Fruits of ‘Navelate’ (C. sinensis L. Osbeck) orange and its spontaneous ABA-deficient mutant ‘Pinalate’ were randomly harvested at six different ripening stages from adult trees grown at ‘The Spanish Citrus Germoplasm Bank’ at the Instituto Valenciano de Investigaciones Agrarias (Moncada, Valencia, Spain), and immediately delivered to the laboratory. The trees were the same age, grown in the same experimental orchard, and subjected to the same standard cultural practices. The six sampling periods were chosen based on previous reports describing colour evolution in citrus fruit during ripening (Rodrigo et al., 2004) and were defined as: immature green, IG; mature green I, MI; mature green II, MII; breaker, Bk; coloured, C; and full coloured, FC. Thus, fruits of both cultivars were hand harvested on the same day and their colour was measured (Supplementay Table S1 available at JXB online) using a Minolta CR-330 on three locations around the equatorial plane of the fruit and expressed as the a/b Hunter ratio (Stewart and Wheaton, 1972), which is classically used for colour measurement in citrus fruit. This ratio is negative for green fruit and positive for orange fruit, while a zero value corresponds to yellow fruit at the midpoint of the colour break period. Flavedo (the outer coloured part of the peel) tissue samples were collected from the total surface of fruits, frozen and homogenized to a fine powder in liquid nitrogen, and kept at –80 ºC for later analysis. Three biological replicates of five fruits each were collected at each sampling period.
In addition, water stress experiments in vegetative tissue were carried out in detached mature leaves. To that end, leaves were collected, weighed, and allowed to dehydrate in storage chambers under continuous light at 22 ºC. Control non-stressed leaves were kept in the chambers at 90% relative humidity (RH) with petioles in distilled water, whereas stressed leaves were dehydrated by placing them on filter paper at 50–55% RH. The weight of the leaves was monitored periodically and tissue was collected after 0.5, 1, 3, 6, and 24h. Three biological replicates of four leaves were used for each time period. Leaves were frozen in liquid nitrogen, ground to a fine powder, and stored at –80 ºC until analysis.
RNA isolation and qRT-PCR analysis
Total RNA was extracted from frozen flavedo and leaf samples by a method modified from that previously described by Rodrigo et al. (2004) and Ballester et al. (2006), as reported in Romero et al. (2012). Total RNA was treated with RNase-free DNase (Applied Biosystems) following the manufacturer’s instructions. Thereafter, the amount of RNA was measured by spectrophotometric analysis and its quality was verified by agarose gel electrophoresis and ethidium bromide staining.
Reverse transcription followed by quantitative PCR analysis (qRT-PCR) was performed as described previously by Romero et al. (2012) to examine the time course of gene expression patterns during fruit ripening and leaf dehydration. Briefly, a two-step qRT-PCR assay was designed as suggested by Udvardi et al. (2008). The cDNAs from all biological replicates were synthesized from 2 µg of total RNA by using SuperScript III RT (Invitrogen) in the presence of an oligo(dT) 20-mer (Invitrogen) and an RNase inhibitor (Invitrogen) according to the manufacturer’s instructions. Gene-specific primers were designed using DNAMAN 4.03 software (Lynnon BioSoft) and incubated, in a LightCycler 480 Instrument (Roche Diagnostics), with the cDNA samples and LightCycler 480 SYBR Green I Master (Roche Diagnostics) at 95 ºC for 10min followed by 40 cycles at 95 ºC for 10 s, 60 ºC for 5 s, and 72 ºC for 10 s. Forward (F) and reverse (R) sequences for specific primers and the amplicon size for each gene are shown in Supplementary Table S2 at JXB online. The occurrence of non-specific amplified products was ruled out after performing a melting curve analysis and sequencing the reaction products. Fluorescent intensity measurements were transformed into relative mRNA levels by using standard curves constructed for all studied genes. The reference genes CsACT, CsGAPDH, and CsTUB (Supplementary Table S2), whose constitutive expression during fruit ripening was confirmed by using the geNorm program (Vandesompele et al., 2002), were used for data normalization. Statistical analysis (pairwise fixed reallocation randomization test) was carried out by the ΔΔCt method using the Relative Expression Software Tool (REST, http://rest.gene-quantification.info) (Pfaffl, 2001). Validation experiments were performed previously to ensure that the efficiency of the target and housekeeping genes was relatively equivalent. Relative expression levels for all flavedo samples were referred to that obtained in MI ‘Navelate’ fruits and those of vegetative samples were relative to that found in freshly harvested ‘Navelate’ leaves. In addition, in order to compare absolute gene expression values, amplicons of each gene were cloned in the pGEMT vector (Promega) and used to generate standard curves by serial dilutions. Data were then normalized by using the above-mentioned housekeeping genes. Three biological samples for each sampling period, tissue, and variety were analysed in duplicate and mean ratios were calculated.
Statistical design
Results are the means of three replicate samples ±SE. A mean comparison using Tukey’s test was performed to determine if means values were significantly different (P ≤ 0.05).
ABA analysis
ABA was extracted from 1g fresh weight (FW) of frozen flavedo and leaves with 80% acetone containing 0.5g l−1 citric acid and 100mg l−1 butylated hydroxytoluene as previously described by Lafuente et al. (1997). After centrifugation, the supernatant was diluted in three serial dilutions in ice-cold TRIS-buffered saline (TBS; 6.05g l–1 TRIS, 8.8g l–1 NaCl, and 0.2mg l–1 MgCl2 at pH 7.8) and three samples for each dilution were analysed by the indirect enzyme-linked immunosorbent assay (ELISA) reported by Walker-Simmons (1987). The ABA-BSA-(4, conjugate) was synthesized as previously reported by Weiler (1980) with some modifications (Norman et al., 1988). The results are the means of three biological replicates of five fruit each ±SE.
Sequence analyses, alignment, and phylogenetics
Sequence similarity comparisons between A. thaliana and C. sinensis proteins were performed by BLASTP in the Phytozome v7.0 database (www.phytozome.org; www.citrusgenomedb.org). A search for amino acid sequences of Arabidopsis PYR/PYL/RCAR, PP2CA, and SnRK2 proteins was carried out using the National Centre for Biotechnology Information (NCBI). Motif prediction was performed using whole protein sequences as input into the PSIPRED secondary structure prediction server. The tertiary structures of CsPYR1, CsPYL2, CsPYL5, CsPYL8, CsABI1, and CsSnRK2.6 proteins were modelled by using the I-Tasser program (Roy et al., 2010), in which their corresponding Arabidopsis homologous crystallographic structures from the PDB database (3K90, 3KL1, 3QRZ, 3UQH, 3UJK, and 3UDB, respectively) were used as templates. Multiple sequence alignments of PYR/PYL/RCAR, PP2CA, and SnRK2 proteins were performed by using the default settings of the CLUSTALX 2.0 software and manually edited in GENEDOC (http://www.nrbsc.org/gfx/genedoc/). Based on these alignments, phylogenetic trees were constructed according to the Neighbor–Joining method using the PhyloWidget program. The reliability of the trees was established by conducting a 1000 bootstrap re-sampling.
Results
The PYR/PYL/RCAR family in Citrus sinensis
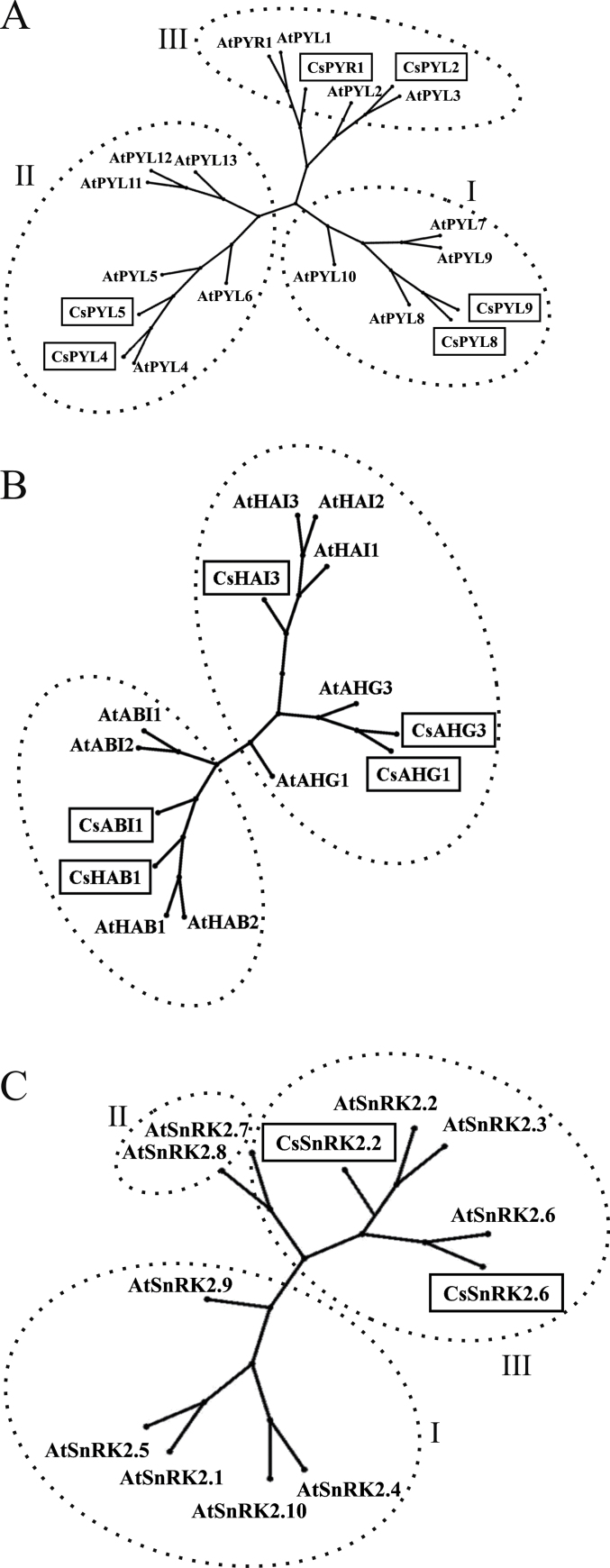
Genes encoding ABA receptors of A. thaliana were used as query to identify the orthologous proteins from C. sinensis. In the orange genome, only six proteins with homology to the 14 Arabidopsis PYR/PYL/RCAR proteins were found (Table 1). The genes AtPYR1 and AtPYL1 shared homology with the same orange locus (orange1.1g046151m) and showed 74% and 73% of identity, respectively, in 175 amino acid residues, which correspond to 84% of the protein length. Likewise, AtPYL2 and AtPYL3 shared homology to the orange1.1g046697m locus and showed 72% and 66% identity, covering 90% and 93% of the protein stretch, respectively. On the other hand, AtPYL4 and AtPYL6 were homologous to the protein encoded by the Citrus gene orange1.1g026007m and showed 78% and 62% identity, respectively. The orange1.1g038201m locus was the most similar to the AtPYL5, AtPYL11, AtPYL12, and AtPYL13 genes and displayed 70, 62, 62, and 56% identity at the protein level, respectively. Genes AtPYL8 and AtPYL10 showed homology to orange1.1g028067m (75% and 74% identity in 183 and 158 amino acid residues, respectively) and AtPYL7 and AtPYL9 to orange1.1g043944m (79% and 86% identity, respectively). Taking into account their highest identity with Arabidopsis proteins, Citrus genes were named CsPYR1, CsPYL2, CsPYL4, CsPYL5, CsPYL8, and CsPYL9, respectively (Table 1). The analysis of the genomic structure of all Citrus PYR/PYL/RCAR genes revealed that only CsPYL8 and CsPYL9 genes showed the predicted introns. This is in concordance with the fact that only Arabidopsis AtPYL7, AtPYL8, AtPYL9, and AtPYL10 contained putative intron regions. The intron number of CsPYL8 and CsPYL9 was also coincident with that of the Arabidopsis homologous genes (Table 1).
