Abstract
Breast cancers commonly become resistant to EGFR–tyrosine kinase inhibitors (EGFR-TKIs); however, the mechanisms of this resistance remain largely unknown. We hypothesized that resistance may originate, at least in part, from molecular alterations that activate signaling downstream of EGFR. Using a screen to measure reversion of malignant cells into phenotypically nonmalignant cells in 3D gels, we identified FAM83A as a candidate cancer-associated gene capable of conferring resistance to EGFR-TKIs. FAM83A overexpression in cancer cells increased proliferation and invasion and imparted EGFR-TKI resistance both in cultured cells and in animals. Tumor cells that survived EGFR-TKI treatment in vivo had upregulated FAM83A levels. Additionally, FAM83A overexpression dramatically increased the number and size of transformed foci in cultured cells and anchorage-independent growth in soft agar. Conversely, FAM83A depletion in cancer cells caused reversion of the malignant phenotype, delayed tumor growth in mice, and rendered cells more sensitive to EGFR-TKI. Analyses of published clinical data revealed a correlation between high FAM83A expression and breast cancer patients’ poor prognosis. We found that FAM83A interacted with and caused phosphorylation of c-RAF and PI3K p85, upstream of MAPK and downstream of EGFR. These data provide an additional mechanism by which tumor cells can become EGFR-TKI resistant.
Introduction
EGFR overexpression is often found in breast carcinomas and correlates with patients’ poor prognosis (1); however, therapeutic use of EGFR–tyrosine kinase inhibitors (EGFR-TKIs) has been hampered by resistance (2–5). In contrast to other types of epithelial cancers, EGFR mutations are rare in breast cancer (6). Thus, it is important to investigate whether there are other alterations activating downstream signals of EGFR that might confer EGFR-TKI resistance in breast cancer (7).
We used a variation of our phenotypic reversion assay in 3D laminin-rich gels (lrECM) (8) using isogenic cell lines of the HMT3522 human breast cancer progression series (9, 10). Reversion of malignant phenotype (depolarized, disorganized, proliferative colonies; ref. 11) to nonmalignant phenotype (growth-arrested, mammary acinus-like structures with basal polarity) by inhibiting a number of pathways, including EGFR signaling (8, 12), decreases tumor growth in animals (8, 13). Hence, this 3D assay provided a robust model with relevance to in vivo response to screen for genes capable of conferring EGFR-TKI resistance. We transfected the malignant cells with a cDNA library made from the same cells and screened genes that disrupted the ability of breast cancer cells to revert in response to the EGFR-TKI AG1478 and identified FAM83A.
Here, we demonstrated that FAM83A (a) had oncogenic properties, (b) conferred EGFR-TKI resistance when overexpressed, (c) correlated with breast cancer patients’ poor prognosis, and (d) promoted tumorigenicity through its putative interactions with c-RAF and PI3K p85. These observations suggest that FAM83A dysregulation could account for some of the observed clinical EGFR-TKI resistance in breast cancers.
Results
Upregulated EGFR signaling disrupts tissue polarity and induces breast cancer cell proliferation and invasion (12, 14). Treatment with an EGFR-TKI, AG1478, caused phenotypic reversion of malignant HMT3522 T4-2 cells into growth-arrested, polarized structures resembling nonmalignant S1 cells in 3D lrECM (Figure 1A and refs. 12, 15). These 2 observations allowed us to screen for genes whose overexpression is responsible for EGFR-TKI resistance by transducing T4-2 cells with an autologous cDNA library, then screening for colonies that had failed to revert in 3D lrECM when treated with AG1478 (Figure 1A). We isolated half a dozen candidate gene sequences and obtained a list of 5 genes conferring the higher resistance to AG1478 (Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI60498DS1). Among these, the sequence showing the highest degree of resistance was a partial open reading frame of the gene family with sequence similarity 83, member A (FAM83A). Here, we characterized this gene (Supplemental Figure 1, A and B) after demonstrating that the overexpression of the full-length protein similarly rendered T4-2 cells resistant to AG1478 (Supplemental Figure 1C).
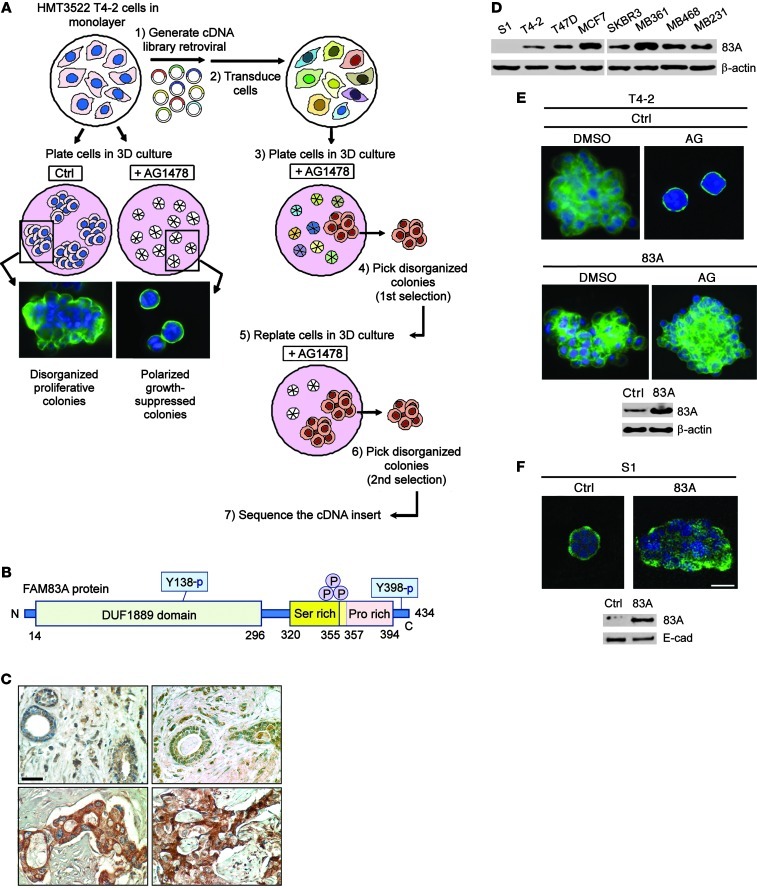
Figure 1. Discovery of FAM83A, a protein upregulated in breast cancer that confers resistance to EGFR-TKI AG1478.
