Abstract
Pichia guilliermondii is a representative of yeast species that overproduce riboflavin (vitamin B2) in response to iron deprivation. P. guilliermondii YFH1 gene coding for frataxin homologue, eukaryotic mito-chondrial protein involved in iron trafficking and storage, was identified and deleted. Constructed P. guilliermondii Δyfh1 mutant grew very poorly in a sucrose-containing synthetic medium supplemented with sulfate or sulfite as a sole sulfur source. Addition of sodium sulfide, glutathione, cysteine, methionine, N-acetyl-L-cysteine partially restored growth rate of the mutant suggesting that it is impaired in sulfate assimilation. Cellular iron content in Δyfh1 mutant was ~3–3.5 times higher as compared to the parental strain. It produced 50–70 times more riboflavin in iron sufficient synthetic media relative to the parental wild-type strain. Biomass yield of the mutant in the synthetic glutathione containing medium supplemented with glycerol as a sole carbon source was 1.4- and 2.6-fold increased as compared to sucrose and succinate containing media, respectively. Oxygen uptake of the Δyfh1 mutant on sucrose, glycerol or succinate, when compared to the parental strain, was decreased 5.5-, 1.7- and 1.5-fold, respectively. Substitution of sucrose or glycerol in the synthetic iron sufficient medium with succinate completely abolished riboflavin overproduction by the mutants. Deletion of the YFH1 gene caused hypersensitivity to hydrogen peroxide and exogenously added riboflavin and led to alterations in superoxide dismutase activities. Thus, deletion of the gene coding for yeast frataxin homologue has pleiotropic effect on metabolism in P. guilliermondii.
Keywords: Frataxin, Iron, Riboflavin, Yeast, Sulfate, Respiration
Introduction
Frataxin is a conserved mitochondrial protein that is universally required for iron metabolism in human, yeast and bacterial cells (Pohl et al. 2007). Mutations in the human gene encoding frataxin are responsible for neurodegenerative disease called Friedreich’s ataxia (Campuzano et al. 1996). Saccharomyces cerevisiae strains lacking the gene coding for yeast frataxin are unable to grow on non-fermentable carbon sources (Babcock et al. 1997). Also they hyper-accumulated iron in mitochondria and are sensitive to oxidative stress (Babcock et al. 1997; Foury and Cazzalini 1997). Phenotype of Δyfh1 mutant of Candida albicans is similar to that of S. cerevisiae counterpart. Frataxin deficient strains of C. albicans have severe growth defects, reduced respiration and lack aconitase and succinate dehydrogenase activities, while over-accumulate iron in mitochondria. Certain features of these mutants resemble those of iron-deprived wild-type cells such as constitutively induced uptake of all forms of iron (elemental, siderophore-bound and heme iron) and increased excretion of flavins (Santos et al. 2004).
It is known that in certain yeast species, iron deprivation, in addition to activation of iron transport, causes activation of riboflavin synthesis (Tanner et al. 1945; Shavlovskii and Logvinenko 1988). Besides C. albicans, this group includes P. guilliermondii, Schwanniomyces occidentalis, Debaryomyces subglobosus, and the industrially important species Debaryomyces hansenii (the anamorph is known as Candida famata) (Shavlovskii and Logvinenko 1988; Voronovsky et al. 2002; Santos et al. 2004). Although this phenomenon was first described in 1945 (Tanner et al. 1945), metabolic advantage gained by the coordinated expression of genes involved in riboflavin biosynthesis and iron uptake as well as the mechanism by which yeasts control these processes are not known. There are no such mutual interrelationships between iron and riboflavin metabolism in the best studied yeast S. cerevisiae (Philpott and Protchenko 2008).
P. guilliermondii (anamorph is also known as Candida guilliermondii) is a convenient model organism for studying interrelationships between iron and flavin metabolisms. This facultative aerobic yeast species possess complex I of the respiratory chain (Zviagil’skaia et al. 1978). Genome of this yeast species is publicly available at http://www.broad.mit.edu (Candida guilliermondii Sequencing Project. Broad Institute of Harvard and MIT). In contrast to other riboflavin producing yeasts, P. guilliermondii can be stimulated to both mate and sporulate (Sibirny 1996) and efficient methods of gene manipulation have been developed for this species (Boretsky et al. 2007a). In the past years, large collection of P. guilliermondii mutants defective in the regulation of riboflavin biosynthesis has been obtained. It was demonstrated that P. guilliermondii mutants constitutively overproducing riboflavin (rib80, rib81, hit1, red1-6) also exhibit increased ferrireductase activity and high levels of iron transport. The above mentioned mutations were shown to be recessive, monogenic, and non-linked to the structural genes of the riboflavin biosynthetic pathway (Shavlovskii et al. 1990; Fedorovich et al. 1999; Stenchuk and Kapustiak 2003). However, corresponding genes were not isolated up to now, mostly due to the absence of useful phenotype in the mutants for gene cloning. To prove interrelationships between iron and riboflavin metabolism, we constructed Δyfh1 frataxin-deficient mutant of P. guilliermondii and studied its properties. Besides iron hyper-accumulation and riboflavin overproduction known for other yeasts (Babcock et al. 1997; Foury and Cazzalini 1997; Santos et al. 2004), P. guilliermondii Δyfh1 mutant exhibited also sulfate assimilation defect. Complementation analysis revealed that Δyfh1 mutant is distinct from previously reported riboflavin-producing mutants of this yeast.
Materials and methods
Strains, growth conditions and media
For plasmid construction and propagation Escherichia coli strain DH5α (lacZΔM15 recA1 endA1 gyrA96 thi-1 supE44 relA1 deoR Δ(lacZYA-argF)U169) was used. Orotidine 5′-mono-phosphate decarboxylase deficient E. coli strain (pyrF cys B) kindly provided by Dr. Beckerich J. M. (Laboratoire de Microbiologie et Genetique Moleculaire, CNRS-Institut National Agronomique Paris-Grignon-INRA, 78850 Thiverval-Grignon, France) was used to propagate plasmid bearing a deletion cassette. E. coli strains were grown in Luria-Bertani medium (LB) at 37°C supplemented with ampicillin (100 μg/ml) if necessary.
P. guilliermondii strains used in this study are listed in Table 1. The recipient uracil deficient strain that possessed wild type regulation of riboflavin biosynthesis, R-66 was selected as haploid meiotic segregant of the diploid obtained previously by crossing hit1 ura3 cytX MAT+ strain with HIT1 URA3 hisX MAT− strain (Boretsky et al. 2007a, b).
Table 1.