Table 1.
Comparison of PYR/PYL/RCAR, clade-A PP2C, and subclass III SnRK2 genes between Arabidopsis thaliana and Citrus sinensis
| Arabidopsis thaliana | Citrus sinensis | |||||||||
| Gene | Code | Introns | Amino acids | Gene | Genome code | Introns | Amino acids | Comparison with A. thaliana | ||
| Most similar | Identity | Match/aligned | ||||||||
| AtPYR1 | AT4G17870 | 0 | 191 | CsPYR1 | orange1.1g046151m | 0 | 209 | PYR1 | 74.0% | 130/175 |
| AtPYL1 | AT5G46790 | 0 | 221 | orange1.1g046151m | ||||||
| AtPYL2 | AT2G26040 | 0 | 190 | CsPYL2 | orange1.1g046697m | 0 | 187 | PYL2 | 72.0% | 121/168 |
| AtPYL3 | AT1G73000 | 0 | 209 | orange1.1g046697m | ||||||
| AtPYL4 | AT2G38310 | 0 | 207 | CsPYL4 | orange1.1g026007m | 0 | 245 | PYL4 | 78.0% | 136/174 |
| AtPYL5 | AT5G05440 | 0 | 203 | CsPYL5 | orange1.1g038201m | 0 | 201 | PYL5 | 70.0% | 111/158 |
| AtPYL6 | AT2G40330 | 0 | 215 | orange1.1g026007m | ||||||
| AtPYL7 | AT4G01026 | 2 | 211 | orange1.1g043944m | ||||||
| AtPYL8 | AT5G53160 | 2 | 188 | CsPYL8 | orange1.1g028067m | 2 | 214 | PYL8 | 75.0% | 137/183 |
| AtPYL9 | AT1G01360 | 2 | 187 | CsPYL9 | orange1.1g043944m | 2 | 186 | PYL9 | 86.0% | 143/167 |
| AtPYL10 | AT4G27920 | 2 | 183 | orange1.1g028067m | ||||||
| AtPYL11 | AT5G45860 | 0 | 161 | orange1.1g038201m | ||||||
| AtPYL12 | AT5G45870 | 0 | 159 | orange1.1g038201m | ||||||
| AtPYL13 | AT4G18620 | 0 | 164 | orange1.1g038201m | ||||||
| AtABI1 | AT4G26080 | 3 | 434 | CsABI1 | orange1.1g008880m | 4 | 550 | ABI1 | 68.0% | 227/391 |
| AtABI2 | AT5G57050 | 3 | 383 | orange1.1g008880m | ||||||
| AtAHG1 | AT5G51760 | 3 | 416 | CsAHG1 | orange1.1g013591m | 3 | 440 | AHG1 | 40.0% | 170/424 |
| AtAHG3 | AT3G11410 | 3 | 399 | CsAHG3 | orange1.1g015135m | 2 | 412 | AHG3 | 66.0% | 258/390 |
| AtHAB1 | AT1G72770 | 4 | 406 | CsHAB1 | orange1.1g009083m | 4 | 544 | HAB1 | 57.0% | 206/362 |
| AtHAB2 | AT1G17550 | 3 | 511 | orange1.1g009083m | ||||||
| AtHAI1 | AT5G59220 | 3 | 413 | orange1.1g036852m | ||||||
| AtHAI2 | AT1G07430 | 2 | 442 | orange1.1g036852m | ||||||
| AtHAI3 | AT2G29380 | 2 | 362 | CsHAI3 | orange1.1g036852m | 3 | 408 | HAI3 | 64.0% | 223/348 |
| AtSnRK2.2 | AT3G50500 | 8 | 369 | CsSnRK2.2 | orange1.1g017860m | 8 | 365 | SnRK2.2 | 82.0% | 297/362 |
| AtSnRK2.3 | AT5G66880 | 8 | 361 | orange1.1g017936m | ||||||
| AtSnRK2.6 | AT4G33950 | 9 | 362 | CsSnRK2.6 | orange1.1g017936m | 8 | 363 | SnRK2.6 | 88.9% | 317/352 |
In order to assess the degree of conservation of the ABA receptors in Citrus, amino acid sequences were aligned and the START-like domain was compared. Sequences were, in general, highly conserved between proteins of both species (Supplementary Fig. S1A at JXB online). The latch and gate loops of Citrus proteins were identical to those described in Arabidopsis, and the functional sites for ABA binding and interaction with PP2Cs were also perfectly conserved in all Citrus proteins. No important differences between Citrus ABA-binding regions and those of Arabidopsis were found, with the exception of an insert of 17 amino acids inside ABA-binding region 2 of the CsPYL8 protein. Furthermore, the alignment of the predicted secondary structure of AtPYR1 with the Citrus sequences showed that most of the elements described in this protein matched with the highly conserved regions of the Citrus homologues. High similarity in the number and location of α-helices and strands forming β-sheets was also found between all Citrus PYR/PYL/RCAR proteins and their Arabidopsis homologues (data not shown). In addition, the predicted tertiary structure of CsPYR1, CsPYL2, CsPYL5, and CsPYL8 showed that two helical segments and seven strands forming a β-sheet formed a cavity for ligand binding highly similar to that found in their respective Arabidopsis homologues (Supplementary Fig. S2A–D). Phylogenetic analysis further showed that the Citrus ABA receptors were distributed in the three main subfamilies proposed by Ma et al. (2009) in Arabidopsis, and two representative Citrus proteins were included in each subfamily (Fig. 1A): CsPYL8 and CsPYL9 belong to subfamily I, CsPYL4 and CsPYL5 to subfamily II, and CsPYR1 and CsPYL2 to subfamily III. In addition, a similarity matrix of the deduced amino acid sequences confirmed that proteins clustered into the same subfamily shared the highest percentage similarity among Citrus proteins (Supplementary Table S3).
Fig. 1.
Unrooted phylogenetic trees containing Citrus sinensis and Arabidopsis thaliana PYR/PYL/RCAR ABA receptors (A), clade-A PP2Cs (B), and SnRK2 protein kinases (C) obtained by using the Neighbor–Joining method in the PhyloWidget software and based on the protein sequence alignments. The full name for each protein is detailed in Table 1.
Family of clade-A PP2C proteins in Citrus sinensis
In the C. sinensis genome, five proteins were identified with significant homology to the nine members of the clade-A PP2C family of Arabidopsis (Table 1). The Citrus gene orange1.1g008880m was the most similar to both components of the ABA-insensitive (ABI) subfamily PP2Cs, AtABI1 and AtABI2. Nevertheless, since AtABI1 showed higher identity (68%) than AtABI2 (58%) to the Citrus protein, the gene was named CsABI1. The members of the ABA-hypersensitive germination (AHG) subfamily, AtAHG1 and AtAHG3, showed homology (40% and 66% identity, respectively) to different loci of the Citrus genome, which were named CsAHG1 (orange1.1g013591m) and CsAHG3 (orange1.1g015135m), respectively. Both components of the Arabidopsis homologous to ABI subfamily, also named hypersensitive to ABA (AtHAB1 and AtHAB2), shared homology to the same Citrus locus (orange1.1g009083m) and showed a very similar percentage identity (57% and 55%, respectively); therefore, the Citrus gene was named CsHAB1. Likewise, the three members of the Arabidopsis highly ABA-induced (HAI) PP2CA subfamily (AtHAI1, AtHAI2, and AtHAI3) shared homology to the same locus of Citrus sinensis and displayed 62, 57, and 64% identity, respectively, to the protein encoded by orange1.1g036852m, which consequently was named CsHAI3.