(A) Process for the screen to identify FAM83A. Treatment of T4-2 cells with a small-molecule EGFR inhibitor, AG1478, induced reversion to a polarized acinar phenotype in 3D lrECM culture, as detected by α6 integrin staining (green) to delineate basal polarity (left); after transduction with the cDNA library, a subpopulation of cells disorganized in 3D culture after AG1478 treatment was isolated, expanded, and searched for the cDNA insert (right). (B) Protein domain structure of FAM83A. (C) Immunohistochemical analysis of FAM83A expression in normal (top) and malignant (bottom) breast tissue specimens. Whereas 0 of 16 normal cells were strongly positive for FAM83A staining, 45 of 48 malignant cells (94%) were positive. Scale bar: 50 μm. (D) FAM83A (83A) protein expression in a panel of breast cancer cell lines. (E and F) Effect of FAM83A overexpression in inhibiting AG1478-induced (AG) reversion of T4-2 cells (E) and ablating the polarized structure of S1 cells (F). Generation of T4-2 and S1 cell lines overexpressing FAM83A was confirmed by Western analyses. Scale bars: 50 μm.
FAM83A was originally identified as BJ-TSA-9, highly expressed in lung cancer, without known function(s) (16). This 434–amino-acid protein contains DUF1669, serine-rich domains, and proline-rich domains (PRDs) (Figure 1B). A conserved PxxP motif in the PRD interacts with Src homology 3 (SH3) domain–containing proteins (17, 18). In the DUF1669 domain, FAM83A contains an arginine (R) (glutamine [Q] in mouse) rather than the key histidine (H) residue of the phospholipase D (PLD) motif (Supplemental Figure 1D), making it unlikely that this domain has PLD function. Indeed, we could not detect PLD activity in the in vitro transcribed/translated FAM83A protein (Supplemental Figure 2, A and B).
After raising a FAM83A antibody, we assessed FAM83A expression in breast tissues by immunohistochemistry (IHC). Examination of human breast tissue samples by IHC revealed a highly significant staining difference between normal and malignant tissues. In normal tissues, FAM83A staining was essentially negative (0 of 16 positive), whereas in malignant breast tumor sections, 94% (45 of 48 positive) showed strong cytosolic staining (P < 0.0001, Fisher exact test; Figure 1C). We compared FAM83A expression in normal versus malignant breast tissues using a published gene expression profiling dataset on clinical samples (Supplemental Figure 3A and ref. 19). FAM83A expression was found to be upregulated in all analyzed breast carcinomas compared with normal breast tissues and was dramatically overexpressed in a fraction of breast cancers. We then examined FAM83A levels in a panel of breast epithelial cell lines: FAM83A again was expressed highly in all breast cancer cell lines tested, including weakly invasive (MCF-7 and T47D) and more invasive (SKBR3, MDA-MB-361, MDA-MB-468, and MDA-MB-231) cancer cells (Figure 1D). FAM83A overexpression in these cancer cell lines was attributable to the amplification of the gene locus (Supplemental Figure 3B and ref. 19). The breast cancer cell lines with higher FAM83A expression (T47D, MCF7, MDA-MB361, MDA-MB468, and MDA-MB231; Figure 1D) were more resistant to EGFR-TKI than cell lines with moderate expression (SKBR and T4-2; refs. 20, 21). In the HMT-3522 series, FAM83A levels correlated with the degree of progression to malignancy; it was almost undetectable in S1 cells, but higher in T4-2 cells, although still lower than other aggressive breast cancer cell lines examined (Figure 1D). Overexpressing FAM83A in T4-2 cells to a level comparable to other breast cancer cell lines rendered them resistant to reversion mediated by AG1478 (Figure 1E), whereas overexpressing FAM83A in S1 cells ablated basal polarity and caused disorganized growth in 3D lrECM (Figure 1F). These data indicate that FAM83A is expressed in primary breast cancer specimens as well as in breast cancer cell lines, at least in part due to the amplification of the gene copy number, and that it contributes to impaired tissue organization and to EGFR-TKI resistance.
FAM83A depletion by siRNAs (Supplemental Figure 4A) and shRNA (Figure 2A) resulted in reversion of T4-2 cells, leading to formation of essentially quiescent tissue-like structures with basal polarity (Figure 2, B and C, and Supplemental Figure 4B). FAM83A depletion also caused actin stress fibers to become primarily cortical (Supplemental Figure 4C) and led to reduced invasiveness (Figure 2D and Supplemental Figure 4D), whereas FAM83A overexpression led to elevated invasiveness (Figure 2D). It should be noted that increased invasiveness as a result of FAM83A overexpression was not caused by elevated T4-2 growth rate (Supplemental Figure 5A). In complementary experiments using MDA-MB468 cells, which expressed a high level of endogenous FAM83A (Figure 1D) and are resistant to EGFR-TKI (20, 21), we depleted the protein by shRNA (Figure 2E). FAM83A depletion reduced proliferation rate by half and dramatically reduced the clonogenic potential (Figure 2, I–K). In 3D, FAM83A-depleted cells showed apoptotic phenotype and markedly reduced viable cell number (Figure 2, F and G). The invasiveness was almost fully abrogated, which could not be accounted for simply by reduced proliferation of these cells (Figure 2, H–K). Clonogenic assay demonstrated that parental MDA-MB468 cells expressing a high level of FAM83A were resistant to treatment with AG1478, whereas downmodulation of FAM83A rendered MDA-MB468 cells significantly more sensitive to the drug (Figure 3F). Complementarily, proliferation assay also showed the similar trend that FAM83A-depleted cells were more sensitive to AG1478 than control (Supplemental Figure 5B), despite the fact that this assay is reported to be less sensitive than the clonogenic assay under continuous drug treatment (22, 23). These data indicate that FAM83A expression is important for proliferation, invasion, and EGFR-TKI resistance.
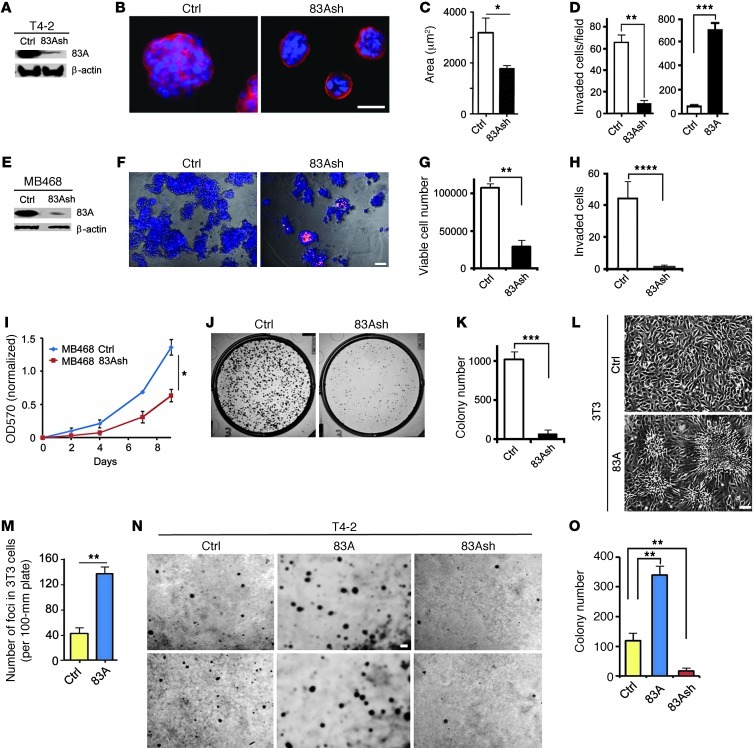
Figure 2. FAM83A levels positively determine cell growth and tumorigenic potential.