P. guilliermodii strains used in this study
| Strains | Genotype | Source or reference |
|---|---|---|
| R-66 | MAT− hisX ura3 | This study |
| Δyfh1 | MAT− YFH1Δ::URA3 hisX | This study |
| L1 | MAT+ adeX | Sibirnyi et al. (1977) |
| rib80 | MAT+ rib80 metX | Shavlovskii et al. (1990) |
| rib81 | MAT+ rib81 metX | Shavlovskii et al. (1993) |
| hit 1 | MAT+ cytX hit1 | Fedorovich et al. (1999) |
| red6 | MAT+ red6 adeX | Stenchuk and Kapustiak (2003) |
adeX, cytX, hisX, metX—unidentified mutations causing adenine, cytosine, histidine and methionine deficiency in P. guilliermondii, respectively
Yeast cells were grown using complete medium YPS (10 g yeast extract, 20 g peptone, 20 g sucrose, 20 g agar per 1 l) at 30°C or synthetic Burkholder medium supplemented with amino acids (50 mg/l), adenine (50 mg/l) or uridine (400 mg/l) if required (Shavlovskii et al. 1990). Sulfur free medium B was used to study assimilation of different sources of sulfur (Cherest and Surdin-Kerjan 1992). Iron-deficient media contained about 0.18 μM of iron. Iron was removed from the medium with 8-hydroxyquin-oline as described earlier (Shavlovskii et al. 1990). Iron supplemented media contained 3.6 μM iron added as ammonium ferrous sulfate hexahydrate. Yeast cells were grown in Erlenmeyer flasks on a gyro shaker (200 rpm) at 30°C. The yeast biomass was determined turbidimetrically with a Helios Gamma UVG-100105 spectrometer at 600 nm (one optical unit corresponds to 0.47 × 108 of cells per 1 ml).
Plasmid construction and analysis
DNA manipulation and transformation of E. coli were carried out according to previously published procedures (Sambrook and Russell 2001).
To provide a high level of expression of the cassette born modified URA3 gene of S. cerevisiae, it was placed under control of P. guilliermondii strong constitutive promoter of phosphoglycerate kinase gene (PGK1). Using plasmid pAGU34 as a template, a 0.9 kb DNA fragment bearing the URA3 gene was amplified by PCR with primers JB 25 and Ura32r, thereby introducing 5′ PstI and 3′ BamHI terminal sites (Table 2) (Boretsky et al. 2007a). Using P. guilliermondii chromosomal DNA, a 0.6 kb DNA fragment carrying promoter region of PGK1 gene was amplified with primers JB3 and JB4, thereby introducing 5′ BamHI and 3′ PstI terminal sites (Table 2). Both PCR products were purified, digested with BamHI and PstI restriction endonucleases and cloned into the BamHI site of the pUC19 vector. Resulted plasmid pPGKURA3 was used to generate BamHI 1.5 kb DNA fragment carrying the modified URA3 gene of S. cerevisiae under control of P. guilliermondii PGK1 gene promoter.
Table 2.
Primers used for this study
| N | Primer | Sequence (5′–3′) |
|---|---|---|
| 1 | JB 25 | ACCTGCAGGAAACGAAGATAAATC |
| 2 | Ura32r | CGGGATCCGGTAATAACTGATATAATT |
| 3 | JB3 | AAGGATCCTTCGGGATGACGAAG |
| 4 | JB4 | CCCTGCAGGGTAATTCTAGCAATCGATC |
| 5 | yfh1 Fw | ACTCTAGAGTAGTCGACGATACGAC |
| 6 | yfh1 Rev | TATCTAGATATAGTGGTTTTATC |
| 7 | JB10 | CCAGATCTGGAAAATGACGTG |
| 8 | JB11 | TGTAGATCTTCCGTGTCTTAC |
| 9 | JB 26 | CACATTTCCATCGAACAAGGTTC |
| 10 | act1-736f | TTGTTCCGTCCTTCCGACTT |
| 11 | act1-736r | CGAGTTGTAGGTGGTTTGGTCAA |
| 12 | act1-997f | TCCTTGTCCACTTTCCAACAAAT |
| 13 | act1-997r | GAAGGTCCGGACTCGTCGTA |
| 14 | rib1-84f | TCCTACATTGACACCATCCCATAT |
| 15 | rib1-84r | TGGCACTTCCGGAGGAATT |
| 16 | rib1-687f | GCAAGACTTGGGAGCGGATA |
| 17 | rib1-687r | GCATCAGCAGGATGTCGTAACA |
A 2.6 kb DNA fragment of P. guilliermondii chromosomal DNA bearing the YFH1 gene was amplified by PCR using primers yfh1 Fw and yfh1 Rev (Table 2) thereby introducing XbaI sites at the ends. The PCR product was purified, digested with XbaI restriction endonuclease and cloned into the XbaI site of the pUC19 vector. The constructed pYfh1 plasmid carried the YFH1 structural gene flanked with 1 and 0.9 kb of promoter and terminator sequences, respectively. This plasmid was used to substitute YFH1 structural gene with the S. cerevisiae URA3 gene. Almost the entire sequence of the pYfh1 plasmid, except of the YFH1 structural gene was amplified with primers JB10 and JB11 (Table 2) thereby introducing BglII sites at the ends of the PCR product. The PCR product was purified, digested with BglII restriction endonuclease and ligated with the 1.5 kb BamHI fragment of pPGKURA3 plasmid carrying the modified S. cerevisiae URA3 gene. The resulting plasmid pYFH1URA3 carried the modified S. cerevisiae URA3 gene inserted between 1.0 and 0.9 kb of promoter and terminator sequences of P. guilliermondii YFH1 gene, respectively. Then plasmid pYFH1URA3 was digested with XbaI endonuclease yielding a yfh1::URA3 deletion cassette which was used for transformation of P. guilliermondii R-66 strain.
All recombinant plasmids were sequenced. Homology search and alignments were performed with the aid of the BLAST and ClustalW 1.8 programs (available at http://www.ncbi.nlm.nih.gov/BLAST/index.html and http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html).
Cell respiration
Cells grown in Burkholder medium containing sucrose, glycerol, or succinate were pelleted (3,000g for 10 min) and re-suspended in the equal volume of the medium without carbohydrates. Cells were incubated for 15 h at 30°C aerobically, chilled on ice, harvested, washed twice with ice-cold distilled water and re-suspended in ice-cold carbohydrate free Burkholder medium. The respiratory rate was measured at 30°C using a Clark-type electrode (Biological Oxygen Monitor YSI Model 5300) in a reaction vessel with 5 ml of air-saturated respiration mixture containing 10 mM of appropriate carbohydrate according to the method described (Ferrero et al. 1981). After short (5 min) pre-incubation of cells in the reaction vessel the reactions were started by addition of the appropriate substrate.
