Abstract
Background
Prostate cancer is generally believed to have a strong inherited component, but the search for susceptibility genes has been hindered by the effects of genetic heterogeneity. The recently developed sumLINK and sumLOD statistics are powerful tools for linkage analysis in the presence of heterogeneity.
Methods
We performed a secondary analysis of 1233 prostate cancer pedigrees from the International Consortium for Prostate Cancer Genetics (ICPCG) using two novel statistics, the sumLINK and sumLOD. For both statistics, dominant and recessive genetic models were considered. False discovery rate (FDR) analysis was conducted to assess the effects of multiple testing.
Results
Our analysis identified significant linkage evidence at chromosome 22q12, confirming previous findings by the initial conventional analyses of the same ICPCG data. Twelve other regions were identified with genomewide suggestive evidence for linkage. Seven regions (1q23, 5q11, 5q35, 6p21, 8q12, 11q13, 20p11-q11) are near loci previously identified in the initial ICPCG pooled data analysis or the subset of aggressive prostate cancer (PC) pedigrees. Three other regions (1p12, 8p23, 19q13) confirm loci reported by others, and two (2p24, 6q27) are novel susceptibility loci. FDR testing indicates that over 70% of these results are likely true positive findings. Statistical recombinant mapping narrowed regions to an average of 9 cM.
Conclusions
Our results represent genomic regions with the greatest consistency of positive linkage evidence across a very large collection of high-risk prostate cancer pedigrees using new statistical tests that deal powerfully with heterogeneity. These regions are excellent candidates for further study to identify prostate cancer predisposition genes.
Introduction
Prostate cancer (PC) is believed to have a complex environmental and genetic etiology potentially involving numerous genes (1). The identification of PCa genes has proven to be very difficult; genetic heterogeneity is a major issue that hinders progress (2). Confirmations of reported PC susceptibility loci are infrequent and some of the loci that have been confirmed by multiple researchers are in chromosomal regions with very few promising candidate genes (3,4). Luo and Yu reported in 2003 that evidence for PC susceptibility variants had been reported on all but two human chromosomes (5). These two remaining chromosomes, 21 (6,7) and 22 (8,9), have subsequently both been implicated. The International Consortium for Prostate Cancer Genetics (ICPCG) was formed by a large and diverse group of researchers who have pooled their resources with the intent of deciphering the principal genetic factors underlying this pervasive disease (10). The ICPCG published the findings of a conventional linkage analysis using the well-known heterogeneity LOD (HLOD) statistic and multiple subset analyses based on 1233 high-risk prostate cancer pedigrees. The study identified several susceptibility loci for further study (8).
Here we present the results of a secondary analysis of the ICPCG pooled pedigree resource using new genome-wide linkage-based statistics, the sumLINK and sumLOD, to identify PC susceptibility loci. These new statistics have been shown in simulation studies to be powerful and robust tools for identifying susceptibility loci in the presence of genetic heterogeneity (11). The sumLINK/sumLOD approach is well-suited to analysis of pooled data resources such as this, because it requires only summary data from each constituent group which is logistically easier to attain (there are often data privacy and confidentiality concerns associated with sharing individual raw genotype data and pedigree structures). Secondary analyses of existing data that are more powerful at addressing genetic heterogeneity have the potential to refine the original analyses, and identify additional evidence for PC predispostion genes.
Methods
The sumLINK approach focuses on ‘linked’ pedigrees, which we define to be a pedigree-specific LOD≥0.588 (p≤0.05). The aim is to identify regions with extreme consistency of linkage evidence across pedigrees. The sumLINK statistic is the sum of multipoint LOD scores for all pedigrees that meet the threshold of LOD≥0.588 at a given point in the genome. This value is computed at intervals of one centimorgan throughout the genome. We assess the significance of the sumLINK empirically using a unique genome randomization and shuffling method that simulates the expected consistency of linked pedigrees under null conditions.(11) Briefly, for each pedigree, the vectors of LOD scores for each chromosome are connected in random order, with the first and last values connected to form a ‘loop’, and the loop is broken at a random position to create a randomized, shuffled ‘genomewide’ vector of LOD scores. These vectors are then aligned across pedigrees and values of the sumLINK statistic are calculated. This procedure is designed to maintain each pedigree’s potential for linkage signals across the genome, but randomizes consistency of linkage evidence across pedigrees. Observed peaks are compared with peaks occurring in 1000 iterations of the randomized data in order to establish the expected frequency of peaks with a similar or greater magnitude for the data in question. This expected frequency may be called a false positive rate, or FPR.
The sumLOD statistic is a complimentary companion to the sumLINK statistic. The sumLOD statistic is similar to the sumLINK statistic, but with a reduced inclusion threshold; all positive pedigree LOD scores at each point in the genome are summed to calculate the sumLOD statistic. Significance of the sumLOD is determined empirically by the same genome randomization procedure that is used for the sumLINK. In accordance with the standards for significant linkage evidence set by Lander and Kruglyak (12), peak sumLINK and sumLOD values are considered to represent significant evidence of linkage if the expected frequency of peaks of similar magnitude under null conditions is less than 0.05 per genome. Peak values are considered to be suggestive evidence of linkage if the expected frequency is less than one per genome.