(A) Generation of a T4-2 cell line stably depleted of FAM83A by shRNA (83Ash). (B) FAM83A shRNA expression reverted T4-2 cells to an acinar-like phenotype in 3D lrECM cultures. α6 integrin (red) was used to indicate basal polarity; blue, DAPI. (C) Colony size. Luciferase shRNA was used as control. (D) Number of T4-2 cells expressing either FAM83A shRNA or FAM83A-overexpressing construct versus control (vector) invaded through lrECM-coated Transwell filters after 48 hours (n = 4). (E) Generation of an MDA-MB468 cell line stably depleted of FAM83A by shRNA. (F) Control versus FAM83A-depleted MDA-MB468 cells grown in 3D lrECM cultures for 5 days. Cells were stained with ethidium bromide to indicate apoptotic cells (blue, Hoechst 33258). (G) Number of viable cells after growth in 3D lrECM for 5 days (n = 4). (H) Number of cells invaded through lrECM-coated Transwell filters after 24 hours (n = 3). (I) Growth of control versus FAM83A-depleted MDA-MB468 cells, measured by MTT assay for 9 days (n = 12). P < 0.05, 2-tailed ANOVA. (J) Clonogenic potential of control versus FAM83A-depleted MDA-MB468 cells measured by colony formation assay for 10 days. Shown are representative images of colonies formed. (K) Number of colonies formed (n = 6). (L) Tumorigenic foci formed by 3T3 fibroblasts overexpressing FAM83A through loss of contact inhibition of growth. Representative images of 3T3 cells transduced by control and FAM83A-overexpressing lentivirus and cultured for 2 weeks. (M) Quantification of 3T3 foci shown in L (n = 4). (N) Representative images of T4-2 cells in duplicates in 2 rows transduced by empty vector, FAM83A-overexpressing, or FAM83A shRNA lentivirus and cultured on soft agar for 6 weeks. (O) Number of colonies formed on soft agar (n = 6). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, Student’s t test. Scale bars: 50 μm.
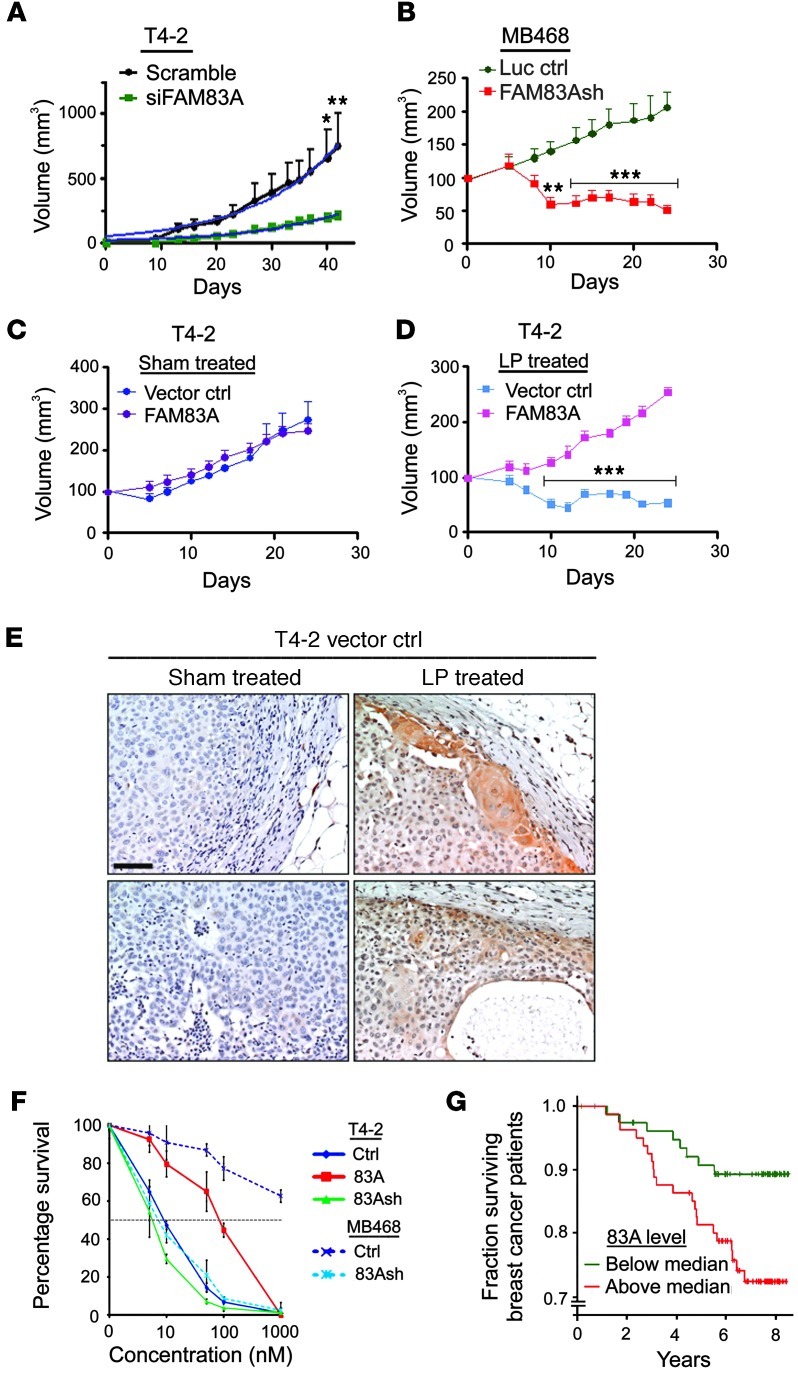
Figure 3. FAM83A levels correlate directly with tumor growth rates and EGFR-TKI resistance in cultures and in vivo, as well as with patients’ prognosis.