Staining for detection of SOD activity
SOD activity on a nondenaturing polyacrylamide gel was detected by negative staining (Ito-Kuwa et al. 1999). The gel was washed twice in 50 mM phosphate buffer (pH 7.8) for 20 min. Then it was soaked in 50 mM phosphate buffer pH 7.5, 0.064 mg/ml of riboflavin, 3.2 μl/ml N,N,N′,N′-tetramethylethylenediamine, 0.3 mg/ml of 2,3,5-triphenyltetrazoliumchloride (TTC) for 20 min. Area of SOD activity remained clear when the gel was exposed to the UV light.
Miscellaneous procedures
Yeast transformation, PCR analysis and Southern blot analysis of transformants were done as previously described (Boretsky et al. 2007a, b). Transformants were selected on an agar medium containing 0.67% yeast nitrogen base (YNB), 2% sucrose and 0.5% casamino acids (Difco) without uridine. Yeast strain hybridization and subsequent spore progeny analysis were performed as described (Sibirnyi et al. 1977). Riboflavin was assayed fluorometrically using solution of synthetic riboflavin as a standard with an EF-3M fluorometer. Thin-layer chromatography was carried out on Silufol (Chemapol) plates with systems n-butanol: acetic acid: water (10:3:7 v/v) or 3% NH4Cl. Cellular iron content was determined with 2,2′-dipiridyl as described earlier (Fedorovich et al. 1999). The ferrireductase activity of washed cells was measured spectrophotometrically with ferric citrate as a substrate (Fedorovich et al. 1999).
Cells were disrupted by grinding with 0.4–0.5 mm glass beads. Protein concentration was determined after dialysis by the Lowry method (Lowry et al. 1951). Activity of GTP cyclohydrolase II was determined by a fluorometric method as described earlier (Shavlovskii et al. 1983). Activity of citrate (isocitrate) hydrolyase (EC 4.2.1.3; aconitase) in cell free extracts was determined by the rate of isocitrate transformation into cis-aconitate according to the method described (Fansler and Lowenstein 1969). Assay of succinate dehydrogenase activity was performed as described earlier (Saliola et al. 2004). Real-time PCR analysis was done as described previously (Protchenko et al. 2008).
Results
The gene PGUG_03715.1 encoding a putative frataxin homologue has been identified in the P. guilliermondii genome using homology search against S. cerevisiae frataxin. This P. guilliermondii gene designated as PgYFH1 encodes a 172-residue protein with a theoretical Mr of 19.36 kDa. Alignment of the PgYFH1p protein sequence with other frataxin sequences revealed that the predicted mature form of PgYfh1p (amino acids 65–172) is 46 and 49% identical to mature human and S. cerevisiae frataxin homologues, respectively (data not shown).
Chromosomal DNA fragment (2.6 kb) of P. guilliermondii genome bearing the identified gene together with 1 kb flanking regions was PCR ampli-fied and cloned as described in M&M section. Obtained plasmid was used as a template to substitute PgYFH1 gene with the modified URA3 gene of S. cerevisiae as described in M&M section. The resulting plasmid pYFH1URA3 was digested with XbaI endonuclease yielding the yfh1::URA3 deletion cassette that was used to transform the P. guilliermondii recipient strain R-66. Nine of 200 transformants were found to bear yfh1::URA3 deletion cassettes integrated into the genome by homologous recombination that lead to a knock-out of the PgYFH1 structural gene as verified by PCR analysis. Integration of a single copy of the cassette into genome of selected transformants was confirmed by Southern blot analysis. All of them exhibited identical mutant phenotype as detailed below.
When grown in YNB medium supplemented with 3.6 μM of iron and 0.5% of casamino acids, Δyfh1 mutant excreted significant amounts of a yellow substance identified as riboflavin by means of both thin-layer chromatography and absorption spectra analysis (data not shown). Its colonies were stained red in the medium supplemented with 40 mg/l of 2,3,5-triphenyltetrazoliumchloride (TTC), due to reduction of TTC to insoluble triphenylformazan that has red color (Bernas and Dobrucki 2000). These fatures of Δyfh1 mutant resemble phenotype of previously characterized riboflavin producing mutant strains of P. guilliermondii rib80, rib81, hit1 and red6 (Shavlovskii et al. 1993; Fedorovich et al. 1999; Stenchuk and Kapustiak 2003). All diploid hybrids resulting from crossing of Δyfh1 mutant with mutants rib80, rib81, hit1 and red6 possessed wild phenotype: they did not overproduce riboflavin in iron-sufficient medium and did not reduce TTC (not shown). Thus, neither of the mutations mentioned above impaired the PgYFH1 gene.
In contrast to the parental strain, Δyfh1 mutant grew very weakly in a synthetic medium though grew well in the same medium supplemented with 0.5% casein hydrolysate or casamino acids, sources of organic sulfur. So, we assayed growth rate of the mutant Δyfh1 using sulfur free iron sufficient medium (Cherest and Surdin-Kerjan 1992) supplemented with different sulfur-containing compounds. It was observed that both ammonium sulfate and sodium sulfite did not support growth of the Δyfh1 mutant whereas glutathione, methionine, cysteine, N-acetyl-L-cysteine and sodium sulfide restored the growth rate. Parental strain grew well in all media except those supplemented with sodium sulfite or sodium sulfide (Fig. 1a). Without source of organic sulfur, the mutant cells could divide 5–6 times. So, organic sulfur auxotrophy could be observed easily when reduced rate of inoculation (A600 less than 0.003) was used, whereas more massive inoculation (A600 more than 0.03) resulted in slightly decreased final cell density when compared to the organic sulfur containing medium. In subsequent experiments 0.2 mM glutathione (which provided the best restoration of the growth) was added to synthetic media to promote growth of the constructed mutant. Increasing glutathione concentration up to 1 mM in the medium did not enhance growth of the mutant (data not shown). Cells of Δyfh1 mutant grown in glutathione supplemented iron sufficient media exhibited a decrease (sixfold to sevenfold less as compared to the parental strain) of activity of Fe/S containing enzymes, aconitase and succinate dehydrogenase (Table 3).