It is interesting to note that all genes of the clade-A PP2C from both Arabidopsis and Citrus contained introns, although the intron number for most of the genes was different between species (Table 1). Concerning the protein alignments, the PP2C-like domain was highly conserved throughout all proteins sequences although the length of all Citrus PP2CA proteins was longer than that of the Arabidopsis homologues (Supplementary Fig. S1B at JXB online). Metal-binding sites described in Arabidopsis were also identified in Citrus proteins, and phosphatase activity regulatory sequences were identical for all proteins analysed. The predicted secondary structure of AtABI1 matched the most conserved regions of the alignment. Secondary structures were also predicted for the Citrus clade-A PP2Cs, and similar sizes and location of the different motifs were observed when each protein was compared with its Arabidopsis homologue (data not shown). Prediction of the tertiary structure of CsABI1 was performed by using the crystallographic structure of AtABI1 as template, and it revealed a high degree of similarity in protein folding between species (Supplementary Fig. S2E). In addition, the phylogenetic tree constructed with Arabidopsis and Citrus PP2CAs showed that Citrus proteins fitted into the two groups described by Schweighofer et al. (2004) for these Arabidopsis proteins (Fig. 1B). Accordingly, the highest percentage similarity among Citrus protein sequences was found between CsABI1 and CsHAB1, and among CsAHG1, CsAHG3, and CsHAI3 proteins (Supplementary Table S4). Furthermore, representative genes of each group were identified and, as expected, each Citrus protein was clustered near to its corresponding Arabidopsis homologue.
The ABA-related subclass III SnRK2 proteins in Citrus
Among SnRK2s of Arabidopsis, the proteins belonging to subclass III, SnRK2.2, SnRK2.3, and SnRK2.6, have been found to be related to ABA signalling. A BLAST search in the C. sinensis genome revealed that two different loci (orange1.1g017860m and orange1.1g017936m) shared homology with these ABA-related SnRK2s (Table 1). The protein encoded by the gene orange1.1g017936m showed the highest identity (90%) to AtSnRK2.6, whereas orange1.1g017860m protein showed 82% identity to AtSnRK2.2. Therefore, these Citrus genes were named CsSnRK2.6 and CsSnRK2.2, respectively. Gene structure analysis revealed that the number of introns in CsSnRK2.6 and CsSnRK2.2 genes was very similar to that found in their Arabidopsis homologues. Amino acid alignment of the Arabidopsis subclass III SnRK2s and their corresponding Citrus homologues showed a kinase domain highly conserved between both species (Supplementary Fig. S1C at JXB online). Furthermore, the ATP-binding and the activation loop regions as well as the ATP-binding and the proton acceptor active sites were identical. In contrast, osmotic stress and ABA-responsive domains were less conserved, even among Arabidopsis proteins. The secondary structure predicted for AtSnRK2.6 showed that α-helices and β-strands matched with the most conserved regions in the protein alignment (Supplementary Fig. S1C). Additionally, the secondary structure predicted for CsSnRK2.2 revealed a high consensus in the number and location of the putative functional motifs when compared with its respective homologue (data not shown). Likewise, the tertiary structure of CsSnRK2.6 was predicted by using the crystallographic structure of AtSnRK2.6 as template, and protein folding was highly conserved between the species (Supplementary Fig. S2F). Phylogenetic analysis further revealed that Arabidopsis ABA-related SnRK2s proteins (AtSnRK2.2, AtSnRK2.3, and AtSnRK2.6) grouped in a branch (subclass III) independent from the other proteins belonging to this family, and the Citrus homologues (CsSnRK2.6 and CsSnRK2.2) were also clustered into this group (Fig. 1C). It should be also mentioned that these Citrus proteins displayed a high percentage (82%) similarity when their sequences were compared (Supplementary Table S5).
Transcriptional regulation of PYR/PYL/RCAR, PP2CA, and subclass III SnRK2 genes during orange fruit development and ripening: influence of endogenous ABA levels
In order to investigate the regulation of the ABA signalling core during citrus fruit development and ripening, and its relationship with endogenous ABA levels, the expression analysis of the six Citrus PYR/PYL/RCAR, five clade-A PP2C, and two SnRK2 genes was carried out together with the ABA measurement in the flavedo of fruits of ‘Navelate’ and its ABA-deficient mutant ‘Pinalate’.
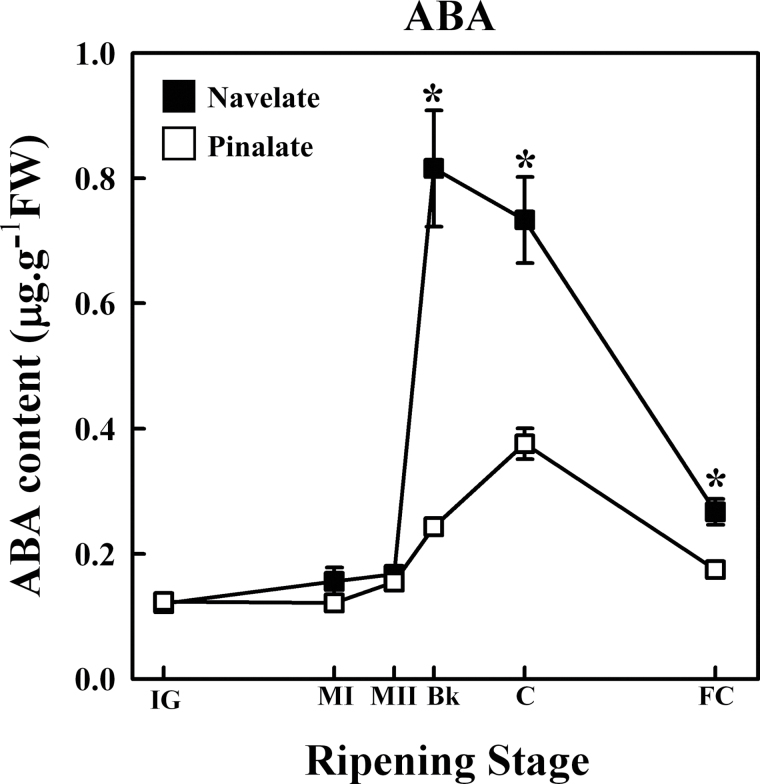
Six ripening stages were selected covering from IG to FC fruits. As expected, no difference in ABA content between ‘Navelate’ and the ‘Pinalate’ mutant was found while the fruits remained green (IG, MI, and MII stages), whereas the differences between parental and mutant fruit increased thereafter with fruit ripening (Fig. 2). The flavedo of parental fruits reached the highest ABA levels at the Bk stage, and the ABA content in the mutant was >3-fold lower. The ABA levels in ‘Pinalate’ fruit peaked at the C stage but the concentration was half of that reached in the parental fruits at the same ripening stage. In the flavedo of FC fruits from both varieties, an important decrease in ABA content was observed, but levels in the parental fruit remained higher than in the mutant (Fig. 2).
Fig. 2.
ABA content in the flavedo of ‘Navelate’ (black) and ‘Pinalate’ (white) fruit during development and ripening (immature green, IG; mature green I, MI; mature green II, MII; breaker, Bk; coloured, C; full coloured, FC). The results are the means of three biological replicates of five fruits each ±SE. Significant differences (P ≤ 0.05) in ABA content between ‘Navelate’ and ‘Pinalate’ flavedo samples for the same maturity stage are indicated by an asterisk.
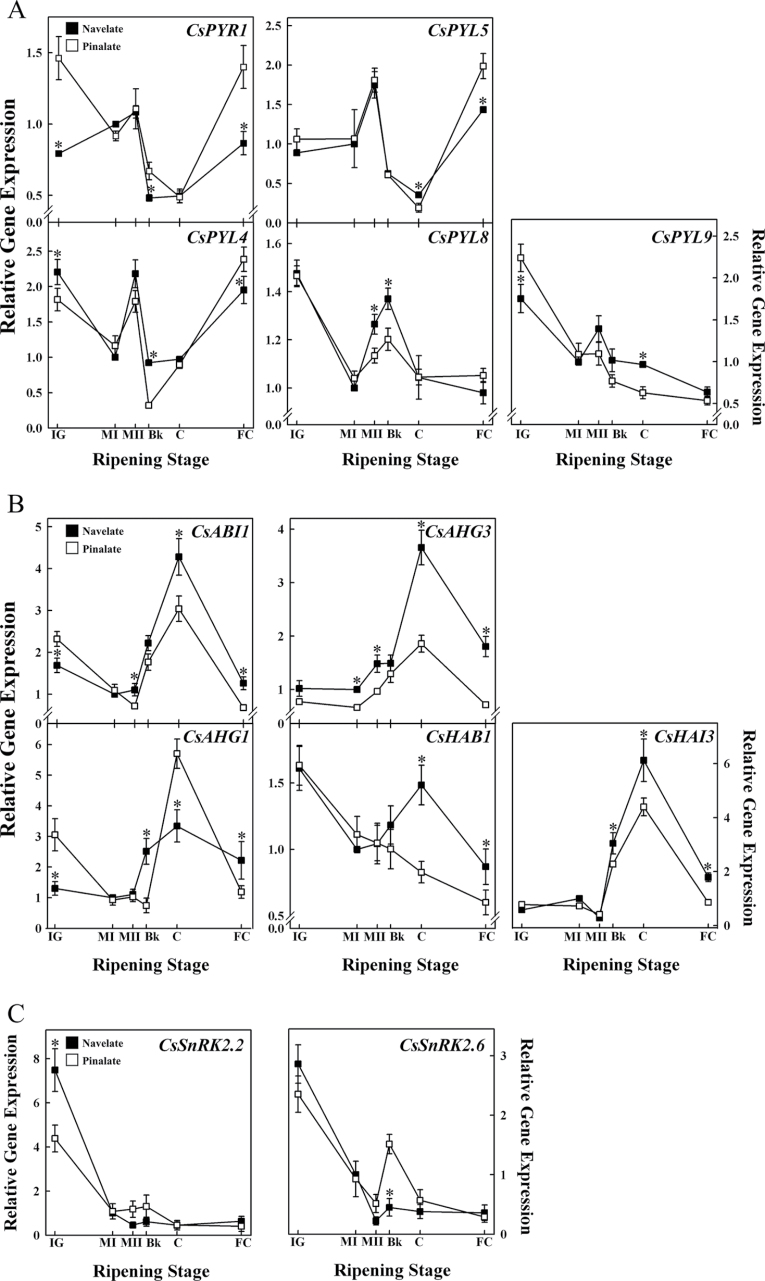
In spite of the differential ABA accumulation in ‘Navelate’ and ‘Pinalate’ flavedo during ripening, only minor differences were observed in the expression pattern of most of the PYR/PYL/RCAR genes between both cultivars (Fig. 3A) and remarkable differences were only observed in the expression profile of CsPYR1. In parental fruit, CsPYR1 transcript levels fluctuated during ripening, reaching a maximum at MII and a minimum at the Bk and C stages, and increased again at the FC stage to levels similar to those at IG. In the mutant fruit, the maximum expression levels of CsPYR1 were found at the IG and FC stages, reaching levels almost 2-fold higher than in ‘Navelate’. Nevertheless, the CsPYR1 transcript level and profile of mutant fruit was similar to that of the parent at intermediate ripening fruit stages (MI, MII, Bk, and C) and showed a minimum at the C stage (Fig. 3A). The evolution of CsPYL4 and CsPYL5 transcripts was similar to that described above since the expression of both genes peaked at MII in ‘Navelate’ and ‘Pinalate’ fruit, decreased dramatically to minimum levels at Bk and C, respectively, and then increased again to higher levels at FC. It should be mentioned that the repression of CsPYL4 at Bk was 2-fold higher in the ABA-deficient mutant. Overall, in spite of the differences observed between varieties, CsPYR1, CsPYL4, and CsPYL5 showed a consistent pattern in which the minimum transcript levels were coincident with the highest ABA levels. On the other hand, CsPYL8 and CsPYL9 displayed the maximum expression at the IG stage. A transient increase in the CsPYL8 expression levels occurred at the Bk stage, which was higher in ‘Navelate’ fruit, and accumulation of CsPYL8 and CsPYL9 decreased to reach minimum levels at the FC stage (Fig 3A). Moreover, absolute gene expression analysis further revealed similar levels of CsPYL4 and CsPYL5 transcripts, whereas the gene most expressed during fruit development and ripening was CsPYL9. It is also interesting that expression of CsPYR1 and CsPYL8 remained at very low levels and CsPYL2 was not detected in any of the fruit samples analysed (Supplementary Table S6 at JXB online).