(A) Growth of tumors derived from T4-2 cells treated with scrambled or FAM83A siRNA and xenografted in nude mice (n = 8). (B) Growth of tumors derived from MDA-MB468 cells treated with control (luciferase) or FAM83A shRNA and xenografted in nude mice (n = 8). (C and D) Tumor growth of vector control and FAM83A-overexpressing T4-2 cells treated with oral gavage of sham (vehicle; B) or 30 mg/kg lapatinib (LP; C). FAM83A-overexpressing T4-2 cells were resistant to lapatinib treatment (n = 8). (E) FAM83A staining of sham- or lapatinib-treated vector control tumors as in C and D. Note the upregulation of endogenous FAM83A in lapatinib-treated tumors. (F) Survival of vector control, FAM83A-overexpressing, and FAM83A-depleted T4-2 cells and luciferase-sh control or FAM83A-depleted MDA-MB468 cells treated with AG1478 at different concentrations and measured by clonogenic assay after 10 days. Values are expressed relative to untreated. IC50 of AG1478 was as follows: T4-2 Ctrl, 10 nM; T4-2 FAM83A, 80 nM; T4-2 FAM83Ash, 6 nM; MDA-MB468 Ctrl, >1,000 nM; MDA-MB468 FAM83Ash, 7.5 nM. (G) Kaplan-Meier curve for a cohort of 159 breast cancer patient samples. High FAM83A expression correlated with poor prognosis (P < 0.05). *P < 0.05, **P < 0.01, ***P < 0.001, 2-way ANOVA with Bonferroni post-test.
In a classic assay of oncogenic potential, we tested FAM83A-overexpressing 3T3 fibroblasts for contact-independent growth (24, 25). FAM83A overexpression caused a dramatic increase in foci formation (Figure 2, L and M). Growing FAM83A-overexpressing and -depleted T4-2 cells in soft agar yielded 3-fold more colonies than control, whereas FAM83A-depleted cells yielded 5-fold fewer colonies than control (Figure 2, N and O). These observations support the oncogenic potential of FAM83A overexpression for both fibroblasts and breast cancer cells.
To characterize FAM83A function in vivo, we xenografted control or FAM83A siRNA-treated T4-2 cells into mice as described previously (26). Tumor take was not affected (100% for both groups); however, growth of the FAM83A siRNA T4-2 tumors was significantly delayed and also slower (Figure 3A). Similarly, xenografting MDA-MB468 cells (control versus FAM83A silenced via shRNA) revealed that FAM83A depletion resulted in dramatic inhibition in the rate of tumor growth (Figure 3B). Indeed, upon pathological examination, we found no surviving tumor cells derived from FAM83A-depleted cells after 3 weeks (data not shown). Thus, the regression of tumors most likely is due to the apoptotic phenotype observed in culture (Figure 2, E–G).
To demonstrate the ability of FAM83A to confer resistance to clinical EGFR-TKIs, we first tested effects of lapatinib (27) and gefitinib (28) on control and FAM83A-overexpressing T4-2 cells in 3D cultures. Both drugs reverted wild-type cells to a degree comparable to AG1478-induced reversion, whereas FAM83A-overexpressing cells remained resistant to reversion (Figure 1E and Supplemental Figure 6A). T4-2 tumors subcutaneously grown in mice were sensitive to lapatinib treatment, and sensitivity was dose independent above 30 mg/kg (Supplemental Figure 6B). Overexpression of FAM83A in these cells did not alter tumor growth (Figure 3C), but rendered cells resistant to lapatinib in vivo (Figure 3D). Whereas lapatinib can inhibit via both EGFR and HER2 (27), its tumor-inhibitory effect observed here was presumed to have occurred almost exclusively via EGFR. We know from previous work that HER2 is absent or undetectable in T4-2 cells in culture (29–31), although we did not measure whether the HER2 pathway is reactivated in these cells in vivo. Pathological examination of residual T4-2 lapatinib-treated vector control tumors showed them to be benign, well circumscribed, and distinct from the stromal regions. In contrast, lapatinib-treated FAM83A-overexpressing tumors did not shrink, were more aggressive, and showed stromal invasion (Supplemental Figure 6C), which suggests that FAM83A overexpression allows resistance to the antitumor function of lapatinib in vivo. Importantly, IHC staining of sham- and lapatinib-treated T4-2 tumors (Figure 3, C and D) revealed higher FAM83A levels in the latter (Figure 3E), which indicates that there may be some selection or upregulation for the FAM83A-high, lapatinib-resistant cells during treatment in vivo. The IC50 of AG1478 for MDA-MB468 and T4-2 cell cultures (controls and derivatives) correlated directly with their respective FAM83A protein levels (Figure 3F), further demonstrating the role of FAM83A in EGFR-TKI resistance.
Given this correlation of FAM83A levels and tumor growth potential, we asked whether FAM83A expression correlates with clinical survival. Using a published breast cancer gene expression dataset (32), we found that patients with tumors expressing above-median levels of FAM83A exhibited significantly poorer clinical outcome than did patients with lower levels (P = 0.022, log-rank test; Figure 3G). Hierarchical clustering of 159 primary breast cancers for the expression of genes at 8q24 identified 17 samples that strongly expressed genes associated with amplification of locus 8q24 (Supplemental Figure 7 and ref. 33). Association of FAM83A expression with poor outcome was found in the remaining 142 samples with low/normal 8q24 copy number (P = 0.031), which suggests that the linkage is independent of 8q24 copy number. Regardless of whether the elevated FAM83A is the result of gene amplification or its upregulation, these findings are suggestive of the clinical importance and potential therapeutic relevance of FAM83A.
We also examined the literature to determine whether FAM83A overexpression also correlates with EGFR-TKI resistance in a different type of cancer. FAM83A was amplified in a number of subtypes of lung cancer (Supplemental Figure 8A and refs. 16, 34). Lung cancers that were resistant to gefitinib treatment were found to have higher FAM83A expression than the sensitive cancers (Supplemental Figure 8B, Supplemental Table 2, and ref. 35). FAM83A expression levels, however, did not correlate with KRAS and EGFR mutations in lung cancer (Supplemental Figure 8, C and D, and ref. 35). These results suggest an additional role for FAM83A in gefitinib resistance of lung cancer.
We have reported previously that EGFR-TKI–mediated reversion of T4-2 cells suppresses the downstream MAPK pathway (12, 14, 15). Inhibition of PI3K, which is activated by EGFR in a divergent pathway (36), also reverts T4-2 cells (37). To elucidate the mechanism(s) by which FAM83A exerts its effects in these 2 pathways, we tested whether FAM83A-overexpressing cells are resistant to the MEK inhibitor PD98059 or the PI3K inhibitor LY294002, as they are to the EGFR inhibitor AG1478. Importantly, LY294002 was also unable to revert FAM83A-overexpressing T4-2 cells, whereas PD98059 could (Figure 4, A and B), which suggests that FAM83A lies downstream of EGFR/PI3K and upstream of MEK.