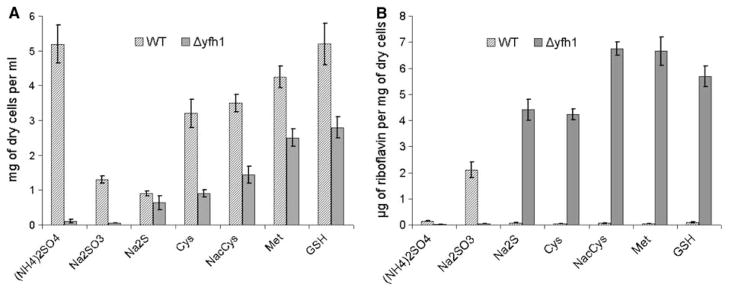
Fig. 1.
Growth (a) and riboflavin productivity (b) of P. guilliermondii WT and Δyfh1 mutant in synthetic sulfur free medium B supplemented with different sulfur containing compounds. Cultures of P. guilliermondii wild-type strain R-66 (WT) and Δyfh1 mutant were grown aerobically in YPS medium for 16 h. Cells were pelleted, washed with water and resuspended in water to an optical density OD600 of 0.2. Aliquots of 0.3 ml were inoculated in 100 ml Erlenmeyer flasks containing 20 ml of the synthetic medium B containing 3.6 μM of iron added as ammonium ferrous sulfate and supplemented with: (NH4)2SO4—40 mM of ammonium sulfate, Na2SO3—2.5 mM of sodium sulfite, Met—0.2 mM of methionine, GSH—0.2 mM of glutathione, Cys—0.2 mM of cysteine, NACys—0.1 mM N-Acetyl-L-cysteine, Na2S—5 mM sodium sulfide. Cell density and concentration of riboflavin were determined after 5 days of incubation at 30°C on gyro shaker at 200 rpm. Values are means ± SE of three independent experiments
Table 3.
Activities of ferrireductase, GTP-cyclohydrolase II, aconitase and succinate dehydrogenase in P. guilliermondii Δyfh1 mutant and wild-type strain
| Strains | Ferrireductase activity, nM of Fe min−1/mg of dry cells | GTP-cyclohydrolase II activity, U min−1/mg of protein | Aconitase activity, U min−1/mg of protein | Succinate dehydrogenase activity, U min−1/mg of protein |
|---|---|---|---|---|
| WT | 2.22 ± 0.15 | 0.76 ± 0.09 | 0.107 ± 0.009 | 0.033 ± 0.0009 |
| Δyfh1 | 104.35 ± 9.47 | 5.12 ± 0.73 | 0.016 ± 0.002 | 0.0055 ± 0.0006 |
Depending on a sulfur source, production of riboflavin by the mutant varied from 4.2 to 7.0 mg/g of dry cells weight, what is 50–70-folds higher as compared to the parental wild-type strain grown in the same iron sufficient media. Wild-type strain enhanced production of riboflavin only in the medium that contained sodium sulfite (Fig. 1b).
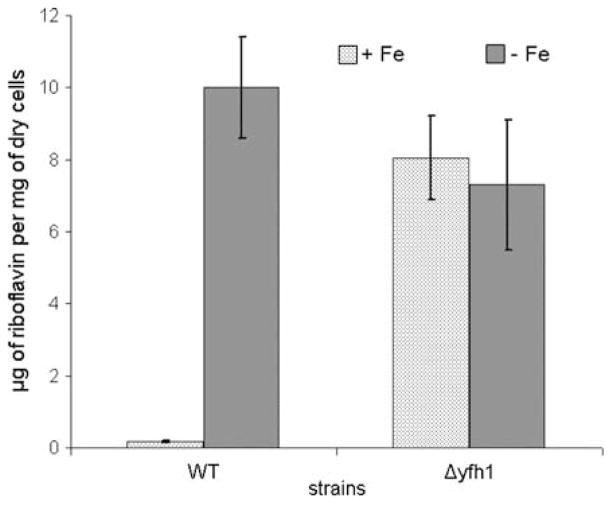
Similar results were obtained using another type of synthetic medium, namely Burkholder medium that is commonly used to study regulation of riboflavin biosynthesis in P. guilliermondii. Δyfh1 mutants grown in the synthetic Burkholder medium supplemented with 3.6 μM of iron (iron repletion conditions) and 0.2 mM of glutathione accumulated 3–3.5 times more iron than the parental wild-type strain (Fig. 2). Despite iron overload, mutant cells behaved as iron deprived wild-type cells. Ferrireductase activity of the mutant cells was 40–50 times elevated as compared to the parental wild-type strain (Table 3). Relative to the parental strain, riboflavin productivity of Δyfh1 mutant strain was 50–60-folds higher (Fig. 3). Transcription level of the RIB1 gene coding for the key enzyme of riboflavin biosynthesis, GTP cyclohydrolase II was elevated 3–4 times resulting in 5–7 times increase in activity of the enzyme in the mutant cells (Table 3). Iron deprivation did not cause additional increase in riboflavin production by the mutant strain. Under these conditions, Δyfh1 mutant produced approximately 5–8 mg of riboflavin/g of dry cell weight, similarly to the parental wild-type strain.
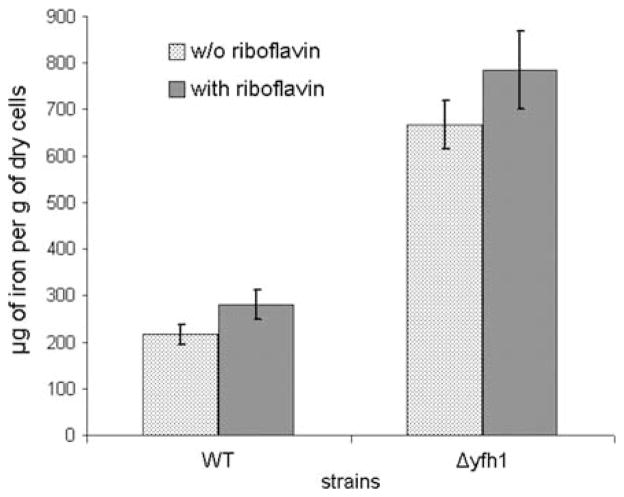
Fig. 2.
Iron content in cells of P. guilliermondii wild-type strain and Δyfh1 mutant. Cultures of P. guilliermondii wild-type strain R-66 (WT) and Δyfh1 mutant were grown aerobically at 30°C in synthetic Burkholder medium supplemented with 3.6 μM of iron added as ammonium ferrous sulfate. Riboflavin supplemented media contained 200 mg/l of this vitamin. Cells from middle exponential growth phase were used to measure iron content. Values are means ± SE of three independent experiments
Fig. 3.