We applied the sumLINK and sumLOD procedures to the 1233 PC pedigrees in the ICPCG pooled pedigree resource. Pedigree characteristics and genotyping details have been described previously (8). The two statistics were computed at 1-cM increments (N=3502) throughout the 22 autosomes based on LOD scores from the dominant and recessive inheritance models that were used in the original ICPCG analysis. The sex chromosomes were not included in the analysis. 572 pedigrees achieved a maximum LOD score of at least 0.588 at some point in the genome under the dominant inheritance model, and 533 pedigrees achieved a LOD score of at least 0.588 under the recessive model. Only these pedigrees contributed to the sumLINK analyses. 1230 pedigrees contributed to the sumLOD analyses under each model. Empirical significance was computed based on 1000 iterations of the genome randomization technique.
False positive rates were calculated based on the empirical distributions for each of the four analyses (dominant and recessive, sumLINK and sumLOD). False discovery rate (FDR) q-values were estimated to account for the effects of multiple testing that are inherent in the usage of multiple models and statistics. Application of FDR methods to multipoint LOD scores have been shown to be valid provided no fine-mapping markers are used (13). This requirement is met in the present analysis. The empirical FDR q-value represents the probability that a given result is a false positive based on the pooled distributions of all four analyses.
Localization
Loci identified with the sumLINK approach have natural potential for subsequent gene localization using statistical recombinant mapping (14), as, by definition, there exist a statistical excess of linked pedigrees contributing to each peak. Hence, for all significant and suggestive sumLINK peaks we will pursue localization using statistical recombinant mapping. The genetic marker sets for which pedigrees were genotyped varied between institutions. Even though the resolution of each separate linkage study map was an average spacing of 10 cM, the disparity of different marker maps helps fine-mapping efforts. If pedigrees from different resources are linked to the same region, they can identify regions smaller than the resolutions of each independent marker map. These genomic segments are the most probable locations for finding a PC susceptibility gene.
Given the linkage evidence for each pedigree is based on a 10 cM map, most pedigrees will have a genotyped marker within 5 cM of any given cM position on the genetic map. Hence, when selecting pedigrees to consider ‘linked’ to a significant or suggestive region, we identified all pedigrees that achieved LOD≥0.588 within 5 cM of the observed sumLINK peak. We then examined the LOD score curves for each of these pedigrees and determined the probable location of recombination events that mark the outer limits of the segregating chromosomal segment within each pedigree. Recombinant events are estimated to be at the outer point of an abrupt drop in LOD score, as these positions are statistical evidence for a loss of genetic sharing by affected pedigree members. A shared chromosomal region bounded by two recombinant events on each side is an approximate 95% confidence interval for the consensus region (14).
Results
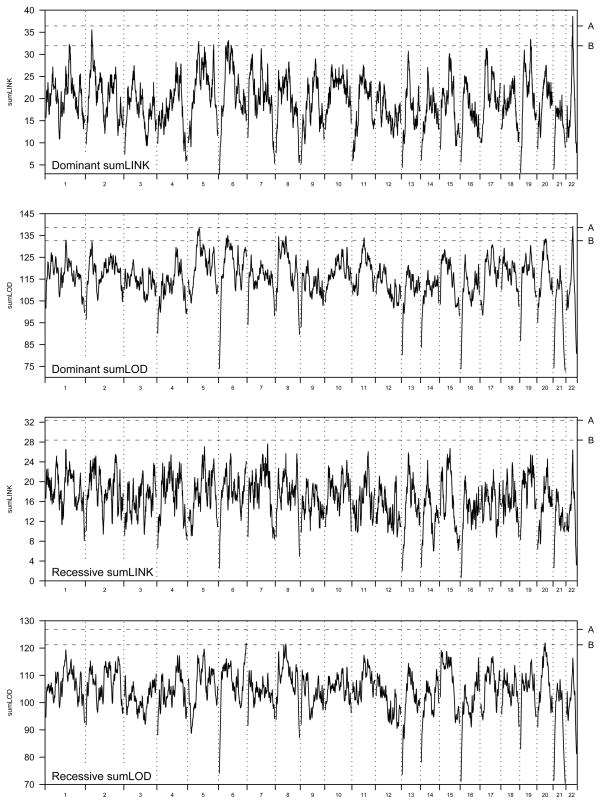
Figure I shows the genome-wide sumLINK and sumLOD statistics for each model, together with lines representing the thresholds for significant and suggestive linkage as determined by the randomization procedure. Results are summarized in Table 1. We identified one locus with significant linkage evidence, and twelve loci with suggestive linkage evidence. There were no significant or suggestive linkage peaks identified by the recessive sumLINK analysis.
Figure I.
Genome-wide sumLINK and sumLOD values for dominant and recessive inheritance models. The line marked “A” in each figure represents the threshold for significant linkage evidence determined by the genome shuffling process. The line marked “B” shows the threshold for suggestive linkage evidence.
Table 1.