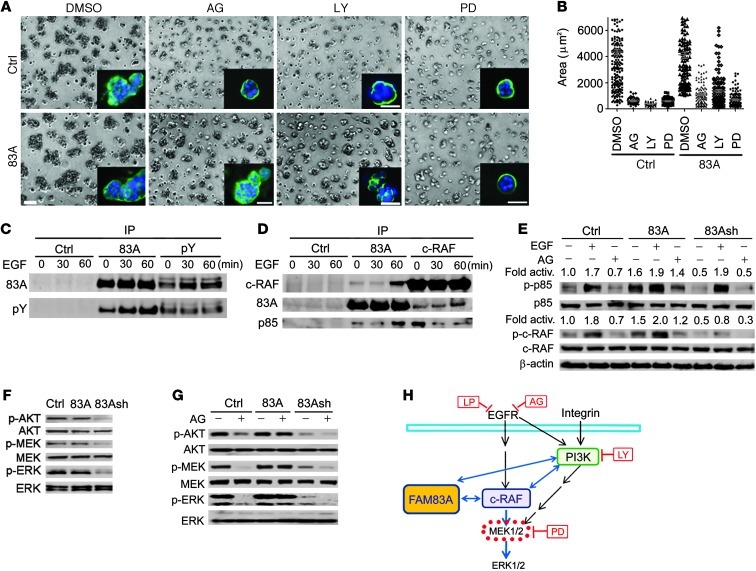
Figure 4. FAM83A interacts with c-RAF and PI3K upstream of MEK1/2 activation.
(A) Control and FAM83A-overexpressing T4-2 cells were tested for response to AG1478 (300 nM), LY294002 (LY; 8 μM), or PD98059 (PD; 20 μM) in 3D lrECM cultures in comparison to vehicle (DMSO) treatment. α6 integrin staining (green) was used to indicate basal polarity; blue, DAPI. Scale bars: 50 μm. (B) Size of colonies formed by control versus FAM83A-overexpressing cells after drug treatment as in A (P < 0.05, Kruskal-Wallis test). (C) Detection of tyrosine-phosphorylated FAM83A after EGF stimulation (0, 30, or 60 minutes) by reciprocal IP with an antibody against FAM83A or phosphotyrosine (pY). Rabbit preimmune serum was used as control. (D) Interaction of endogenous FAM83A with c-RAF and PI3K p85 subunit after EGF stimulation (0, 30, or 60 minutes) by reciprocal co-IP with an antibody against FAM83A or c-RAF. Preimmune serum was used as control. (E) Levels of phosphorylated PI3K p85 subunit (p-Y458) and c-RAF (p-S338) in control, FAM83A-overexpressing, and FAM83Ash-expressing T4-2 cells after treatment with EGF (1 hour) or AG1478 (2 hours). (F) Levels of phosphorylated AKT (p-S473), MEK1/2 (p-S217/221), and ERK1/2 (p-T202/Y204) in control, FAM83A-overexpressing, and FAM83Ash-expressing T4-2 cells after growth in 3D lrECM culture for 5 days. (G) Levels of phosphorylated AKT, MEK1/2, and ERK1/2 in control, FAM83A-overexpressing, and FAM83Ash-expressing T4-2 cells after treatment with AG1478 for 2 hours. (H) FAM83A interacts with c-RAF and PI3K upstream of MEK activation.
To explore the connection between FAM83A and EGFR signaling, we treated T4-2 cells with EGF and monitored the phosphorylation status of endogenous FAM83A. We observed increasing tyrosine phosphorylation of FAM83A as a function of time (Figure 4C). Since EGFR/Ras signaling activates c-RAF and leads to MEK activation (38, 39), and FAM83A-overexpressing cells were resistant to the PI3K inhibitor (Figure 4, A and B), we tested whether EGF treatment induces interaction of FAM83A with c-RAF and PI3K. Co-IP analysis revealed that EGF treatment caused endogenous FAM83A to interact with c-RAF and PI3K p85 subunit on a similar time scale (Figure 4D). c-RAF also interacted with PI3K p85; however, EGF treatment increased the interaction of these proteins with FAM83A, while reducing the interaction of c-RAF with PI3K p85 (Figure 4, D and E). This interaction of FAM83A with c-Raf and PI3K suggests strongly that FAM83A interacts with Ras, because Ras binding to c-Raf and PI3K is essential for its role in mitogenic/oncogenic signal transduction (38, 40, 41) and Ras binding is essential for c-Raf activation (38, 41).
To evaluate the role of FAM83A’s interactions with c-RAF and PI3K p85, we assessed the activation status of c-RAF and PI3K p85 in FAM83-overexpressing and -depleted T4-2 cells in response to treatment with EGF or AG1478. Phosphorylation of c-RAF (p-S338) and PI3K p85 subunit (p-Y458) leads directly to phosphorylation of the downstream proteins, ERK and AKT, respectively (42–44). In FAM83A-overexpressing cells, we found that PI3K p85 and c-RAF were highly phosphorylated even in the absence of EGF or in the presence of AG1478 (Figure 4E), suggestive of EGF/EGFR-independent activation. In agreement, in FAM83A-depleted cells, the basal levels of PI3K p85 and c-RAF phosphorylation were reduced, and c-RAF phosphorylation was inhibited even in the presence of EGF (Figure 4E). These results suggest that FAM83A is essential for c-RAF activation upon EGF stimulation and that FAM83A overexpression is sufficient to activate c-RAF and PI3K p85 in the absence of EGF/EGFR. Importantly, FAM83A-depleted T4-2 cells in 3D cultures exhibited decreased phosphorylation of the downstream AKT, MEK, and ERK, which was further exacerbated by treatment with AG1478 (Figure 4, F and G). A similar phenomenon was also observed in MDA-MB468 cells depleted of FAM83A (Supplemental Figure 9A). These observations suggest that FAM83A knockdown enhances the cells’ sensitivity to AG1478. Conversely, FAM83A-overexpressing T4-2 cells exhibited EGFR-independent activation of these 3 proteins in the presence of AG1478 treatment (Figure 4G). Analogous to AG1478, LY294002 treatment failed to inhibit phosphorylation of AKT, MEK, and ERK in FAM83A-overexpressing T4-2 cells, whereas it severely inhibited the 3 proteins in FAM83A-depleted cells (Supplemental Figure 9B), further suggesting that FAM83A lies downstream of EGFR/PI3K. These results suggest that FAM83A association with c-RAF and PI3K is activated upon EGFR signaling, leading to activation of the downstream MEK/ERK pathway (Figure 4H). Such a biochemical function of FAM83A appears to be the basis for its oncogenic role and its resistance to AG1478.