Riboflavin productivity of P. guilliermondii wild-type strain and Δyfh1 mutant. Cultures of P. guilliermondii wild-type strain R-66 (WT) and Δyfh1 mutant were grown for 5 days aerobically at 30°C in synthetic Burkholder medium supplemented with 3.6 μM of iron added as ammonium ferrous sulfate (+Fe). Iron deficient medium contained approximately 0.18 μM of iron (−Fe). Values are means ± SE of three independent experiments
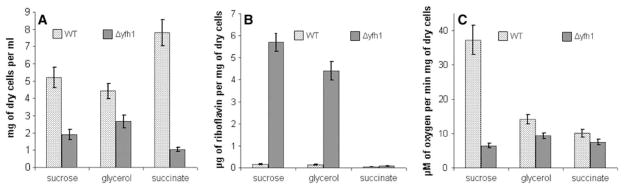
Growth of the mutant in the synthetic glutathione containing medium supplemented with glycerol as a sole carbon source was 1.4- and 2.6-fold increased as compared to sucrose and succinate containing media, respectively (Fig. 4a). Substitution of sucrose with glycerol in the Burkholder synthetic medium decreased riboflavin production by the mutant for 1.3-fold. No riboflavin over-production by the mutant was observed in the succinate supplemented medium (Fig. 4b). Δyfh1 mutant and its parental strain both possessed 1.5-fold increased cellular iron content when sucrose was substituted with glycerol in the medium.
Fig. 4.
Influence of carbon sources on growth (a), riboflavin productivity (b) and oxygen uptake (c) of P. guilliermondii wild-type strain and Δyfh1 mutant. Cultures of P. guilliermondii wild-type strain R-66 (WT) and Δyfh1 mutant were grown aerobically at 30°C in synthetic Burkholder medium containing 0.2 mM of glutathione, 3.6 μM of iron added as ammonium ferrous sulfate and supplemented with sucrose or glycerol or succinate as a sole carbon source. a, b Cells were grown for 5 days. c Exponentially growing cells (20–40 h) were used to measure respiration activity. Values are means ± SE of three independent experiments
Oxygen uptake by the mutant cells with sucrose, glycerol or succinate used as respiratory substrates was, respectively, 5.5-, 1.7- and 1.5-fold decreased as compared to the parental strain (Fig. 4c). Additional threefold to sevenfold decrease of oxygen uptake was observed after 16 h incubation of the mutant cells in media without glutathione. No effect of glutathione supplementation on oxygen consumption by the parental strain was found (data not shown).
Constructed Δyfh1 mutant of P. guilliermondii is hypersensitive to oxidative stress like other iron accumulating mutants of this yeast species (Protchenko et al. 2000; Boretsky et al. 2007b). Only 2–7% of the Δyfh1 mutant cells survived during 1 h exposure to 1 mM hydrogen peroxide, while viability of the parental strain remained ~100% under the same conditions (Fig. 5). The observed rearrangement in superoxide dismutase (SOD) activities (Fig. 6), namely, decreased activity of fast migrating form and significantly increased activity of the second form, may also suggest that cells of Δyfh1 mutant are under oxidative stress. In favor of this assumption, the growth rate of Δyfh1 mutant (but not of the parental wild-type strain) is significantly decreased in Burkholder synthetic medium supplemented with 200 mg/l of riboflavin which is a prooxidant under certain conditions (Ito-Kuwa et al. 1999; Reddy et al. 2008) (Fig. 7). This effect did not depend on concentration of available iron in the media. Supplementation of the riboflavin containing media with 100 mM of iron chelator, ferrozine, or with 3.6 μM of iron did not affect growth rate of both strains (data not shown). Both strains accumulated ~1.3-fold more iron in the cells when grown in the medium containing 3.6 μM of iron and supplemented with riboflavin (Fig. 2). Under such conditions, colonies of mutant strain stained rose, possibly due to their high reducing activity that caused formation of a colored partially reduced form (apparently rhodoflavin) of riboflavin (Berezovskii 1973).
Fig. 5.
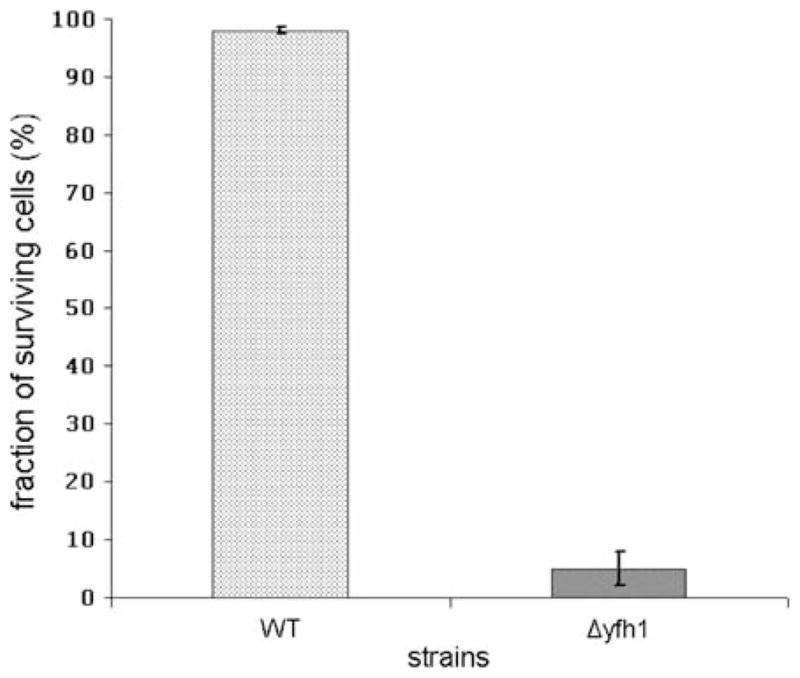
Sensitivity of P. guilliermondii wild-type strain and Δyfh1 mutant to hydrogen peroxide. Cultures of P. guilliermondii wild-type strain R-66 (WT) and Δyfh1 mutant were grown aerobically in YPD medium for 16 h, diluted to an OD600 of 0.2 and allowed to grow for 3.5 h. Aliquots (2 ml) were treated with 1 mM H2O2 for 1.5 h at 30°C. Cells were pelleted at 3,000g for 10 min and re-suspended in fresh YPD medium. Suspensions were diluted 100–1,000-fold in complete medium and plated on YPD agar plates. Colonies were counted after 3 days of incubation at 30°C. Quantities of colonies obtained with untreated cultures were assumed as a 100%. Values are means ± SE of three independent experiments
Fig. 6.