Chromosomal regions with at least suggestive linkage evidence
| FDR | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Chr | Nearest marker | cM | # peds contributing | Analysis | Model | FPR | q-val | Obs peaks | Exp peaks |
| 22q12 | D22S283 | 42 | 46 | sumLINK | Dom | 0.010 | 0.115 | 1 | 0.1 |
| D22S283 | 42 | 454 | sumLOD | Dom | 0.032 | 0.186 | 2 | 0.4 | |
| 5q11 | D5S2500 | 75 | 507 | sumLOD | Dom | 0.059 | 0.200 | 4 | 0.8 |
| D5S407 | 72 | 43 | sumLINK | Dom | 0.529 | 0.259 | 17 | 4.4 | |
| 2p24 | D2S1360 | 39 | 45 | sumLINK | Dom | 0.089 | 0.200 | 4 | 0.8 |
| 6p21 | D6S2427 | 59 | 495 | sumLOD | Dom | 0.350 | 0.259 | 17 | 4.4 |
| D6S1017 | 64 | 40 | sumLINK | Dom | 0.445 | 0.259 | 17 | 4.4 | |
| 19q13 | D19S900 | 70 | 43 | sumLINK | Dom | 0.379 | 0.259 | 17 | 4.4 |
| 8q12 | D8S285 | 68 | 487 | sumLOD | Dom | 0.393 | 0.259 | 17 | 4.4 |
| D8S285 | 68 | 449 | sumLOD | Rec | 0.851 | 0.259 | 17 | 4.4 | |
| 8p23 | D8S1130 | 18 | 467 | sumLOD | Dom | 0.442 | 0.259 | 17 | 4.4 |
| 11q13 | D11S1314 | 79 | 491 | sumLOD | Dom | 0.558 | 0.259 | 17 | 4.4 |
| 20p11–q11 | D20S912 | 51 | 484 | sumLOD | Rec | 0.688 | 0.259 | 17 | 4.4 |
| D20S195 | 58 | 489 | sumLOD | Dom | 0.736 | 0.259 | 17 | 4.4 | |
| 6q27 | D6S281 | 189 | 450 | sumLOD | Rec | 0.740 | 0.259 | 17 | 4.4 |
| 1q23 | D1S2628 | 164 | 44 | sumLINK | Dom | 0.822 | 0.259 | 17 | 4.4 |
| 5q35 | D5S400 | 177 | 43 | sumLINK | Dom | 0.852 | 0.259 | 17 | 4.4 |
| 1p12 | D1S534 | 140 | 491 | sumLOD | Dom | 0.935 | 0.262 | 18 | 4.7 |
False positive rate (FPR) refers to the expected frequency of peaks of similar or greater magnitude based on the results of 1000 repetitions of the genome randomization procedure. False discovery rate (FDR) results are based on the cumulative distribution of null p-values from all analyses. The q-value indicates the proportion of all results of similar or greater significance that are expected to be false positives.
Significant linkage evidence was observed at chromosome 22q12 by both the dominant sumLINK (FPR=0.010, 46 contributing pedigrees) and the dominant sumLOD (FPR=0.032, 454 contributing pedigrees). In addition to both of these findings being genome-wide significant in their respective single genomewide screens (FPRs < 0.05), after correction for all four genomewide analyses, the FDR was 0.186. This indicates that under the null hypothesis, the expected number of peaks at least as extreme as these two is only 0.4 (=0.186×2), and therefore that 1.6 of these 2 peaks are not likely to be from the null distribution. Since both peaks are at 22q12, this indicates that even after correction for the four genomewide analyses performed here, there is excellent evidence that the 22q12 locus is a true positive.
Suggestive peaks are those that in a single genomewide screen would only be expected once per genome under the null hypothesis. Twelve loci were identified within their respective single genomewide analyses to have suggestive evidence for linkage. In decreasing order of significance, these regions were at chromosomes 5q11 (dominant sumLOD and sumLINK), 2p24 (dominant sumLINK), 6p21 (dominant sumLOD and sumLINK), 19q13 (dominant sumLINK), 8q12 (dominant and recessive sumLOD), 8p23 (dominant sumLOD), 11q13 (dominant sumLOD), 20p11-q11 (dominant and recessive sumLOD), 6q27 (recessive sumLOD), 1q23 (dominant sumLINK), 5q35 (dominant sumLINK), and 1p12 (dominant sumLOD). Loci at 5q11 and 2p24, are perhaps worthy of particular note because although strictly only suggestive, both were borderline significant (FPRs of 0.059 and 0.089, respectively). Accounting for the four genomewide analyses, the FDR value associated with these 18 suggestive and significant peaks (distributed across 13 regions) was 0.262, indicating that only 4.7 (18×0.262) peaks would have been expected under the null. That is, we observed 13.3 more peaks than expected and thus 13.3 are likely not from the null. Hence, there is good evidence that many, although not all, of these loci with suggestive evidence for linkage are also true positive findings.
Table 2 shows the results of our localization analysis for the seven significant and suggestive regions identified with the sumLINK analyses. Estimated regions are based on the observation of two recombination events at each end, indicating an approximate 95% support interval. The microsatellite markers flanking the two-recombinant region are also reported. These two-recombinant localization intervals range from 5 to 17 cM, with a mean of 9.1 cM. Since we included information from all pedigrees with a LOD≥0.588 within 5 cM of the peak, there were some instances where pedigrees showed conflicting evidence about the location of the shared chromosomal region. In these instances, we selected the region where the greatest number of pedigrees agreed, and reported the number of conflicting pedigrees in the table together with the number of supporting pedigrees.