Discussion
Amplification and/or overexpression of EGFR is observed in many cancers, including 30% of breast cancers (1–5). In lung cancer, activating mutations in the kinase domain (exons 18–21) are predictive of a response to specific therapies, such as those using the EGFR antibody cetuximab and EGFR-TKI’s lapatinib, erlotinib, or gefitinib, but amplification and overexpression assays are not predictive (45). In breast cancer, EGFR mutations are rare, and when they are described, the mutation rate varies among different datasets (6). Kinase domain mutations similar to those found in lung cancer have been reported in certain breast cancer cohorts. However, whereas the predictive utility is well documented in lung cancer (46), this is not true for breast cancer (47), which points to the need for further exploration of alternative mechanisms for EGFR-TKI resistance. Despite the fact that EGFR-TKI leads to inhibition of EGFR phosphorylation in cell culture studies, clinical response rates are somewhat disappointing and activation of downstream pathways has been suspected (48, 49). This activation could explain the lack of relative efficacy of EGFR-TKIs in breast cancer patients (48). In some EGFR-TKI–resistant breast cancers, Met and c-Src tyrosine kinases are overexpressed, hyperactivating EGFR even in the presence of the inhibitor (5). Furthermore, in EGFR-TKI–resistant breast cancer cell lines, EGFR is localized at lipid rafts even in the presence of the drug, leading to hyperactivation of the downstream Akt signaling (50). In the present study, we examined whether there were additional molecular modulations that confer EGFR-TKI resistance in breast cancer. Such molecular mechanisms might provide a basis for improved predictive diagnostics and could also provide novel drug targets for independent activity or combinatorial therapy.
Here, we used our 3D lrECM culture system (11) to screen for genes involved in EGFR-TKI resistance. In this model, inhibition of the EGFR pathway with EGFR-TKI reverts most of the malignant T4-2 cells to a nonmalignant phenotype, which indicates that these cells are EGFR-TKI sensitive (12). We often observe that a very small population of treated cells does not revert, i.e., these cells appear to be EGFR-TKI resistant. We reasoned that this intrinsic mechanism could be exploited and used as the basis for a screen to find additional targets involved in resistance. We transduced T4-2 cells with an autologous cDNA library and identified a number of molecules, including FAM83A, as possible targets of EGFR-TKI resistance (Supplemental Table 1). We chose FAM83A for further characterization as a gene related to the EGFR pathway that could mediate breast cancer aggressiveness because it showed the best resistance to EGFR-TKI. Downmodulation of FAM83A in breast cancer cells using RNAi led to decreased proliferation and invasiveness in cell culture as well as to decreased tumor growth in vivo. Conversely, overexpression of FAM83A conferred resistance to EGFR-TKIs both in culture and in vivo.
We tested other breast cancer cell lines known to be resistant to EGFR-TKI (20, 21), such as MDA-MB468, and determined that these cells also exhibited high levels of FAM83A (Figure 1D). Downmodulation of FAM83A in these cells decreased proliferation and, importantly, also rendered them sensitive to EGFR-TKIs (Figure 2I and Figure 3F). These data are indicative of the potential clinical applicability of our findings. Indeed, breast cancer patients exhibiting high levels of FAM83A expression had significantly lower survival than did patients with low levels of FAM83A (Figure 3G). Therefore, targeting FAM83A may be of benefit to breast cancer patients who are resistant to EGFR-TKI; additionally, it may also increase EGFR-TKI efficacy (Figure 3F).
Mechanistically, we showed that FAM83A interacted with c-RAF and PI3K, leading to activation of the protein complex. Cipriano et al. have identified FAM83B, another member of the FAM83 family, using a completely different screen to identify genes that could replace the RAS oncogene for anchorage-independent growth of mammary epithelial cells in soft agar (51). They report complementary findings: FAM83B also operates upstream of MEK to activate MAPK signaling, interacts with c-RAF, and is upregulated in breast cancers, and its overexpression impairs AG1478 sensitivity (51). Their and our studies have identified a family of breast cancer–associated genes or a possible family of oncogenes and support the contention that FAM83A and FAM83B (and possibly other family members; see Supplemental Figure 1D and Supplemental Figure 10) are involved in therapeutic resistance in breast cancer and other cancer types. Our findings suggest the importance of FAM83 family members as potential drug targets for therapy as well as for sensitization to EGFR-TKIs. We are currently examining the possibility that FAM83A is localized to lipid rafts when interacting with c-RAF and PI3K, which are also associated with lipid rafts during activation and signal transduction (50, 52, 53).
Methods
Cell culture.
The isogenic cell lines of HMT3522 human breast cancer progression series, nonmalignant S1 cells and malignant T4-2 cells, were maintained as described previously (9). This cell line series was established in an attempt to recapitulate the stochastic and prolonged nature of breast cancer progression by continuously culturing S1 cells derived from reduction mammoplasty, in the absence of serum, followed by EGF removal and injection into mice, to give rise to T4-2 cells (9, 10, 29). For 3D culture experiments, S1 and T4-2 cells were seeded at densities of 2.5 × 104 and 1.8 × 104 cells/cm2, respectively, in growth factor reduced Matrigel (BD Biosciences) and maintained for 4 days with addition of fresh medium on alternate days. For inhibitor studies, cells were treated with 350 nM AG1478 (Calbiochem Corp.), 20 μM PD98059 (Biomol), 8 μM LY294002 (Biomol), or the appropriate vehicle controls.
cDNA library construction.
The retroviral pESY-Neo vector used for cDNA library construction was modified from the pEYK 3.1 vector (gift of G. Daley, Whitehead Institute for Biomedical Research, Cambridge, Massachusetts, USA; ref. 54) by inserting the G418 resistance gene (Neo) into a multiple-cloning site to allow for directional cloning of cDNA inserts. cDNA was synthesized from mRNA isolated from T4-2 cells in 2D cultures following the guidelines of the SuperScript Plasmid System with Gateway technology for cDNA Synthesis and Cloning manual (Invitrogen). The first-strand cDNA was synthesized using Oligo dT18-Not I (underlined) unidirectional primer 5′-AACCCGGCTCGAGCGGCCGC(T)18-3′ (Invitrogen) for constructing directional libraries. cDNA was blunted enzymatically and ligated to BamHI adapters. cDNA was digested with BamHI/NotI restriction endonucleases, then size fractionated on a 0.7% agarose gel. cDNAs 0.5–5 kb in size were extracted and ligated into BamHI/NotI–digested pESY–Neo. The complexity of the library (5 × 104 independent clones) was estimated by counting the number of transformed E. coli colonies on agar plates. The plates were gently scraped, and plasmid DNA was isolated using Qiagen plasmid Mega prep Kit (Qiagen).