Fig. 3.
Relative gene expression analysis by qRT-PCR of Citrus PYR/PYL/RCAR ABA receptors (A), clade-A PP2Cs (B), and subclass III SnRK2s (C) in ‘Navelate’ (black) and ‘Pinalate’ (white) fruits during fruit development and ripening (immature green, IG; mature green I, MI; mature green II, MII; breaker, Bk; coloured, C; full coloured, FC). Expression values are relative to transcript levels obtained in MI ‘Navelate’ fruits. Values are mean ratios ±SE from three biological samples for each sampling period and variety analysed in duplicate. Significant differences (P ≤ 0.05) in gene expression between ‘Navelate’ and ‘Pinalate’ flavedo samples for the same maturity stage are indicated by an asterisk.
The analysis of the Citrus clade-A PP2C genes revealed a differential regulation between both varieties. Although CsABI1, CsAHG3, and CsHAI3 transcript accumulation followed a similar pattern, peaking at the C stage in both ‘Navelate’ and ‘Pinalate’ fruit, the relative expression levels reached by the parental fruit were higher than those reached by the ABA-deficient mutant (Fig. 3B). Interestingly, CsAHG1 showed a similar expression profile to that described above for Citrus PP2CA genes, but the transcript levels at the C stage were 2-fold higher in ‘Pinalate’ than in ‘Navelate’. On the other hand, CsHAB1 was the only PP2CA gene whose expression decreased continuously in ‘Pinalate’ during fruit ripening while in ‘Navelate’ it displayed a transient increase at the C stage. In general, the highest expression levels of the Citrus PP2CA genes were observed at the C stage (Fig. 3B; Supplementary Table S6 at JXB online), agreeing with higher levels of ABA in both varieties. Interestingly, CsHAB1 showed the highest transcript accumulation in both varieties at the beginning of the experiment, followed by CsAHG3 and CsHAI3. However, only in ‘Navelate fruit were the transcript levels of CsAHG3 at the C stage almost double those of CsHAB1 and CsHAI3, and showed >14-fold accumulation compared with the other genes of this family (Supplementary Table S6).
Transcriptional analysis of Citrus SnRK2 genes revealed similar expression patterns between CsSnRK2.2 and CsSnRK2.6 genes, although the CsSnRK2.6 transcript accumulation was at least 8-fold higher than that of CsSnRK2.2 (Supplementary Table S6 at JXB online). The highest transcript levels were found at the IG stage in fruits of both cultivars and decreased thereafter as ripening progressed (Fig. 3C). Differences between cultivars in CsSnRK2.2 transcript accumulation were found at the IG stage, in which parental fruit showed 2-fold higher levels than the mutant. In contrast, similar relative transcript levels were found in CsSnRK2.6 at this stage, although gene expression peaked at the Bk stage in ‘Pinalate’ fruit but not in ‘Navelate’ (Fig. 3C). It should be mentioned that the expression level of these genes bottomed out in both varieties at the MII stage, which was concomitant with the inductions in several PYR/PYL/RCAR genes.
Water stress-induced changes in ABA content and transcriptional regulation of ABA signalosome components in leaves
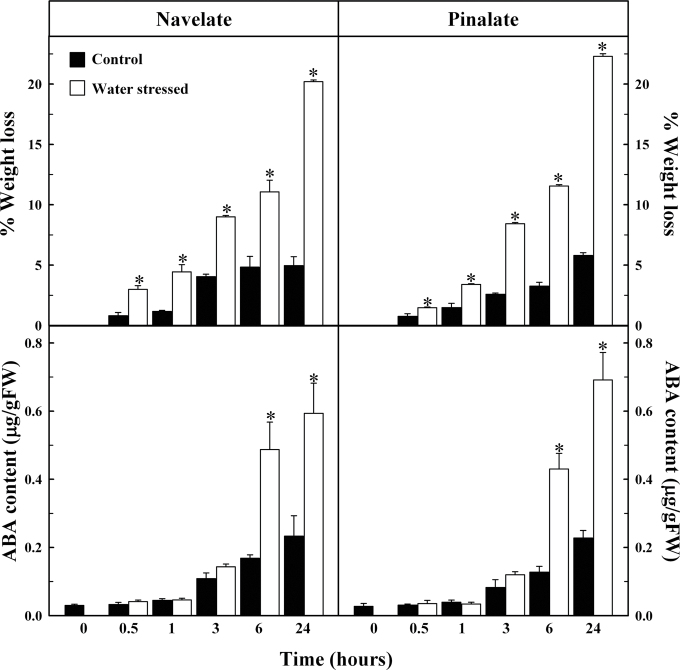
The evolution of ABA content and weight loss in ‘Pinalate’ and ‘Navelate’ leaves during the course of a water stress experiment was very similar for both genotypes. The ABA content increased ~20-fold in response to water stress by 24h (Fig. 4), whereas a minor increase was observed in detached control leaves of both genotypes. Significant differences in ABA content between dehydrated and control leaves were observed after 6h. Differences in weight loss between control and dehydrated leaves were observed from the beginning of the experiment (0.5h). Therefore, the increase in weight loss preceded that of ABA, and significant increases in ABA in response to dehydration only occurred when the leaves reached a 10% weight loss (Fig. 4).
Fig. 4.
Effect of water stress on weight loss and ABA content in ‘Navelate’ and ‘Pinalate’ detached leaves. Changes in control samples are represented as black bars and in water-stressed leaves as white bars. The results are the means of three biological replicates of four leaves each ±SE. Significant differences (P ≤ 0.05) in weight loss and ABA content between samples for the same analysed period are indicated by an asterisk.
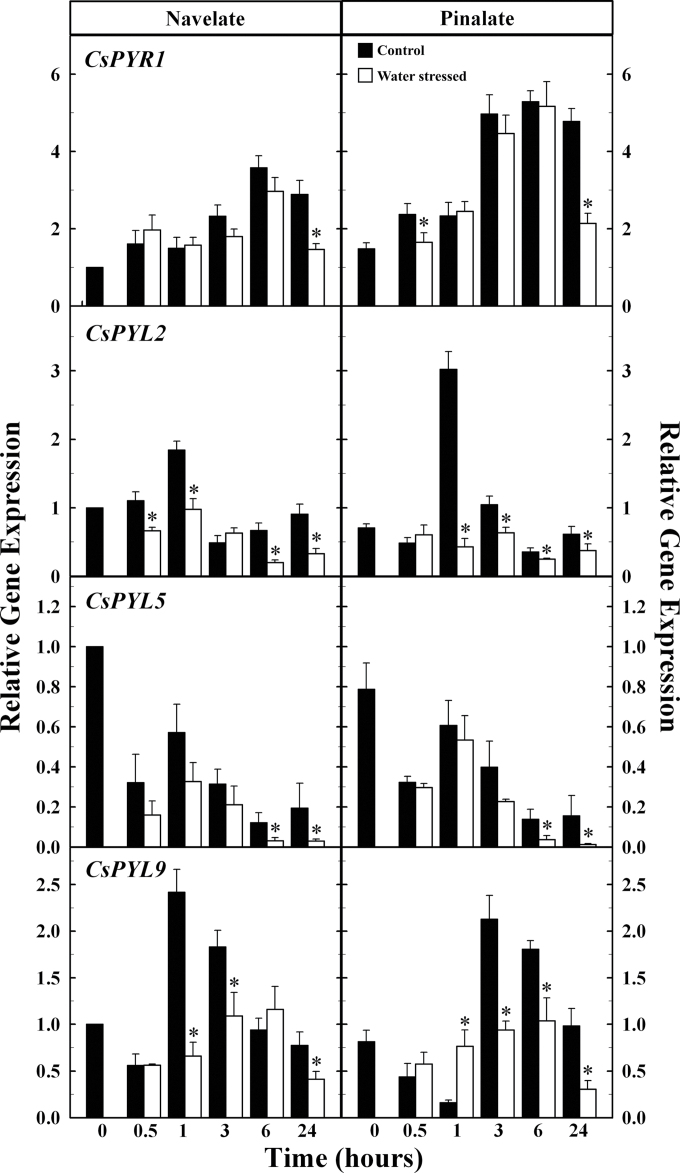
The accumulation of CsPYR1 increased in both ‘Pinalate’ and ‘Navelate’ leaves after detachment, but no significant differences were found between control and water-stressed leaves until the end of the experiment (24h) (Fig. 5). The most important increase occurred by 6h, and 3- and 5-fold increases were found in ‘Navelate’ and ‘Pinalate’ leaves, respectively. Thereafter, the expression level remained almost constant in the control leaves but significantly decreased in water-stressed ‘Pinalate’ and ‘Navelate’ samples (Fig. 5). In contrast to that found in flavedo samples, CsPYL2 expression was detected in leaves, and the results showed that this gene was down-regulated by water stress. The relative gene expression of CsPYL5 decreased rapidly (0.5h) after leaf detachment, and significant differences between control and water-stressed leaves were only found by 6h and 24h. On the other hand, CsPYL9 gene expression sharply increased and reached a maximum by 1h and 3h in control ‘Navelate’ and ‘Pinalate’ leaves, respectively. The transcript level of this gene was, in general, lower in water-stressed leaves, and changes were less relevant. Moreover, CsPYL4 and CsPYL8 transcripts were not detected in fresh, detached, or water-stressed ‘Pinalate’ and ‘Navelate’ leaves. Interestingly, absolute expression showed that CsPYL2 and CsPYL9 were the most highly expressed genes in leaves, whereas CsPYR1 and CsPYL5 transcript accumulation remained at very low levels during the whole experiment (Supplementary Table S6 at JXB online).