Table 2.
Localization intervals for sumLINK regions
| Locus | Peak (cM) | 2-recomb. Interval (cM) | Supporting Pedigrees* | Conflicting Pedigrees | Flanking markers |
|---|---|---|---|---|---|
| 1q23 | 164 | 161—170 | 59 | 0 | D1S1677—D1S452 |
| 2p24 | 39 | 29—40 | 53 | 0 | D2S1400—D2S1360 |
| 5q11 | 72 | 72—79 | 52 | 2 | D5S407—D5S647 |
| 5q35 | 177 | 168—185 | 52 | 0 | D5S422—D5S1960 |
| 6p21 | 64 | 65—74 | 50 | 0 | D6S1582—D6S1280 |
| 19q13 | 70 | 63—69 | 55 | 2 | D19S570—D19S420 |
| 22q12 | 42 | 37—42 | 57 | 0 | D22S280—D22S683 |
Number of pedigrees with LOD ≥0.588 within 5cM of the peak.
Discussion
We have performed a secondary analysis of data from the largest collection of high-risk prostate cancer pedigrees ever assembled with new multipoint linkage-based statistics, sumLINK and sumLOD, which are specifically designed to address genetic heterogeneity. Three of the thirteen loci that we identified in the present analysis (5q11, 5q35 and 22q12) correspond directly to peaks that were reported in the original ICPCG analysis using the conventional HLOD statistic (8). In that analysis, a dominant LOD score of 1.95 was observed at 22q12, which increased to 3.57 in the subset of pedigrees with at least five affected family members. Additionally, a non-parametric LOD of 2.28 was reported at 5q12, and a dominant LOD of 2.05 was reported at 5q35 in the subset of families with mean age at diagnosis ≤65 years. Two other loci (1q23 and 8q12) are near peaks that were reported in the first analysis (8). The loci on chromosomes 6p21, 11q13 and 20p11-q11 correspond to susceptibility loci previously identified in the ICPCG data resource in linkage scans for aggressive prostate cancer (11,15). The remaining loci have not previously been identified in pooled ICPCG data, though many of them correspond to findings reported elsewhere in linkage studies by individual institutions.
The dominant and recessive sumLOD peaks on chromosome 20 appear to be supportive of the HPC20 locus (16), although it should be noted that the original HPC20 linkage peak was at 20q13, about 20–30 cM downstream from the peaks we report here. Our tentative replication of HPC20 is in contrast to an earlier ICPCG study using the same data and a conventional HLOD approach that failed to replicate this locus (10), although a later ICPCG study concentrating on aggressive prostate cancer pedigrees did find linkage evidence (15). The ICPCG aggressive PC linkage study found a dominant LOD score of 2.49 midway between the dominant and recessive sumLOD peaks that we report here. The observed LOD score increased to 2.65 in the subset of pedigrees with mean age at onset >65 years. The present study includes data from most of the pedigrees that were included in the ICPCG aggressiveness analysis, but the difference in phenotype definition prevents a direct comparison of the pedigrees that contribute to the results. HPC20 was originally identified by the Mayo Clinic site (16,17); however, of the 45 pedigrees that exhibited LOD≥0.588 within 5 cM of the dominant sumLOD peak, only 6 were from Mayo Clinic. As seen from these comparisons, one distinct advantage of the sumLINK and sumLOD statistics is that the approach inherently identifies subgroups of pedigrees that are genetically alike, and hence one analysis can encompass what in conventional analyses may take many subset analyses and multiple testing corrections. It is therefore perhaps not surprising that our results more closely align with linkage findings for subset-based analyses such as aggressive prostate cancer (15).
In addition to the findings discussed above, three of the other suggestive linkage regions reported here support previously identified loci. Our peak at 1p12 falls within a region of interest reported by other ICPCG member-sites (17). The peak at chromosome 1q23 approximates the HPC1 susceptibility region (19), although the RNASEL candidate gene proposed as the HPC1 gene (20) is located about 20 Mb beyond the boundary of our support interval. An ICPCG member-site previously reported linkage at 8p23 (21), a finding that was recently replicated and refined by combined somatic deletion and fine linkage mapping (22). The suggestive sumLOD peak at 8p23 is about 4 Mb from the MSR1 PC candidate gene. Our 19q13 region also corresponds to previously reported linkage for aggressive PC (23,24).
Our suggestive regions on chromosomes 2p24 and 6q27 appear to be new. Of particular interest of these new loci is perhaps 2p24. Statistical evidence for 2p24 was borderline significant, and recently a germline copy number variant at the 2p24 locus has been associated with aggressive prostate cancer (25). Other notable association studies have focused on regions identified in this report. Copy number variations at 8p23 and 11q13 have been implicated in aggressive PC and PC recurrence, respectively (26). Kallikrein genes KLK2 and KLK3 at chromosome 19q13 have been identified as PC candidate genes (27).