Retrovirus production.
Retrovirus production and transduction of target cells were performed according to the Nolan lab protocol ( http://www.stanford.edu/group/nolan/protocols/pro_helper_dep.html). Briefly, Phoenix-amphotropic retrovirus packaging cells (gift of G.P. Nolan, Stanford University, Stanford, California, USA) were plated 24 hours prior to transfection and transfected with retroviral plasmid using Lipofectamine 2000 (Invitrogen). After 48 hours, medium was harvested and filtered through a 0.45-μm syringe filter. The titer of collected retrovirus was determined using NIH3T3 cells at a density of 2 × 105 cells per 60 mm plate with serial dilution of virus supernatant.
FAM83A expression constructs.
The full-length human FAM83A cDNA clone was obtained from Open Biosystems (Clone ID: 6421272). The coding region was PCR-amplified using the following primers: forward, 5′-CCCAGGATCCGCCACCATGAGCCGGTCAAGGCACCTGGGCAAA-3′ (BamH1 site underlined); reverse, 5′-CTTTGGAATTCAGGGAGTGCCCAGCCCTGGGCCTGCGGAGG-3′ (EcoR1 site underlined). The PCR product was ligated into the BamH1/EcoR1 site of PCDF1-MCS2-EF1-puro lentiviral vector (System Biosciences) or pcDNA3.1+ vector (Invitrogen).
FAM83A polyclonal antibody generation.
Abie Pro3.0 Peptide Antibody Design program (ChangBioscience) was used to predict peptide sequences that have a high probability of antigenic epitope. The FAM83A antibody (amino acid sequence N-DSHLKNISIRSVEG-C) was raised by and obtained from Biosynthesis Inc.
FAM83A knockdown by RNA interference.
Silencer Pre-Designed siRNA oligos for human FAM83A (nos. 215708, 149024, and 149025) were purchased from Ambion. siRNA was transfected into 4 × 105 cells per 35-mm dish to a final concentration of 100 nM using Lipofectamine 2000. Cells were then grown in 3D lrECM culture for 4 days. A Silencer siRNA for a fluorescent Cy3 dye (Ambion) was used as negative control. The target sequences of the siRNAs were as follows: 5′-GCCCUACCUGAAGGAAAAA-3′, 5′-GGAGAGAUAUACUGUGCCA-3′, and 5′-GGAAAUUCGCUGGCCAAAU-3′. For FAM83A shRNA production, a double-stranded DNA oligonucleotide was generated from the following sequences: sense, 5′-GATCCGTGTGGAAGGAGAGATATACTTCCTGTCAGATATATCTCTCCTTCCACACTTTTTG-3′; antisense, 5′-AATTCAAAAAGTGTGGAAGGAGAGATATATCTGACAGGAAGTATATCTCTCCTTCCACACG-3′ (target sequence underlined; BamH1/EcoR1 cohesive ends italicized). Both oligonucleotides were annealed and ligated into BamH1/EcoR1 site of pGreen puro lentiviral vector (System Biosciences).
Lentivirus production and transduction.
Lentivirus production and transduction of target cells were conducted according to the manufacturer’s instructions (System Biosciences). Briefly, lentivirus vector and packaging plasmid mix (System Biosciences) were transfected into 293FT cells (Invitrogen) using Lipofectamine 2000. After 48 hours, medium was harvested, filtered, and used to infect target cells with the addition of 10 μg/ml polybrene. After 24 hours, medium was replaced. At 72 hours after infection, 0.5 μg/ml puromycin was added for selection and maintained throughout the culturing period.
Immunoprecipitation.
Cells were lysed in Triton lysis buffer (25 mM Tris, pH 7.6; 150 mM NaCl; 1% Triton X-100; 1 mM EDTA; 1% glycerol) freshly supplemented with 1 mM PMSF, phosphatase inhibitors (1 mM sodium orthovanadate, 30 mM sodium fluoride, 2 mM sodium pyrophosphate, and 10 mM β-glycerophosphate) and protease inhibitor cocktail (0.5 mM Pefa-bloc, 150 nM aprotinin, 1 μM E-64 protease inhibitor, and 1 μM leupeptin; Calbiochem) on ice for 30 minutes. Lysates were centrifuged at 4°C for 10 minutes at 10,000 g, and the protein concentration was normalized. 1 mg protein (1 mg/ml) was precleared with 50 μl protein A/G (1:1) agarose beads (Roche) at 4°C for 1 hour, then incubated with 2 μg primary antibody at 4°C overnight and subsequently with 50 μl protein A/G agarose beads for 2 hours at 4°C. Beads were washed 3 times with lysis buffer. The immunoprecipitates were boiled in 100 μl SDS sample buffer for 10 minutes, and half the sample volume was analyzed by immunoblotting.
Immunofluorescence staining.
Immunofluorescence was performed as described previously (8). Samples were incubated with primary antibody for 2 hours at room temperature in a humidified chamber. After intensive washing (3 times for 15 minutes each) in 0.1% BSA, 0.2% Triton-X 100, 0.05% Tween 20, and 0.05% NaN3 in PBS, fluorescence-conjugated secondary antibodies (Molecular Probes) were added for 1 hour at room temperature. Nuclei were stained with 0.5 ng/ml DAPI.
Immunohistochemistry.
Breast cancer tissue arrays containing paraffin-embedded sections of normal and malignant (stages II and III) tissues were obtained from US Biomax Inc. Slides were deparaffinized, hydrated, and treated with antigen unmasking solutions (Vector Laboratories Inc.) After being blocked with 0.3% H2O2 and nonimmune goat serum, sections were incubated at room temperature with a rabbit anti-FAM83A antibody and link antibodies, followed by peroxidase-conjugated streptavidin complex and diaminobenzidine tetrahydroxychloride solution as the peroxidase substrate (Vector Laboratories). Sections were counterstained with hematoxylin. Photomicrographs were taken with Zeiss Axioskop Imaging platform and SPOT Basic software (version 4.7).
Cell proliferation assay.
Cells were plated at a density of 1 × 103 cells per well in 96-well plates in DMEM plus 10% FBS and incubated at 37°C. 24 hours before each time point, the medium was replaced. At each time point, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT; Sigma-Aldrich) was added to cells to a final concentration of 1.6 mg/ml, and the reaction was incubated at 37°C for 4 hours. Then, the medium was removed, and the precipitated reaction product was dissolved in MTT solvent (4 mM HCl, 0.1% NP-40 in isopropanol). Absorbance was measured at 570 nm.
Clonogenic assay.