Fig. 5.
Relative gene expression analysis by qRT-PCR of Citrus PYR/PYL/RCAR ABA receptors in control (black) and water-stressed (white) ‘Navelate’ and ‘Pinalate’ leaves. The results are the means of three biological replicates of four leaves each ±SE. Significant differences (P ≤ 0.05) in gene expression between samples for the same analysed period are indicated by an asterisk.
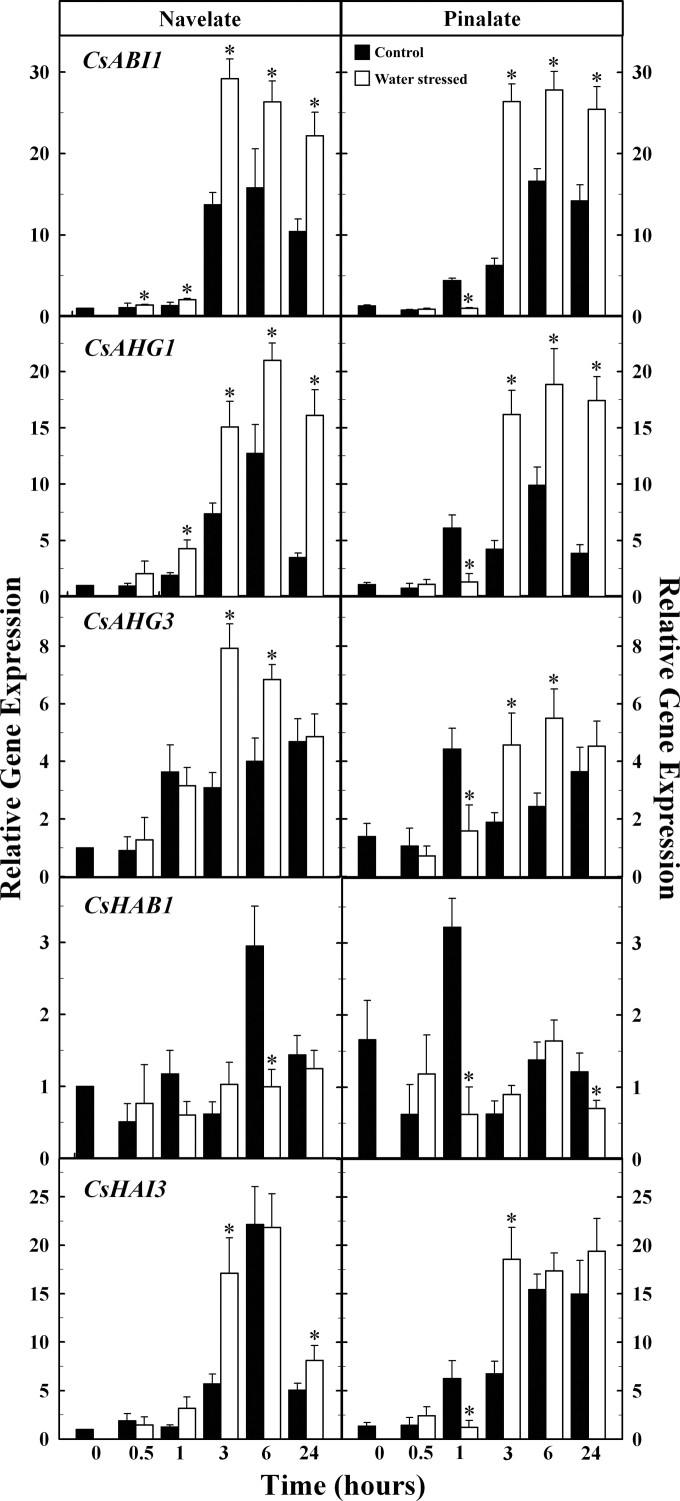
The expression of the CsPP2CA genes increased after detachment in both control and stressed ‘Navelate’ and ‘Pinalate’ leaves, but such increases were, in general, substantially higher in the water-stressed leaves (Fig. 6). As shown in Fig. 6, dehydration had an important impact, up-regulating the expression of both CsABI1 and CsAHG1 genes, which reached maximum levels by 3h in water-stressed leaves. The effect of dehydration on CsAHG3 gene expression was also evident, and important differences between control and stressed leaves were found by 3h and 6h after detachment. In contrast, dehydration had little effect on CsHAB1 and CsHAI3 transcript levels. Absolute gene expression analysis revealed that CsHAB1 was the most expressed CsPP2CA in freshly harvested leaves, followed by CsAHG3, CsHAI3, and CsABI1. In contrast, the most expressed genes during dehydration of ‘Navelate’ and ‘Pinalate’ leaves were CsAHG3, CsHAI3, and CsABI1 (Supplementary Table S6 at JXB online). As occurred in fruit, CsAHG1 transcript accumulation remained at much lower levels in both varieties.
Fig. 6.
Relative gene expression analysis by qRT-PCR of Citrus clade-A PP2Cs in control (black) and water-stressed (white) ‘Navelate’ and ‘Pinalate’ leaves. The results are the means of three biological replicates of four leaves each ±SE. Significant differences (P ≤ 0.05) in gene expression between samples for the same analysed period are indicated by an asterisk.
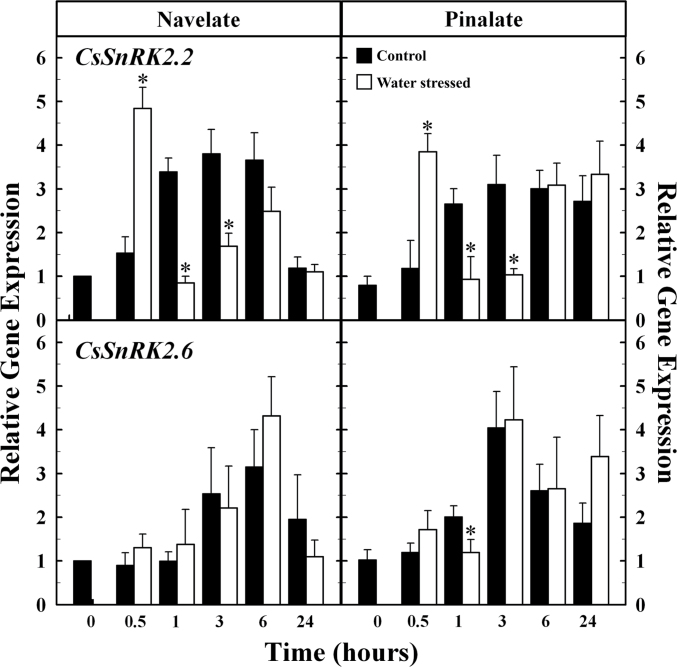
The CsSnRK2.2 gene showed a different gene expression profile in dehydrated and control leaves (Fig. 7). The transcript level of this gene transiently peaked by 0.5h in ‘Navelate’ and ‘Pinalate’ water-stressed leaves, whereas the expression continuously increased from 0h to 3h in the control leaves. Interestingly, CsSnRK2.2 expression was similar in control and stressed ‘Navelate’ and ‘Pinalate’ leaves by 6h, but transcript accumulation was higher in leaves of the mutant at the end of the experiment. The CsSnRK2.6 expression pattern barely differed between control and dehydrated leaves and was very similar in ‘Navelate’ and ‘Pinalate’. The transcript levels of this gene continuously increased after detachment and reached a maximum by 3h in ‘Pinalate’ and by 6h in ‘Navelate’ leaves. It is also interesting to note that absolute gene expression of CsSnRK2.6 was substantially higher than that of the CsSnRK2.2 gene during the whole experiment in both varieties (Supplementary Table S6 at JXB online).
Fig. 7.
Relative gene expression analysis by qRT-PCR of Citrus subclass III SnRK2s in control (black) and water-stressed (white) ‘Navelate’ and ‘Pinalate’ leaves. The results are the means of three biological replicates of four leaves each ±SE. Significant differences (P ≤ 0.05) in gene expression between samples for the same analysed period are indicated by an asterisk.
Discussion
The homologous genes of the ABA signalosome have been identified in this work for the first time in the Citrus genome in order to explore the relationship between the regulation of these components and the changes in the endogenous ABA levels occurring in citrus fruit during natural fruit ripening and in dehydrated leaves. A comparative transcriptional analysis of these genes has been performed between ‘Navelate’ orange fruit and its spontaneous fruit-specific ABA-deficient mutant ‘Pinalate’. In this context, it is noteworthy that the link between ABA and the ripening process has been reported in non-climacteric fruits such as strawberry (Chai et al., 2011), grapevine (Giribaldi et al., 2010), sweet cherry (Ren et al., 2011), and citrus (Lafuente et al., 1997; Alférez and Zacarías, 1999; Rodrigo et al., 2003; Gambetta et al., 2011), although the molecular mechanism of how ABA regulates this process has not been fully established.