We did not identify linkage evidence to regions that have recently received much attention due to highly significant and replicable association evidence with PC in genome-wide association studies. The most compelling of these results are located on chromosomes 8q24, 17q12, and 10q11 (3). It is perhaps not surprising that we did not find any evidence to support these regions because these SNPs have common minor allele frequencies and very small effect sizes. The sumLINK and sumLOD are linkage-based statistics, and linkage is most powerful for finding rarer, more highly-penetrant variants.
The localization procedure we used here to delimit support intervals generated much more concise intervals than the 1-LOD drop regions reported previously by ICPCG for the four sumLINK peaks that overlapped with previous findings (8). The intervals reported previously ranged from 12 to 30 cM with a mean length of 21.2 cM, substantially longer than the mean length of 9.5 cM we report here for the same 4 regions. A particularly interesting example of the narrower intervals can be seen in the putative susceptibility locus at chromosome 5q11–12. The previous analysis of this data identified a suggestive HLOD peak at 77 cM, with a reported 1-LOD support interval extending from 66—96 cM. In the present analysis, the sumLINK statistic identified a suggestive linkage peak at 72 cM and a 2-recombinant support interval of only 7 cM which includes the original HLOD peak. This ability to more narrowly define regions using statistical recombinant mapping was also illustrated by an earlier candidate region localization study for the chromosome 22q12 susceptibility locus (9). That report had the advantage of LOD score data from several large pedigrees with fine-mapping markers which were not included in the present results. Nonetheless, and as expected, the 2-recombinant localization region we report here supports the region previously reported in that paper.
Conclusion
A secondary reanalysis of 1233 PC pedigrees using novel linkage statistics identified 13 regions with at least genomewide suggestive evidence for linkage. Eight regions provide confirmation of loci previously identified by conventional linkage analyses in the same ICPCG data (8) or the subset of aggressive PC pedigrees (15), three are regions that confirm loci not seen in the original analyses, but are reported in other linkage studies (18,22–24), and two are novel loci. One distinct benefit of the sumLINK and sumLOD approach is that the statistics are based on the identification of pedigrees that are genetically alike at a locus, and the constituent set of pedigrees may change from locus-to-locus. This both addresses genetic heterogeneity directly and largely circumvents the need for subset and stratification analyses that are costly in terms of multiple testing. This is illustrated by the fact that several of the regions identified here replicate results that were originally found in stratification analyses. The second advantage for the sumLINK statistic is the natural progression to statistical recombinant mapping, which appears to hold much promise for narrowing linkage regions. Furthermore, the FDR approach for correction of multiple genomewide analyses can better guide interpretation and aid prioritization of findings. Evidence here suggests that these statistics have the potential to further refine the results of original analyses, and provide new directions in the pursuit for PC susceptibility genes.
Acknowledgments
We would like to express our gratitude to the many families who participated in the many studies involved in the International Consortium for Prostate Cancer Genetics (ICPCG). The ICPCG, including the consortium’s Data Coordinating Center (DCC), is made possible by a grant from the National Institutes of Health U01 CA89600 (to W.B.I.). G.B.C. was supported by National Library of Medicine training grant NLM T15 LM07124. N.J.C. was supported in part by USPHS CA98364 (to N.J.C.). Additional support to participating groups, or members within groups, is as follows:
AAHPC Group: The authors would like to express their gratitude to the African-American Hereditary Prostate Cancer Study (AAHPC) families and study participants for their continued involvement in this research. We specifically name, C. Ahaghotu, J. Bennett, W. Boykin, G. Hoke, T. Mason, C. Pettaway, S. Vijayakumar, S. Weinrich, M. Franklin, P. Roberson, J. Frost, E. Johnson, L. Faison-Smith, C. Meegan, M. Johnson, L. Kososki, C. Jones and R. Mejia. We would also like to thank members of the National Human Genome Center (NHGC) at Howard University namely R. Kittles, G.M. Dunston, P. Furbert-Harris and C. Royal. We would also like to acknowledge the contribution of the National Human Genome Research Institute (NHGRI) and TGen genotyping staff including E. Gillanders and C. Robbins. The AAHPC study would not have been possible without F. Collins (Director of NHGRI) and J. Trent (Director of TGen). This research was funded primarily through the NIH Center for Minority and Health Disparities (1-HG-75418). A.B.B-B and A.G also received support from USPHS CA-06927 and an appropriation from the Commonwealth of Pennsylvania. This research was also supported in part by the Intramural Research Program of the NIH (NHGRI) and USPHS RR03048 from the National Center for Research Institute and USPHS RR03048 from the National Center for Research Resources. ACTANE Group: Genotyping and statistical analysis for this study, and recruitment of U.K. families, was supported by Cancer Research U.K (CR-UK). Additional support was provided by The Prostate Cancer Research Foundation, The Times Christmas Appeal and the Institute of Cancer Research. Genotyping was conducted in the ‘Jean Rook Gene Cloning Laboratory’ which is supported by BREAKTHROUGH Breast Cancer - Charity No. 328323. The funds for the ABI 377 used in this study were generously provided by the legacy of the late Marion Silcock. We thank S. Seal and A. Hall for kindly storing and logging the samples that were provided. D.F.E is a Principal Research Fellow of CR-UK. Funding in Australia was obtained from The Cancer Council Victoria, The National Health and Medical Research Council (grants 940934, 251533, 209057, 126402, 396407), Tattersall’s and The Whitten Foundation. We would like to acknowledge the work of the study coordinator M. Staples and the Research Team B. McCudden, J. Connal, R. Thorowgood, C. Costa, M. Kevan, and S. Palmer, and to J. Karpowicz for DNA extractions. The Texas study of familial prostate cancer was initiated by the Department of Epidemiology, M.D. Anderson Cancer Center. M.B. was supported by an NCI Post-doctoral Fellowship in Cancer Prevention (R25). We would also like to specifically thank the following members of ACTANE: S. Edwards, M. Guy, Q. Hope, S. Bullock, S. Bryant, S. Mulholland, S. Jugurnauth, N. Garcia, A. Ardern-Jones, A. Hall, L. O’Brien, B. Gehr-Swain, R. Wilkinson, D. Dearnaley, The UKGPCS Collaborators, British Association of Urological Surgeons’ Section of Oncology (UK Sutton); Chris Evans (UK Cambridge); M. Southey (Australia); N. Hamel, S. Narod, J. Simard (Canada); C. Amos (USA Texas); N. Wessel, T. Andersen (Norway); D.T. Bishop (EU Biomed). BC/CA/HI Group: USPHS CA67044. FHCRC Group: USPHS CA80122 (to J.L.S.) which supports the family collection; USPHS CA78836 (to E.A.O). E.A.O was supported in part by the NHGRI. JHU Group: Genotyping for the JHU, University of Michigan, University of Tampere, and University of Umeå groups’ pedigrees was provided by NHGRI and TGen genotyping staff including E. Gillanders, MP Jones, D. Gildea, E. Riedesel, J. Albertus, D. Freas-Lutz, C. Markey, J. Carpten and J. Trent. Mayo Clinic Group: USPHS CA72818. Michigan Group: USPHS CA079596. University of Tampere Group: The Competitive Research Funding of the Pirkanmaa Hospital District, Reino Lahtikari Foundation, Finnish Cancer Organisations, Sigrid Juselius Foundation, and Academy of Finland grant 211123. University of Ulm Group: Deutsche Krebshilfe, grant number 70–3111-V03. University of Umea Group: Work was supported by the Swedish Cancer Society and a Spear grant from the Umeå University Hospital, Umeå, Sweden. University of Utah Group: Data collection was supported by USPHS CA90752 (to L.A.C.-A.) and by the Utah Cancer Registry, which is funded by Contract #N01-PC-35141 from the National Cancer Institute’s Surveillance, Epidemiology, and End-Results Program with additional support from the Utah State Department of Heath and the University of Utah. Partial support for all datasets within the Utah Population Database was provided by the University of Utah Huntsman Cancer Institute and also by the USPHS M01-RR00064 from the National Center for Research Resources. Genotyping services were provided by the Center for Inherited Disease Research (N01-HG-65403). DCC: The study is partially supported by USPHS CA106523 (to J.X.), USPHS CA95052 (to J.X.), and Department of Defense grant PC051264 (to J.X.).
References
- 1.Schaid D. The Complex Genetic Epidemiology of Prostate Cancer. Human Molecular Genetics. 2004;13(Supp 1):R103–121. doi: 10.1093/hmg/ddh072. [DOI] [PubMed] [Google Scholar]