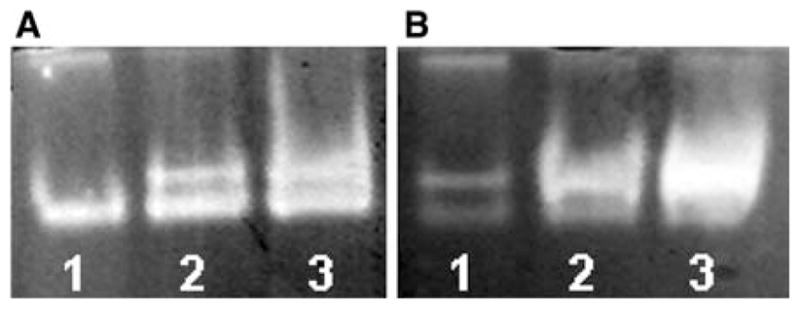
SOD activity in P. guilliermondii wild-type strain (A) and Δyfh1 mutant (B) correspondingly. Gels were stained for SOD activity following electrophoresis under non-denaturing conditions as described in the M&M section. Each lane was loaded with 0.04 mg of protein of cell free extract. Cells from early (1), middle (2), and late (3) exponential growth phase were used for the analysis
Fig. 7.

Inhibition of growth of P. guilliermondii wild-type strain and Δyfh1 mutant by riboflavin. Cultures of P. guilliermondii wild-type strain R-66 (WT) and Δyfh1 mutant were grown aerobically in YPD medium for 48 h. Cells were harvested, washed with water and re-suspended in water to an optical density A600 = 0.2. Serial five times dilutions were made. Five microliter aliquots of each dilutions were plated onto synthetic YNB medium containing 0.5% of casamino acids, 400 mg/l of uridine without riboflavin and supplemented with 200 mg/l of riboflavin. Plates were incubated at 30°C for 4 days. Results of a typical representative experiment are shown
Discussion
As compared to the wild-type strain, the described in current work P. guilliermondii Δyfh1 mutant exhibited approximately 3.5- and 30–50-fold increased cellular iron content and riboflavin productivity, respectively. This phenotype is similar to that of Δyfh1 mutants reported in C. albicans (Santos et al. 2004). Obtained results further support existence of common mechanisms of regulation of riboflavin biosynthesis and iron assimilation in P. guilliermondii and, possibly, in other so called “flavinogenic” yeasts that overproduce riboflavin in response to iron deprivation.
P. guilliermondii Δyfh1 mutant could not grow in synthetic medium with sulfate or sulfite as the sole sulfur sources, but grew in the same medium supplemented with sulfide, cysteine, methionine, glutathione, etc. Among compounds tested, glutathione was found to provide the best growth restoration of the mutant. One may suggest that this compound could serve as a source of sulfur for synthesis of Fe/S clusters and simultaneously as a protective agent for them. In favour of this assumption, P. guilliermondii Δyfh1 mutant exhibited moderate (6–7 times lower as compared to the wild-type parental strain) decrease of activity of Fe/S containing enzymes aconitase and succinate dehydrogenase, whereas activity of aconitase in C. albicans Δyfh1 mutants was almost zero (Santos et al. 2004). Activity of both enzymes in mutant cells was additionally decreased (twofold to sixfold) after incubation of the mutant cells in glutathione free medium (data not shown). Earlier glutathione was shown to be required for maturation of cytosolic Fe/S proteins and for maintaining redox status in wild-type and frataxin-deficient yeast cells (Sipos et al. 2002; Auchere et al. 2008). Stimulation of growth of P. guilliermondii Δyfh1 mutant by other S2− containing compound can suggest that deletion of the PgYFH1 gene interrupted sulfate assimilation pathway. Mechanisms of sulfur metabolism impairment are not known and will be subject of our future investigations.
In contrast to the wild-type strain, substitution of sucrose by glycerol in the synthetic culture medium facilitated growth of the P. guilliermondii Δyfh1 mutant. Glycerol grown mutant cells displayed 1.7-fold reduction of respiratory activity as compared to the wild-type cells grown under the same conditions. Respiratory activity of the sucrose grown mutant cells was 5.5-fold lower as compared to the parental strain. Based on these data, it could be supposed that sucrose itself decreased respiratory activity of the mutant; in other words, deletion of the frataxin gene renders P. guillermondii more “Crabtree positive”.
Apparently, the main function of frataxin in P. guilliermondii is to convert redox-active iron to an inert mineral, thereby preventing oxidative damage of labile Fe/S clusters and of the cells. This role of frataxin was elucidated recently in S. cerevisiae (Gakh et al. 2008). P. guilliermondii Δyfh1 mutant is hypersensitive to oxidative stress similarly to previously reported rib80, rib81, hit1, red1-6 mutants (Protchenko et al. 2000; Boretsky et al. 2007b). The observed rearrangement in superoxide dismutase activities in the P. guilliermondii Δyfh1 strain can be explained by iron overload since yeast manganese containing SOD is sensitive to increased concentration of iron (Yang et al. 2006; Irazusta et al. 2008). It is difficult to determine which enzyme is inactivated in the Δyfh1 mutant since we found at least six putative homologues of SOD genes in the P. guilliermondii genome. Dramatic changes in SOD activity and increased level of iron accumulation, both could enhance sensitivity of Δyfh1 mutants to riboflavin which is prooxidant when photo- or chemically reduced (Ito-Kuwa et al. 1999; Reddy et al. 2008). We did not illuminate plates or supplement media with a reducing agent in this experiment, since Δyfh1 mutant cells itself possess high non-specific reductive activity toward different compounds including riboflavin which was converted to semi-reduced form (probably rhodoflavin) during growth of colonies (Fig. 7) (Berezovskii 1973). Interestingly, that P. guilliermondii iron deprived wild-type cells also exhibit high reductive activity toward Fe3+, TTC and riboflavin (Fedorovych, unpublished results). It could be suggested that this reductive activity is nonspecific and has a dual role in iron uptake. First of all, reduction of Fe3+ to Fe2+ increases solubility and availability of iron. Also, it was hypothesized that some bacteria can use reduced riboflavin to reduce Fe3+ to Fe2+ and to chelate produced ferrous ions facilitating iron uptake (Marsili et al. 2008). On the other hand, P. guilliermondii wild-type cells do not possess active transport of riboflavin (Sibirny 1996). So, excretion and reduction of riboflavin which can be a source of superoxide anion causing oxidative stress and death of the cells, may be used to inhibit growth of competitor micro-organisms that have active transport of this vitamin (Ito-Kuwa et al. 1999; Reddy et al. 2008). Thus, several hypotheses have to be checked to explain interconnection between iron and riboflavin metabolism in flavinogenic yeasts. At present, metabolic advantage gained by the coordinated regulation of these metabolic branches in flavinogenic yeasts are not known despite extensive studies on regulation of iron acquisition and riboflavin biosynthesis (Santos et al. 2004; Boretsky et al. 2005). Recently, transcriptional factor SEF1p was identified as a main regulator of riboflavin biosynthesis in C. famata (Dmytruk et al. 2006). Maybe studying of its role in regulation of riboflavin biosynthesis and, possibly, iron acquisition by other yeast species will help to answer some of these questions.