In silico analysis of the sweet orange (C. sinensis) genome database has revealed that proteins belonging to the ABA signalosome were less represented in Citrus as compared with Arabidopsis. Only six PYR/PYL/RCAR, five PP2CA, and two subclass III SnRK2 genes were found in Citrus (Table 1), while in Arabidopsis there are 14 PYR/PYL/RCAR ABA receptors, nine clade-A PP2Cs, and several protein kinases, including three of the subclass III SnRK2 (Merlot et al., 2001; Yoshida et al., 2002; R. Yoshida et al., 2006; Ma et al., 2009; Park et al., 2009). This is in concordance with the lower number of PYR/PYL/RCAR and PP2CA genes recently identified in tomato (Sun et al., 2011) and in strawberry (Chai et al., 2011). High percentage identity was observed between Citrus proteins and their homologues in Arabidopsis, as well as similar protein length and genetic structures (Table 1; Supplementary Tables S3–S5 at JXB online). Interestingly, the consensus motifs for functional protein folding, such as the gate and latch regions in PYR/PYL/RCARs (Melcher et al., 2009) (Supplementary Figs S1A, Fig. S2), and for phosphatase activity in PP2CAs (Weiner et al., 2010) (sequences underlined in Supplementary Fig. S1B) were identified in Citrus. D-rich C-terminal domain II, which has been shown to be essential for ABA signal transduction (R. Yoshida et al., 2006), was also fully conserved in Citrus SnRK2s proteins (number 4 in Supplementary Fig. S1C). Phylogenetic analysis revealed that Citrus PYR/PYL/RCARs were clustered together with their homologues in accordance with the distribution proposed by Ma et al. (2009), in which Arabidopsis ABA receptors were divided into three main subfamilies. In fact, two representative genes of each group were identified in the Citrus genome (Fig. 1A). The Citrus clade-A PP2Cs were clustered close to their respective homologues (Fig. 1B) and arranged in two separate branches as previously described by Schweighofer et al. (2004) in the phylogenetic analysis of Arabidopsis PP2Cs. Furthermore the two Citrus kinases, CsSnRK2.2 and CsSnRK2.6, were classified into subclass III of AtSnRK2s (Fig. 1C), whose components have been related to ABA signalling (Fujii and Zhu, 2009). Therefore, the sweet orange proteins encoded by CsPYR/PYL/RCAR, CsPP2CA, and CsSnRK2 genes identified in this work might function as the core elements of the ABA perception and signalling pathway.
The comparative transcriptional analysis between wild-type ‘Navelate’ fruit and its ABA-deficient mutant ‘Pinalate’ revealed no important differences in most of the CsPYR/PYL/RCAR expression profiles, although the transcript level of CsPYR1 in IG and FC fruits was different between genotypes. This result suggests that the expression of this gene family might be developmentally regulated in Citrus and that changes in ABA content found in ‘Pinalate’ fruit during ripening may be sufficient for regulating CsPYR/PYL/RCAR gene expression. Indeed, the expression profiles of CsPYR1, CsPYL4, and CsPYL5 suggest the involvement of ABA in their regulation since the lowest transcript levels of these genes were concomitant with the highest ABA levels in ‘Navelate’ and ‘Pinalate’ fruits, whereas their expressions peaked before the ABA increase (Figs 2, 3A). This result is in agreement with that found in sweet cherry showing the concomitant down-regulation of the plastid ABA receptor magnesium chelatase (CHLH) and the increment in endogenous ABA during fruit ripening (Ren et al., 2011). Overall, these results suggest that the reduction in ABA receptor gene expression may be concomitant with the increase in ABA during non-climacteric fruit ripening. In this context, it should be pointed out that the expression of PYR/PYL/RCAR genes is differentially affected by ABA treatment in seedlings of Arabidopsis (Santiago et al., 2009b; Szostkiewicz et al., 2010), and that the accumulation of PYR/PYL/RCAR transcripts may also parallel the increase in ABA during ripening of strawberry and tomato fruits (Chai et al., 2011; Sun et al., 2011). In this work, three different expression patterns were observed among PYR/PYL/RCAR genes: a first set of genes (CsPYR1, CsPYL4, and CsPYL5) showed their minimum transcript levels when the highest ABA content was detected in the flavedo of ‘Navelate’ and ‘Pinalate’ fruits. Although their expression patterns were similar, transcript accumulation of CsPYR1 was much lower than that of CsPYL4 and CsPYL5 genes, which showed similar values (Supplementary Table S6 at JXB online). Secondly, the CsPYL8 transcript level peaked when ABA increased during fruit ripening and, finally, CsPYL9 continuously decreased as ripening progressed, although it increased slightly before the increment in ABA in both varieties (Fig. 3A). Interestingly, genes whose changes in expression did not mirror ABA accumulation during ripening (CsPYL8 and CsPYL9) were clustered into subfamily I (Fig. 1A). In this context, it is interesting to note that tomato genes belonging to this subfamily (SlPYL1, SlPYL2, and SlPYL3) have been related to ABA changes during fruit development and ripening (Sun et al., 2011), and functional activity for AtPYL8 and AtPYL9 proteins has been demonstrated by Ma et al. (2009) in vegetative tissues. It is noteworthy, however, that the ABA-binding region of CsPYL8 showed an insert of 17 amino acids, which is not present either in Arabidopsis or in tomato sequences and might affect the ability of this protein to bind the hormone (Supplementary Fig. S1A). In addition, CsPYL8 transcript levels were much lower than that of CsPYL9, which showed the highest transcript accumulation among CsPYR/PYL/RCAR genes (Supplementary Table S6). It is also interesting that the expression of the CsPYL2 gene was not detected in fruits of both cultivars during ripening, which suggests that the expression of some ABA receptors could be tissue specific in Citrus. In agreement with this, some tomato genes such as SlPYL5, belonging to the same subfamily as CsPYL2 (subfamily III, Fig. 1A), were almost undetectable during fruit ripening (Sun et al., 2011). Therefore, gene expression levels indicated the relevance of CsPYL4, CsPYL5, and CsPYL9 genes in ABA perception during fruit development and ripening.
Clade-A PP2Cs function as negative regulators of the ABA signalling pathway and their expression is highly induced by ABA in plants (Merlot et al., 2001; Saez et al., 2004; T. Yoshida et al., 2006; Xue et al., 2008; Li et al., 2009). Within this context, a transcriptional negative feedback regulatory mechanism has been proposed for modulating the ABA responses (Merlot et al., 2001; Melcher et al., 2009; Santiago et al., 2009b; Vlad et al., 2009; Weiner et al., 2010). Thus, the initial response to ABA implies the ABA-dependent PYR/PYL/RCAR-mediated inactivation of PP2CAs, which allows the release of SnRKs and hence the phosphorylation of ABA-dependent transcription factors or other proteins. This ABA signal is later attenuated by the up-regulation of PP2CA and the down-regulation of PYR/PYL/RCAR genes in an ABA-dependent manner. Thus, the resetting of the ABA transduction pathway offers a dynamic mechanism to modulate the ABA response (Santiago et al., 2009b). The expression pattern of the CsPP2CA genes analysed in this work mostly paralleled the ABA accumulation in ‘Navelate’ and ‘Pinalate’ fruit during ripening(Figs 2, 3B) and, interestingly, the up-regulation of CsPP2CA genes was also concomitant with the down-regulation of the CsPYL4 and CsPYL5 genes (Fig. 3A). Therefore, these results suggest that a transcriptional negative feedback regulatory mechanism might be modulating the ABA responses during Citrus fruit ripening. In tomato, however, only SlPP2C1 and SlPP2C5 transcripts peaked, with the increment in ABA occurring during fruit ripening, while all of the SlPYR/PYL/RCAR genes analysed were negatively related to the accumulation of those SlPP2C genes (Sun et al., 2011). Therefore, it would be interesting to investigate further the functionality of these proteins through protein–protein interactions, which would help to unravel the involvement of these subfamilies in ABA perception in Citrus.
The availability of the fruit-specific ABA-deficient mutant ‘Pinalate’ has allowed analysis of the relationship between the expression of the CsPP2CA genes and endogenous ABA accumulation during Citrus fruit ripening. Gene expression levels of CsABI1, CsAHG3, and CsHAI3 peaked at the C stage in both ‘Navelate’ and ‘Pinalate’ fruit, but transcript accumulation was always higher in parental fruit. Likewise, CsHAB1 transcript levels peaked in ‘Navelate’ at the C stage, although the level continuously decreased in the ABA-deficient mutant fruit (Fig. 3B). These results, together with the fact that gene expression of CsAHG3 and CsHAI3 increased from 3- to 10-fold (Fig. 3B; Supplementary Table S6 at JXB online), suggest an important effect of ABA content on CsPP2CA gene expression. In agreement with this idea and with the lower differences found between cultivars in the CsPYR/PYL/RCAR transcriptional levels, Szostkiewicz et al. (2010) reported that PP2CA genes were more responsive to ABA compared with ABA receptors, and suggested a higher sensitivity of these negative regulators to ABA changes. Unexpectedly, although CsAHG1 showed an expression pattern similar to that of the other CsPP2CA genes, the transcript level at the C stage was 2-fold higher in the ABA-deficient mutant whereas the ABA content in ‘Navelate’ was double that of ‘Pinalate’ (Figs 2, 3B). The increased expression of this negative regulator supports previous molecular data suggesting the impaired response of this mutant to ABA treatments and dehydration (Romero et al., 2012). Moreover, the expression of well-known ABA-dependent genes is also strongly reduced in the mutant fruit during ripening (Supplementary Fig. S3), which further supports the idea of a reduced sensitivity of ‘Pinalate’ fruit to ABA.