- 2.Ostrander EA, Stanford JL. Genetics of prostate cancer: too many loci, too few genes. Am J Hum Genet. 2000;67(6):1367–1375. doi: 10.1086/316916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Witte JS. Prostate cancer genomics: towards a new understanding. Nature reviews. 2009;10(2):77–82. doi: 10.1038/nrg2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghoussaini M, Song H, Koessler T, Al Olama AA, Kote-Jarai Z, Driver KE, Pooley KA, Ramus SJ, Kjaer SK, Hogdall E, DiCioccio RA, Whittemore AS, Gayther SA, Giles GG, Guy M, Edwards SM, Morrison J, Donovan JL, Hamdy FC, Dearnaley DP, Ardern-Jones AT, Hall AL, O’Brien LT, Gehr-Swain BN, Wilkinson RA, Brown PM, Hopper JL, Neal DE, Pharoah PD, Ponder BA, Eeles RA, Easton DF, Dunning AM. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst. 2008;100(13):962–966. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo JH, Yu YP. Genetic factors underlying prostate cancer. Expert reviews in molecular medicine. 2003;5(12):1–26. doi: 10.1017/S1462399403006057. [DOI] [PubMed] [Google Scholar]
- 6.Yoshimoto M, Joshua AM, Chilton-Macneill S, Bayani J, Selvarajah S, Evans AJ, Zielenska M, Squire JA. Three-color FISH analysis of TMPRSS2/ERG fusions in prostate cancer indicates that genomic microdeletion of chromosome 21 is associated with rearrangement. Neoplasia (New York, NY) 2006;8(6):465–469. doi: 10.1593/neo.06283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu W, Chang B, Sauvageot J, Dimitrov L, Gielzak M, Li T, Yan G, Sun J, Sun J, Adams TS, Turner AR, Kim JW, Meyers DA, Zheng SL, Isaacs WB, Xu J. Comprehensive assessment of DNA copy number alterations in human prostate cancers using Affymetrix 100K SNP mapping array. Genes, chromosomes & cancer. 2006;45(11):1018–1032. doi: 10.1002/gcc.20369. [DOI] [PubMed] [Google Scholar]
- 8.Xu J, Dimitrov L, Chang BL, Adams TS, Turner AR, Meyers DA, Eeles RA, Easton DF, Foulkes WD, Simard J, Giles GG, Hopper JL, Mahle L, Moller P, Bishop T, Evans C, Edwards S, Meitz J, Bullock S, Hope Q, Hsieh CL, Halpern J, Balise RN, Oakley-Girvan I, Whittemore AS, Ewing CM, Gielzak M, Isaacs SD, Walsh PC, Wiley KE, Isaacs WB, Thibodeau SN, McDonnell SK, Cunningham JM, Zarfas KE, Hebbring S, Schaid DJ, Friedrichsen DM, Deutsch K, Kolb S, Badzioch M, Jarvik GP, Janer M, Hood L, Ostrander EA, Stanford JL, Lange EM, Beebe-Dimmer JL, Mohai CE, Cooney KA, Ikonen T, Baffoe-Bonnie A, Fredriksson H, Matikainen MP, Tammela T, Bailey-Wilson J, Schleutker J, Maier C, Herkommer K, Hoegel JJ, Vogel W, Paiss T, Wiklund F, Emanuelsson M, Stenman E, Jonsson BA, Gronberg H, Camp NJ, Farnham J, Cannon-Albright LA, Seminara D. A combined genomewide linkage scan of 1,233 families for prostate cancer-susceptibility genes conducted by the international consortium for prostate cancer genetics. Am J Hum Genet. 2005;77(2):219–229. doi: 10.1086/432377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camp NJ, Cannon-Albright LA, Farnham JM, Baffoe-Bonnie AB, George A, Powell I, Bailey-Wilson JE, Carpten JD, Giles GG, Hopper JL, Severi G, English DR, Foulkes WD, Maehle L, Moller P, Eeles R, Easton D, Badzioch MD, Whittemore AS, Oakley-Girvan I, Hsieh CL, Dimitrov L, Xu J, Stanford JL, Johanneson B, Deutsch K, McIntosh L, Ostrander EA, Wiley KE, Isaacs SD, Walsh PC, Thibodeau SN, McDonnell SK, Hebbring S, Schaid DJ, Lange EM, Cooney KA, Tammela TL, Schleutker J, Paiss T, Maier C, Gronberg H, Wiklund F, Emanuelsson M, Isaacs WB. Compelling evidence for a prostate cancer gene at 22q12.3 by the International Consortium for Prostate Cancer Genetics. Hum Mol Genet. 2007;16(11):1271–1278. doi: 10.1093/hmg/ddm075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaid DJ, Chang BL. Description of the International Consortium For Prostate Cancer Genetics, and failure to replicate linkage of hereditary prostate cancer to 20q13. Prostate. 2005;63(3):276–290. doi: 10.1002/pros.20198. [DOI] [PubMed] [Google Scholar]
- 11.Christensen GB, Knight S, Camp NJ. The sumLINK statistic for genetic linkage analysis in the presence of heterogeneity. Genet Epidemiol. 2009 doi: 10.1002/gepi.20414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11(3):241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 13.Chen L, Storey JD. Relaxed Significance Criteria for Linkage Analysis. Genetics. 2006;173:2371–2381. doi: 10.1534/genetics.105.052506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camp NJ, Farnham JM, Cannon-Albright L. Localization of a Prostate Cancer Predisposition Gene to an 880-kb Region of Chromosome 22q12.3 in Utah High-Risk Pedigrees. Cancer Res. 2006;66(20):10205–10212. doi: 10.1158/0008-5472.CAN-06-1233. [DOI] [PubMed] [Google Scholar]
- 15.Schaid D ICPCG. Pooled genome linkage scan of aggressive prostate cancer: Results from the International Consortium for Prostate Cancer Genetics. Hum Genet. 2006;120(4):471–485. doi: 10.1007/s00439-006-0219-9. [DOI] [PubMed] [Google Scholar]