Acknowledgments
This work was supported by CRDF Grant UKB1-2810--LV-06.
Contributor Information
Yuriy V. Pynyaha, Institute of Cell Biology, NAS of Ukraine, Drahomanov Street 14/16, 79005 Lviv, Ukraine
Yuriy R. Boretsky, Institute of Cell Biology, NAS of Ukraine, Drahomanov Street 14/16, 79005 Lviv, Ukraine
Daria V. Fedorovych, Institute of Cell Biology, NAS of Ukraine, Drahomanov Street 14/16, 79005 Lviv, Ukraine
Lubov R. Fayura, Institute of Cell Biology, NAS of Ukraine, Drahomanov Street 14/16, 79005 Lviv, Ukraine
Andriy I. Levkiv, Institute of Cell Biology, NAS of Ukraine, Drahomanov Street 14/16, 79005 Lviv, Ukraine
Vira M. Ubiyvovk, Institute of Cell Biology, NAS of Ukraine, Drahomanov Street 14/16, 79005 Lviv, Ukraine
Olha V. Protchenko, Liver Diseases Branch, National Institute of Diabetes, Digestive, and Kidney Diseases, National Institutes of Health, Building 10, Room 9B-16, 10 Center Drive, Bethesda, MD 20892-1800, USA
Caroline C. Philpott, Liver Diseases Branch, National Institute of Diabetes, Digestive, and Kidney Diseases, National Institutes of Health, Building 10, Room 9B-16, 10 Center Drive, Bethesda, MD 20892-1800, USA
Andriy A. Sibirny, Email: sibirny@cellbiol.lviv.ua, Institute of Cell Biology, NAS of Ukraine, Drahomanov Street 14/16, 79005 Lviv, Ukraine. Rzeszów University, Ćwiklińskiej 2, 35-601 Rzeszów, Poland
References
- Auchere F, Santos R, Planamente S, Lesuisse E, Camadro JM. Glutathione-dependent redox status of frataxin-deficient cells in a yeast model of Friedreich’s ataxia. Hum Mol Genet. 2008;17:2790–2802. doi: 10.1093/hmg/ddn178. [DOI] [PubMed] [Google Scholar]
- Babcock M, de Silva D, Oaks R, Davis-Kaplan S, Jiralerspong S, Montermini L. Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science. 1997;276:1709–1712. doi: 10.1126/science.276.5319.1709. [DOI] [PubMed] [Google Scholar]
- Berezovskii V. Pishchevaya promyshlennost’. Moscow: 1973. Khimiya vitaminov (chemistry of vitamins) [Google Scholar]
- Bernas T, Dobrucki J. The role of plasma membrane in bioreduction of two tetrazolium salts, MTT, and CTC. Arch Biochem Biophys. 2000;380:108–116. doi: 10.1006/abbi.2000.1907. [DOI] [PubMed] [Google Scholar]
- Boretsky Y, Kapustyak K, Fayura L, Stasyk O, Stenchuk M, Bobak Y, Drobot L, Sibirny A. Positive selection of mutants defective in transcriptional repression of riboflavin synthesis by iron in the flavinogenic yeast Pichia guilliermondii. FEMS Yeast Res. 2005;5:829–837. doi: 10.1016/j.femsyr.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Boretsky Y, Pynyaha Y, Boretsky V, Kutsyaba V, Protchenko O, Philpott C, Sibirny A. Development of a transformation system for gene knock-out in the flavinogenic yeast Pichia guilliermondii. J Microbiol Methods. 2007a;70:13–19. doi: 10.1016/j.mimet.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Boretsky Y, Protchenko O, Prokopiv T, Mukalov I, Fedorovych D, Sibirny A. Mutations and environmental factors affecting regulation of riboflavin synthesis and iron assimilation also cause oxidative stress in the yeast Pichia guilliermondii. J Basic Microbiol. 2007b;47:371–377. doi: 10.1002/jobm.200610279. [DOI] [PubMed] [Google Scholar]
- Campuzano V, Montermini L, Moltò M, Pianese L, Cossée M, Cavalcanti F. Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- Cherest H, Surdin-Kerjan Y. Genetic analysis of a new mutation conferring cysteine auxotrophy in Saccharomyces cerevisiae: updating of the sulfur metabolism pathway. Genetics. 1992;130:51–58. doi: 10.1093/genetics/130.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmytruk K, Voronovsky A, Sibirny A. Insertion mutagenesis of the yeast Candida famata (Debaryomyces hansenii) by random integration of linear DNA fragments. Curr Genet. 2006;3:183–191. doi: 10.1007/s00294-006-0083-0. [DOI] [PubMed] [Google Scholar]
- Fansler B, Lowenstein J. Aconitase from pig heart. Methods Enzymol. 1969;13:26–30. [Google Scholar]
- Fedorovich D, Protchenko O, Lesuisse E. Iron uptake by the yeast Pichia guilliermondii. Flavinogenesis and reductive iron assimilation are co-regulated processes. Biometals. 1999;12:295–300. doi: 10.1023/a:1009298530145. [DOI] [PubMed] [Google Scholar]
- Ferrero I, Viola A, Goffeau A. Induction by glucose of an antimycin-insensitive, azide-sensitive respiration in the yeast Kluyveromyces lactis. Antonie Van Leeuwenhoek. 1981;47:11–24. doi: 10.1007/BF00399063. [DOI] [PubMed] [Google Scholar]
- Foury F, Cazzalini O. Deletion of the yeast homologue of the human gene associated with Friedreich’s ataxia elicits iron accumulation in mitochondria. FEBS Lett. 1997;411:373–377. doi: 10.1016/s0014-5793(97)00734-5. [DOI] [PubMed] [Google Scholar]
- Foury F, Pastore A, Trincal M. Acidic residues of yeast frataxin have an essential role in Fe/S cluster assembly. EMBO Rep. 2007;8:194–199. doi: 10.1038/sj.embor.7400881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gakh O, Smith D, Isaya G. Assembly of the iron-binding protein frataxin in Saccharomyces cerevisiae responds to dynamic changes in mitochondrial iron influx and stress level. J Biol Chem. 2008;283:31500–31510. doi: 10.1074/jbc.M805415200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazusta V, Moreno-Cermeno A, Cabiscol E, Ros J, Tamarit J. Major targets of iron-induced protein oxidative damage in frataxin-deficient yeasts are magnesium-binding proteins. Free Radic Biol Med. 2008;44:1712–1723. doi: 10.1016/j.freeradbiomed.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Ito-Kuwa S, Nakamura K, Aoki S, Osafune T, Vidotto V, Pienthaweechai K. Oxidative stress sensitivity and superoxide dismutase of a wild-type parent strain and a respiratory mutant of Candida albicans. Med Mycol. 1999;37:307–314. doi: 10.1046/j.1365-280x.1999.00224.x. [DOI] [PubMed] [Google Scholar]