It is well known that the release of SnRK2s by PP2CAs after ABA binding to PYR/PYL/RCARs allows these positive effectors to phosphorylate downstream transcription factors and proteins involved in the ABA response (Umezawa et al., 2009; Vlad et al., 2009; Hirayama and Umezawa, 2010). The results obtained in the present work revealed that both CsSnRK2.2 and CsSnRK2.6 genes reached their highest transcript levels at the most immature stages, when the minimum ABA content was detected in both cultivars (Figs 2, 3C), although transcript accumulation of the CsSnRK2.6 gene was much higher than that of CsSnRK2.2 in both varieties during fruit ripening (Supplementary Table S6 at JXB online). As ripening progressed, however, CsSnRK2.2 remained almost unchanged in ‘Navelate’ and ‘Pinalate’ fruits, whereas CsSnRK2.6 showed a transient increase at the Bk stage only in the mutant fruit. Similar expression patterns were found in the climacteric tomato fruit. During ripening of tomato, expression levels of SnRKs were high in the most immature stages and transiently increased with the increase in ABA (Sun et al., 2011). In spite of differences found in CsSnRK2.6 transcript levels between ‘Navelate’ and ‘Pinalate’ fruits, the overall results suggest that the relationship between endogenous ABA content and the transcriptional regulation of these components of the ABA signalosome during Citrus fruit ripening is less relevant than that occurring for the CsPP2CA genes.
In order to gain further insights into the role of the ABA signalosome components and to understand whether the key genes are common or tissue specific in Citrus, expression analysis of these elements has also been performed in leaves exposed to dehydration. As indicated above, the deficiency in ABA of ‘Pinalate’ is fruit specific and, consequently, no relevant differences were found in ABA content or weight loss between ‘Navelate’ and ‘Pinalate’ leaves. Since leaves are very prone to dehydration, special attention was paid to minimizing the water loss in control leaves. Under the experimental conditions used, water loss was always <5% and 4-fold lower than in the water-stressed leaves. Thus, changes observed in ABA levels can be related to changes in weight loss. The attenuated expression profiles of some of the studied genes in control leaves suggest that the response of vegetative tissue to dehydration may depend on the severity of the stress imposed.
Gene expression changes of the three core components of the ABA signalosome in dehydrated leaves (Figs 5–7) were similar to those found in Arabidopsis (Santiago et al., 2009; Szostkiewicz et al., 2009). Overall, transcriptional profiling of these genes suggested that ABA increases caused by dehydration up-regulate the levels of all CsPP2CA and down-regulate some PYR/PYL/RCAR and SnRK2 family members, such as CsPYL2, CsPYL5, CsPYL9, and CsSnRK2.2, whereas the relative levels of other members of these families, such as CsPYR1 and CsSnRK2.6, remain fairly constant. Nevertheless, it is interesting to note that CsPYL2 and CsPYL9 were the most expressed genes in control and dehydrated leaves, suggesting that CsPYL2 might play a key role in ABA responses in leaves but not in fruit, while CsPYL9 could be relevant in both vegetative and reproductive tissues (Supplementary Table S6 at JXB online). Therefore, the results obtained in the leaf are complementary to those found in reproductive tissue. The high transcript accumulation of CsPYL9 was down-regulated as ABA increased during both fruit ripening and leaf dehydration. Likewise, CsPYR1 and CsPYL5 gene expression was lowest when the highest ABA levels were achieved during fruit ripening and leaf dehydration, although transcript accumulation of CsPYL5 in fruit was much higher than in vegetative tissue, suggesting a minor role for this gene in leaves (Supplementary Table S6). Moreover, the expression profile of these genes did not mirror that of ABA accumulation during leaf dehydration as occurred during fruit ripening. This differential regulation under physiological or stress conditions may provide a means for the plant to cope with sustained high levels of ABA or to adjust the sensitivity of ABA perception and signalling. It is interesting to mention that some common responses in the ABA signalosome were observed between fruit and leaf tissues, such as the high sensitivity of the CsPP2CA gene components to ABA changes and CsSnRK2.6 as the major subclass III SnRK2 expressed gene. Moreover, in both tissues, CsPYR1 transcript accumulation was very low compared with the other CsPYR/PYL/RCAR genes, and CsPYL9 was highly expressed. Comparison between fruit and vegetative tissue has also revealed some tissue specificity: the CsPYL2 gene was highly expressed in leaves but no expression was detected in fruit, whereas CsPYL4 and CsPYL8 transcripts were detected during fruit development but not in leaves subjected or not to water stress.
In summary, this work reports for the first time the identification of ABA signalling core components in Citrus comprising six PYR/PYL/RCAR ABA receptors, five PP2CAs, and two subclass III SnRK2s. During sweet orange fruit development and ripening, the expression pattern of some ABA receptors mirrored the ABA content, whereas that of CsPP2CA genes paralleled the hormone accumulation, together modulating ABA perception, downstream signalling, and, consequently, physiological ABA responses. Additionally, transcriptional analysis performed in water-stressed leaves revealed that some members of the PYR/PYL/RCAR family are tissue specific and that sensitivity to ABA changes in the PP2CA genes, which are negative regulators of the ABA signal transduction pathway, was much higher than in other components of the ABA signalosome.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Multiple sequence alignment of the Arabidopsis thaliana and Citrus sinensis ABA signalling core components.
Figure S2. Predicted tertiary structure model of the ABA signalosome components of Citrus.
Figure S3. Gene expression analysis of ABA-dependent downstream signalling genes.
Table S1. Colour evolution of ‘Navelate’ and ‘Pinalate’ fruit during ripening.
Table S2. Primers designed for the ABA signalling core component gene expression analysis by quantitative RT-PCR (qRT-PCR).
Table S3 Similarity matrix between Citrus and Arabidopsis PYR/PYL/RCAR proteins.
Table S4. Similarity matrix between Citrus and Arabidopsis PP2CA proteins.
Table S5. Similarity matrix between Citrus and Arabidopsis SnRK2 proteins.
Table S6. Absolute gene expression levels of ABA signalosome components during fruit ripening and leaf dehydration.
Supplementary Material
Acknowledgements
We thank Dr L. Navarro (IVIA, Spain) for allowing us to use the Spanish Citrus Germplasm Bank. Special thanks are also due to Dr L. Gonzalez-Candelas for his help with absolute gene expression analysis. The technical assistance of M. Sánchez-Hervás is also gratefully acknowledged. This work was supported by the Spanish Ministry of Science and Technology (Research Grants AGL2006-09496, AGL2009-11969, and AGL2009-11558) and by the Generalitat Valenciana (PROMETEO/2010/010). PR was the recipient of a fellowship from the Spanish Ministry of Science and Technology.
References
- Agustí J, Zapater M, Iglesias DJ, Cercós M, Tadeo FR, Talón M. 2007. Differential expression of putative 9-cis-epoxycarotenoid dioxygenases and abscisic acid accumulation in water stressed vegetative and reproductive tissues of citrus Plant Science 172, 85–94. [Google Scholar]
- Alférez F, Zacarías L. 1999. Interaction between ethylene and abscisic acid in the regulation of citrus fruit maturation In: Kanellis AK, Chang C, Klee H, Blecker AB, Pech JC, Grierson D. eds. Biology and biotechnology of the plant hormone ethylene II Amsterdam: Kluwer Academic Publishers; 183 184 [Google Scholar]
- Antoni R, Rodríguez L, González-Guzman M, Pizzio GA, Rodríguez PL. 2011. News on ABA transport, protein degradation, and ABFs/WRKYs in ABA signaling Current Opinion in Plant Biology 14, 547–553. [DOI] [PubMed] [Google Scholar]
- Armstrong F, Leung J, Grabov A, Brearley J, Giraudat J, Blatt MR. 1995. Sensitivity to abscisic acid of guard-cell K+ channels is suppressed by abi1-1, a mutant Arabidopsis gene encoding a putative protein phosphatase Proceedings of the National Academy of Sciences, USA 92, 9520–9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester AR, Lafuente MT, González-Candelas L. 2006. Spatial study of antioxidant enzymes, peroxidase and phenylalanine ammonia-lyase in the citrus fruit–Penicillium digitatum interaction Postharvest Biology and Technology 39, 115–124. [Google Scholar]
- Bartels D, Sunkar R. 2005. Drought and salt tolerance in plants Critical Reviews in Plant Sciences 24, 23–58. [Google Scholar]
- Bastías A, López-Climent M, Valcárcel M, Rosello S, Gómez-Cadenas A, Casaretto JA. 2011. Modulation of organic acids and sugar content in tomato fruits by an abscisic acid-regulated transcription factor Physiologia Plantarum 141, 215–226. [DOI] [PubMed] [Google Scholar]
- Chai YM, Jia HF, Li CL, Dong QH, Shen YY. 2011. FaPYR1 is involved in strawberry fruit ripening Journal of Experimental Botany 62, 5079–5089. [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Gampala SL, Rock CD. 2002. Abscisic acid signaling in seeds and seedlings. The Plant Cell 14, S15–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Zhu JK. 2009. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress Proceedings of the National Academy of Sciences, USA 106, 8380–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galpaz N, Wang Q, Menda N, Zamir D, Hirschberg J. 2008. Abscisic acid deficiency in the tomato mutant high-pigment 3 leading to increased plastid number and higher fruit lycopene content The Plant Journal 53, 717–730. [DOI] [PubMed] [Google Scholar]
- Gambetta G, Martínez-Fuentes A, Bentancur O, Mesejo C, Reig C, Gravina A, Agustí M. 2011. Hormonal and nutritional changes in the flavedo regulating rind color development in sweet orange (Citrus sinensis (L.) Osb.). Journal of Plant Growth Regulation (in press) [Google Scholar]
- Giribaldi M, Gény L, Delrot S, Schubert A. 2010. Proteomic analysis of the effects of ABA treatments on ripening Vitis vinifera berries Journal of Experimental Botany 61, 2447–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Cadenas A, Arbona V, Jacas J, Primo-Millo E, Talon M. 2002. Abscisic acid reduces leaf abscission and increases salt tolerance in citrus plants Journal of Plant Growth Regulation 21, 234–240. [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AAR, Vartanian N, Giraudat J. 1999. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling The Plant Cell 11, 1897–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot SPC, Karssen CM. 1992. Dormancy and germination of abscisic acid-deficient tomato seeds: studies with the sitiens mutant Plant Physiology 99, 952–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Umezawa T. 2010. The PP2C–SnRK2 complex: the central regulator of an abscisic acid signaling pathway Plant Signaling and Behavior 5, 160–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huai J, Wang M, He J, et al. 2008. Cloning and characterization of the SnRK2 gene family from Zea mays Plant Cell Reports 27, 1861–1868 [DOI] [PubMed] [Google Scholar]