- 16.Berry R, Schroeder JJ, French AJ, McDonnell SK, Peterson BJ, Cunningham JM, Thibodeau SN, Schaid DJ. Evidence for a prostate cancer-susceptibility locus on chromosome 20. Am J Hum Genet. 2000;67(1):82–91. doi: 10.1086/302994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cunningham JM, McDonnell SK, Marks A, Hebbring S, Anderson SA, Peterson BJ, Slager S, French A, Blute ML, Schaid DJ, Thibodeau SN. Genome linkage screen for prostate cancer susceptibility loci: results from the Mayo Clinic Familial Prostate Cancer Study. Prostate. 2003;57(4):335–346. doi: 10.1002/pros.10308. [DOI] [PubMed] [Google Scholar]
- 18.Slager S, Zarfas KE, Brown WM, Lange E, McDonnell S, Wojno KJ, Cooney KA. Genome-wide linkage scan for prostate cancer aggressiveness loci using families from the University of Michigan Prostate Cancer Genetics Project. Prostate. 2006;66(2):173–179. doi: 10.1002/pros.20332. [DOI] [PubMed] [Google Scholar]
- 19.Smith JR, Freije D, Carpten JD, Gronberg H, Xu J, Isaacs SD, Brownstein MJ, Bova GS, Guo H, Bujnovszky P, Nusskern DR, Damber JE, Bergh A, Emanuelsson M, Kallioniemi OP, Walker-Daniels J, Bailey-Wilson JE, Beaty TH, Meyers DA, Walsh PC, Collins FS, Trent JM, Isaacs WB. Major susceptibility locus for prostate cancer on chromosome 1 suggested by a genome-wide search. Science. 1996;274(5291):1371–1374. doi: 10.1126/science.274.5291.1371. [DOI] [PubMed] [Google Scholar]
- 20.Carpten J, Nupponen N, Isaacs S, Sood R, Robbins C, Xu J, Faruque M, Moses T, Ewing C, Gillanders E, Hu P, Bujnovszky P, Makalowska I, Baffoe-Bonnie A, Faith D, Smith J, Stephan D, Wiley K, Brownstein M, Gildea D, Kelly B, Jenkins R, Hostetter G, Matikainen M, Schleutker J, Klinger K, Connors T, Xiang Y, Wang Z, De Marzo A, Papadopoulos N, Kallioniemi OP, Burk R, Meyers D, Gronberg H, Meltzer P, Silverman R, Bailey-Wilson J, Walsh P, Isaacs W, Trent J. Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nat Genet. 2002;30(2):181–184. doi: 10.1038/ng823. [DOI] [PubMed] [Google Scholar]
- 21.Xu J, Zheng SL, Hawkins GA, Faith DA, Kelly B, Isaacs SD, Wiley KE, Chang B, Ewing CM, Bujnovszky P, Carpten JD, Bleecker ER, Walsh PC, Trent JM, Meyers DA, Isaacs WB. Linkage and association studies of prostate cancer susceptibility: evidence for linkage at 8p22–23. Am J Hum Genet. 2001;69(2):341–350. doi: 10.1086/321967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang BL, Liu W, Sun J, Dimitrov L, Li T, Turner AR, Zheng SL, Isaacs WB, Xu J. Integration of somatic deletion analysis of prostate cancers and germline linkage analysis of prostate cancer families reveals two small consensus regions for prostate cancer genes at 8p. Cancer Res. 2007;67(9):4098–4103. doi: 10.1158/0008-5472.CAN-06-4570. [DOI] [PubMed] [Google Scholar]
- 23.Neville PJ, Conti DV, Krumroy LM, Catalona WJ, Suarez BK, Witte JS, Casey G. Prostate cancer aggressiveness locus on chromosome segment 19q12-q13.1 identified by linkage and allelic imbalance studies. Genes, chromosomes & cancer. 2003;36(4):332–339. doi: 10.1002/gcc.10165. [DOI] [PubMed] [Google Scholar]
- 24.Slager SL, Schaid DJ, Cunningham JM, McDonnell SK, Marks AF, Peterson BJ, Hebbring SJ, Anderson S, French AJ, Thibodeau SN. Confirmation of linkage of prostate cancer aggressiveness with chromosome 19q. Am J Hum Genet. 2003;72(3):759–762. doi: 10.1086/368230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu W, Sun J, Li G, Zhu Y, Zhang S, Kim ST, Sun J, Wiklund F, Wiley K, Isaacs SD, Stattin P, Xu J, Duggan D, Carpten JD, Isaacs WB, Gronberg H, Zheng SL, Chang BL. Association of a germ-line copy number variation at 2p24.3 and risk for aggressive prostate cancer. Cancer Res. 2009;69(6):2176–2179. doi: 10.1158/0008-5472.CAN-08-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paris PL, Andaya A, Fridlyand J, Jain AN, Weinberg V, Kowbel D, Brebner JH, Simko J, Watson JE, Volik S, Albertson DG, Pinkel D, Alers JC, van der Kwast TH, Vissers KJ, Schroder FH, Wildhagen MF, Febbo PG, Chinnaiyan AM, Pienta KJ, Carroll PR, Rubin MA, Collins C, van Dekken H. Whole genome scanning identifies genotypes associated with recurrence and metastasis in prostate tumors. Hum Mol Genet. 2004;13(13):1303–1313. doi: 10.1093/hmg/ddh155. [DOI] [PubMed] [Google Scholar]
- 27.Pal P, Xi H, Sun G, Kaushal R, Meeks JJ, Thaxton CS, Guha S, Jin CH, Suarez BK, Catalona WJ, Deka R. Tagging SNPs in the kallikrein genes 3 and 2 on 19q13 and their associations with prostate cancer in men of European origin. Hum Genet. 2007;122(3–4):251–259. doi: 10.1007/s00439-007-0394-3. [DOI] [PubMed] [Google Scholar]