- Lowry O, Rosebrough N, Farr A, Randall R. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Marsili E, Baron D, Shikhare I, Coursolle D, Gralnick J, Bond D. Shewanella secretes flavins that mediate extracellular electron transfer. Proc Natl Acad Sci USA. 2008;105:3968–3973. doi: 10.1073/pnas.0710525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott C, Protchenko O. Response to iron deprivation in Saccharomyces cerevisiae. Eukaryot Cell. 2008;7:20–27. doi: 10.1128/EC.00354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl T, Walter J, Stolpe S, Soufo J, Grauman P, Friedrich T. Effects of the deletion of the Escherichia coli frataxin homologue CyaY on the respiratory NADH: ubiquinone oxidoreductase. BMC Biochem. 2007;24:8–13. doi: 10.1186/1471-2091-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protchenko O, Boretsky Y, Romaniuk T, Fedorovych D. Oversynthesis of riboflavin by yeast Pichia guilliermondii in response to oxidative stress. Ukr Biokhim Zh. 2000;72:19–23. [PubMed] [Google Scholar]
- Protchenko O, Shakoury-Elizeh M, Keane P, Storey J, Androphy R, Philpott C. Role of PUG1 in inducible porphyrin and heme transport in Saccharomyces cerevisiae. Eukaryot Cell. 2008;7:859–871. doi: 10.1128/EC.00414-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy H, Dayan A, Cavagnaro J, Gad S, Li J, Goodrich R. Toxicity testing of a novel riboflavin-based technology for pathogen reduction and white blood cell inactivation. Transfus Med Rev. 2008;22:133–153. doi: 10.1016/j.tmrv.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Saliola M, Bartoccioni P, De Maria I, Lodi T, Falcone C. The deletion of the succinate dehydrogenase gene KlSDH1 in Kluyveromyces lactis does not lead to respiratory deficiency. Eukaryot Cell. 2004;3:589–597. doi: 10.1128/EC.3.3.589-597.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Molecular cloning, a laboratory manual. 3. Cold Spring Harbor Laboratory; Cold Spring Harbor: 2001. pp. 14–450. [Google Scholar]
- Santos R, Buisson N, Knight S, Dancis A, Camadro J, Lesuisse E. Candida albicans lacking the frataxin homologue: a relevant yeast model for studying the role of frataxin. Mol Microbiol. 2004;54:507–519. doi: 10.1111/j.1365-2958.2004.04281.x. [DOI] [PubMed] [Google Scholar]
- Shavlovskii G, Logvinenko E. Riboflavin oversynthesis in yeast and mechanisms of its regulation. Prikl Biokhim Mikrobiol. 1988;24:435–447. [PubMed] [Google Scholar]
- Shavlovskii G, Logvinenko E, Zakalskii A. Purification and properties of GTP cyclohydrolase II of the yeast Pichia guilliermondii. Biokhimiia. 1983;48:837–843. [PubMed] [Google Scholar]
- Shavlovskii G, Fedorovich D, Babyak L. The effect of carbon sources on the manifestation of rib80 and rib81 regulatory mutations in Pichia guilliermondii. Mikrobiologiia. 1990;59:404–410. [Google Scholar]
- Shavlovskii G, Fedorovich D, Babyak L. The effect rib81 mutation on riboflavin biosynthesis and iron transport in Pichia guilliermondii yeast. Mikrobiologiia. 1993;62:897–903. [Google Scholar]
- Sibirny A. Chapter VII. Pichia guilliermondii. In: Wolf K, editor. Nonconvential yeasts in biotechnology. Springer; Berlin: 1996. [Google Scholar]
- Sibirnyi A, Zharova V, Kshanovskaia B, Shavlovskii G. Selection of a genetic line of Pichia guilliermondii yeasts capable of forming a significant quantity of spores. Tsitol Genet. 1977;11:330–333. [PubMed] [Google Scholar]
- Sipos K, Lange H, Fekete Z, Ullmann P, Lill R, Kispal G. Maturation of cytosolic iron-sulfur proteins requires glutathione. J Biol Chem. 2002;277:26944–26949. doi: 10.1074/jbc.M200677200. [DOI] [PubMed] [Google Scholar]
- Stenchuk N, Kapustiak K. The red mutations impair the regulation of flavinogenesis and metal homeostasis in yeast Pichia guilliermondii. Genetika. 2003;39:1026–1032. [PubMed] [Google Scholar]
- Tanner F, Vojnovich C, Lanee J. Riboflavin production by Candida species. Science. 1945;101:180–183. doi: 10.1126/science.101.2616.180. [DOI] [PubMed] [Google Scholar]
- Voronovsky A, Abbas C, Fayura L, Kshanovska B, Dmytruk K, Sybirna K, Sibirny A. Development of a transformation system for the flavinogenic yeast Candida famata. FEMS Yeast Res. 2002;2:381–388. doi: 10.1016/S1567-1356(02)00112-5. [DOI] [PubMed] [Google Scholar]
- Yang M, Cobine P, Molik S, Naranuntarat A, Lill R, Winge D, Culotta V. The effects of mitochondrial iron homeostasis on cofactor specificity of superoxide dismutase 2. EMBO J. 2006;25:1775–1783. doi: 10.1038/sj.emboj.7601064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zviagil’skaia R, Fedorovich D, Shavlovskii G. Respiratory system of Pichia guilliermondii yeasts with different levels of flavinogenesis. Mikrobiologiia. 1978;47(6):975–984. [PubMed] [Google Scholar]