- Jia HF, Chai YM, Li CL, Lu D, Luo JJ, Qin L, Shen YY. 2011. Abscisic acid plays an important role in the regulation of strawberry fruit ripening Plant Physiology 157, 188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Hwang JU, Lee M, Kim YY, Assmann SM, Martinoia E, Lee Y. 2010. PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proceedings of the National Academy of Sciences, USA 107, 2355–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karssen CM, Brinkhorst-van der Swan DLC, Breekland AE, Koornneef M. 1983. Induction of dormancy during seed development by endogenous abscisic acid: studies on abscisic acid deficient genotypes of Arabidopsis thaliana (L.) Heynh Planta 157, 158–165. [DOI] [PubMed] [Google Scholar]
- Kim H, Hwang H, Hong JW, et al. 2012. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth Journal of Experimental Botany 63, 1013–1024. [DOI] [PubMed] [Google Scholar]
- Klingler JP, Batelli G, Zhu JK. 2010. ABA receptors: the START of a new paradigm in phytohormone signalling Journal of Experimental Botany 61, 3199–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D. 2004. Naturally occurring genetic variation in Arabidopsis thaliana Annual Review of Plant Biology 55, 141–172. [DOI] [PubMed] [Google Scholar]
- Kuromori T, Miyaji T, Yabuuchi H, et al. 2010. ABC transporter AtABCG25 is involved in abscisic acid transport and responses Proceedings of the National Academy of Sciences, USA 107, 2361–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafuente MT, Martínez-Téllez MA, Zacarías L. 1997. Abscisic acid in the response of ‘Fortune’ mandarins to chilling. Effect of maturity and high-temperature conditioning Journal of the Science of Food Agriculture 73, 494–502. [Google Scholar]
- Li FH, Fu FL, Sha LN, He L, Li WC. 2009. Differential expression of serine/threonine protein phosphatase type-2C under drought stress in maize Plant Molecular Biology Reporter 27, 29–37. [Google Scholar]
- Li G, Xin H, Zheng XF, Li S, Hu Z. 2012.. Identification of the abscisic acid receptor VvPYL1 in Vitis vinifera Plant Biology 14, 244–248. [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Alexander C, Grill E. 2009. Regulators of PP2C phosphatase activity function as abscisic acid sensors Science 324, 1064–1068. [DOI] [PubMed] [Google Scholar]
- Melcher K, Ng LM, Zhou XE, et al. 2009. A gate–latch–lock mechanism for hormone signalling by abscisic acid receptors Nature 462, 602–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J. 2001. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway The Plant Journal 25, 295–303. [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. 2005. Abscisic acid biosynthesis and catabolism Annual Review of Plant Biology 56, 165–185 [DOI] [PubMed] [Google Scholar]
- Nishimura N, Sarkeshik A, Nito K, et al. 2010. PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis The Plant Journal 61, 290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman SM, Poling SM, Maier VP. 1988. An indirect enzyme-linked immunosorbent assay for (+)-abscisic acid in Citrus, Ricinus, and Xanthium leaves Journal of Agricultural and Food Chemistry 36, 225–231 [Google Scholar]
- Park SY, Fung P, Nishimura N, et al. 2009. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins Science 324, 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Cortés H, Sánchez-Serrano JJ, Mertens R, Willmitzer L, Prat S. 1989. Abscisic acid is involved in the wound-induced expression of the proteinase inhibitor II gene in potato and tomato Proceedings of the National Academy of Sciences, USA 86, 9851–9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29,:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Sun L, Wang C, Zhao S, Leng P. 2011. Expression analysis of the cDNA for magnesium chelatase H subunit (CHLH) during sweet cherry fruit ripening and under stress conditions Plant Growth Regulation 63, 301–307. [Google Scholar]
- Rodrigo MJ, Alquézar B, Zacarías L. 2006. Cloning and characterization of two 9-cis-epoxycarotenoid dioxygenase genes, differentially regulated during fruit maturation and under stress conditions, from orange (Citrus sinensis L. Osbeck). Journal of Experimental Botany 57, 633–643. [DOI] [PubMed] [Google Scholar]
- Rodrigo MJ, Marcos JF, Alférez F, Mallent MD, Zacarías L. 2003. Characterization of ‘Pinalate’, a novel Citrus sinensis mutant with a fruit-specific alteration that results in yellow pigmentation and decreased ABA content Journal of Experimental Botany 54, 727–738. [DOI] [PubMed] [Google Scholar]
- Rodrigo MJ, Marcos JF, Zacarías L. 2004. Biochemical and molecular analysis of carotenoid biosynthesis in flavedo of orange (Citrus sinensis L.) during fruit development and maturation Journal of Agricultural and Food Chemistry 52, 6724–6731. [DOI] [PubMed] [Google Scholar]
- Romero P, Rodrigo MJ, Alférez F, Ballester AR, González-Candelas L, Zacarías L, Lafuente MT. 2012. Unravelling molecular responses to moderate dehydration in harvested fruit of sweet orange (Citrus sinensis L. Osbeck) using a fruit-specific ABA-deficient mutant Journal of Experimental Botany 63, 2753–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction Nature Protocols 5, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A, Apostolova N, González-Guzman M, González-García MP, Nicolas C, Lorenzo O, Rodríguez PL. 2004. Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling The Plant Journal 37, 354–369. [DOI] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, et al. 2009a. The abscisic acid receptor PYR1 in complex with abscisic acid Nature 462, 665–668. [DOI] [PubMed] [Google Scholar]
- Santiago J, Rodrigues A, Saez A, et al. 2009b. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs The Plant Journal 60, 575–578. [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Tan BC, Gage DA, Zeevaart JAD, McCarty DR. 1997. Specific oxidative cleavage of carotenoids by VP14 of maize Science 276, 1872–1874 [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I. 2004. Plant PP2C phosphatases: emerging functions in stress signaling Trends in Plant Science 9, 236–243 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. 2007. Gene networks involved in drought stress response and tolerance Journal of Experimental Botany 58, 221–227. [DOI] [PubMed] [Google Scholar]
- Stewart I, Wheaton TA. 1972. Carotenoids in citrus: their accumulation induced by ethylene. Journal of Agricultural and Food Chemistry 20, 448–449. [Google Scholar]
- Sun L, Sun YF, Zhang M, et al. 2012. Suppression of 9-cis-epoxycarotenoid dioxygenase (NCED), which encodes a key enzyme in abscisic acid biosynthesis, alters fruit texture in transgenic tomatoes Plant Physiology 158, 283–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wang YP, Chen P, et al. 2011. Transcriptional regulation of SlPYL, SlPP2C, and SlSnRK2 gene families encoding ABA signal core components during tomato fruit development and drought stress Journal of Experimental Botany 62, 5659–5669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Zhang M, Ren J, Qi J, Zhang G, Leng P. 2010. Reciprocity between abscisic acid and ethylene at the onset of berry ripening and after harvest BMC Plant Biology 10,–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostkiewicz I, Richter K, Kepka M, et al. 2010. Closely related receptor complexes differ in their ABA selectivity and sensitivity The Plant Journal 61, 25–35 [DOI] [PubMed] [Google Scholar]
- Ton J, Flors V, Mauch-Mani B. 2009. The multifaceted role of ABA in disease resistance. Trends in Plant Science 14, 310–317. [DOI] [PubMed] [Google Scholar]
- Udvardi MK, Czechowski T, Scheible WR. 2008. Eleven golden rules of quantitative RT-PCR The Plant Cell 20, 1736–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K. 2010. Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport Plant and Cell Physiology 51, 1821–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, et al. 2009. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis Proceedings of the National Academy of Sciences, USA 106, 17588–17593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, de Preter K, Pattyn F, Poppe B, Van Roy N, de Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes Genome Biology 3, research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Zhu JK. 2007. New developments in abscisic acid perception and metabolism. Current Opinion in Plant Biology 10, 447–452 [DOI] [PubMed] [Google Scholar]
- Vlad F, Rubio S, Rodrigues A, et al. 2009. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis The Plant Cell 21, 3170–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker-Simmons M. 1987. ABA levels and sensitivity in developing wheat embryos of sprouting resistant and susceptible cultivars Plant Physiology 84, 61–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler EW. 1980. Radioimmunoassays for the differential and direct analysis of free and conjugated abscisic acid in plant extracts Planta 148, 262–272. [DOI] [PubMed] [Google Scholar]
- Weiner JJ, Peterson FC, Volkman BF, Cutler SR. 2010. Structural and functional insights into core ABA signaling Current Opinion in Plant Biology 13, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue T, Wang D, Zhang S, et al. 2008. Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis BMC Genomics 9,–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R, Hobo T, Ichimura K, et al. 2002. ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis Plant and Cell Physiology 43, 1473–1483. [DOI] [PubMed] [Google Scholar]
- Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K. 2006. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis Journal of Biological Chemistry 281, 5310–5318. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Nishimura N, Kitahata N, et al. 2006. ABA-Hypersensitive Germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs Plant Physiology 140, 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DP, Chen SW, Peng YB, Shen YY. 2001.. Abscisic acid-specific binding sites in the flesh of developing apple fruit Journal of Experimental Botany 52, 2097–2103 [DOI] [PubMed] [Google Scholar]
- Zhang M, Leng P, Zhang GL, Li XX. 2009a. Cloning and functional analysis of 9-cis-epoxycarotenoid dioxygenase (NCED) genes encoding a key enzyme during abscisic acid biosynthesis from peach and grape fruits Journal of Plant Physiology 166, 1241–1252. [DOI] [PubMed] [Google Scholar]
- Zhang M, Yuan B, Leng P. 2009b. The role of ABA in triggering ethylene biosynthesis and ripening of tomato fruit Journal of Experimental Botany 60, 1579–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.