Cells were plated at a density of 1 × 103 cells per well in 6-well plates in DMEM plus 10% FBS and incubated at 37°C for 10 days. Cells were stained with 0.2% methylene blue in 50% ethanol and destained with tap water. Each well was photographed, and the number of colonies was counted.
Invasion assays.
Invasion assays were performed as described previously (55). 1 × 105 cells were placed on top of a thin Matrigel layer and cultured for 48 hours. They were then fixed with 5% glutaraldehyde and stained with 0.5% toluidine blue solution. Samples were prepared in triplicate, and cells were counted on at least 3 different fields on the Transwell filters.
Soft agar assay.
1% agar was mixed with the equivalent volume of 2× DMEM/F12 medium supplemented with all the additives necessary for culturing T4-2 cells (9) plus 20% FBS and 2% penicillin/streptomycin. 1 ml agar solution was poured into a 35-mm plate in triplicate and solidified. 0.7% agar solution equilibrated to 40°C was mixed with 2× growth medium and breast cancer cells at 7,000 cells/ml and poured onto the base agar at 1 ml/plate. The solidified agar was covered with 500 μl growth medium and maintained in 37°C humidified incubator for 2 weeks. Plates were stained with 0.01% crystal violet for 30 minutes, and colonies were counted under dissecting microscope.
PLD activity analysis.
As a positive control, PLD1 expression vector was generated by PCR-amplifying the coding region of the full-length human PLD1 cDNA (clone ID 606838; Invitrogen) with the following primers: forward, 5′-GACATGCTAGCGCCGCCACCATGTCACTGAAAAACTAGCCACGGGTAAATACCTCTGCAC-3′ (Nhe1 site underlined); reverse, 5′-GACATGTTTAAACGGAAGTCCAAACCTCCATGGGCACTATGGCCTCTTTGGTCCCA-3′ (Pme1 site underlined). The PCR product was ligated into the Nhe1/Pme1 site of pcDNA3.1+ vector (Invitrogen). PLD1 and FAM83A proteins were generated by incubating PLD1/pcDNA3.1+ and FAM83A/pcDNA3.1+ plasmids, respectively, with rabbit reticulocyte lysate using in vitro transcription/translation system (Promega) at 30°C for 90 minutes. Protein products were confirmed by Western blot. PLD activity was measured as described previously (56). Briefly, BODIPY-phosphatidylcholine (BPC; Molecular Probes) was dissolved in ethanol to a final concentration of 1 mM. An aliquot (1.3 μl) of BPC was dried under nitrogen and resuspended in resuspension buffer (0.5 mM octylglucoside; 400 mM NaCl; and 66 mM Mes, pH 6.0) and mixed with 1.6 μl of 10× reaction buffer (100 mM HEPES, pH 7.5; 40 mM β-glycerophosphate, pH 7.5; 1% n-butanol, pH 7.5; 1 mM dithiothreitol; 10 mM EGTA; and 10 mM EDTA). A designated volume of PLD1 (1 U/μl; Sigma-Aldrich) as the standard or in vitro translation product — prepared from an empty vector (control) or from PLD1 or FAM83A plasmid — was diluted in water to 10 μl, mixed with reaction mixture, and incubated at 30°C for 1.5 hours. A 5-μl aliquot of the reaction mixture was spotted on silica gel G60 plate (Merck) and separated by development in methanol/chloroform/water (45:45:10). The plate was photographed under UV light.
Tumor inhibition assay in nude mice.
Exponentially growing T4-2 and MDA-MB-468 cells were suspended at a density of 5 × 106 and 10 × 106 cells, respectively, in 100 μl medium containing 50% DMEM/F12 medium and 50% Matrigel. Cell suspension was subcutaneously injected into the rear flank of 6- to 8-week-old athymic female BALB/c nude mice (nu/nu; n = 8 per group; Charles River Laboratories; ref. 26). For lapatinib treatment, tumors derived from T4-2 cells were grown to 100 mm3 (day 5). Then, mice were randomized into vehicle (0.5% hydroxypropylmethylcellulose with 0.1% Tween 80 in water) and experimental treatment groups (n = 8 per group) to receive oral gavage of lapatinib at 30 mg/kg or 100 mg/kg twice daily for 3 weeks (57). Tumor volumes were measured every other day. At the end of experiments, mice were sacrificed and subjected to pathological examinations.
Genomic and survival analysis.
An association between FAM83A expression and breast cancer survival was evaluated using a previously published gene expression microarray dataset from 159 primary breast cancer patients with longitudinal outcome data (58). Using the NETAFFX database (59), we identified 4 probe sets designed to measure the expression of FAM83A on the U133 arrays used in this study. The CEL files from this dataset were downloaded from GEO (accession no. GSE1456) and processed using robust Multi-array Average (RMA) method in Bioconductor (60) to derive gene expression estimates for all probe sets on the array. We derived a single measure of FAM83A expression for each sample by averaging the RMA values for the 4 probe sets. Patient samples were then dichotomized by whether they had above- or below-median expression of FAM83A, and the difference in breast cancer survival between these 2 classes was assessed using the log-rank statistic. We identified probe sets designed to measure the expression of genes in the 8q24 amplicon (DERL1, WDR67, FAM83A, C8orf76, ZHX1, ATAD2, WDYHV1, FBXO32, ANXA13, FAM91A1, TMEM65, TRMT12, RNF139, TATDN1, NDUFB9, MTSS1, LOC157381, SQLE, KIAA0196, NSMCE2, TRIB1, FAM84B, MYC, PVT1) using the NETAFFX database and used their expression values to organize the samples via hierarchical clustering. A group of samples with relatively low expression of 8q24 genes was identified using the dendrogram from this clustering to divide the samples into 3 groups. These analyses were performed using the base and survival packages in version 2.9.1 of the R Language for Statistical Computing ( http://www.R-project.org).
Statistics.
Unless otherwise indicated, statistical analyses were performed using GraphPad Prism version 5 software and unpaired 2-tailed Student’s t test. For the tumor inhibition assay, 2-way ANOVA with Bonferroni post-test was used. A P value less than 0.05 was considered significant. Average results of multiple experiments are presented as arithmetic means ± SEM.
Study approval.
Animal housing, care, and experiments were performed under federal guidelines (Guide for the Care and Use of Laboratory Animals. NIH publication no. 85-23. Revised 1985) and approved by Institutional Animal Welfare and Research Committee at Lawrence Berkeley National Laboratory. Studies using human cell lines and commercial tissue arrays were exempted, but also reviewed and approved by Human Subjects Committee at Lawrence Berkeley National Laboratory (approval no. 273H01-05MR10).
Supplementary Material
Acknowledgments
We thank Eva Lee for considerable help with tissue culture and technical support, Aileen Lee for IHC, and Alvin Lo for invasion assay. We are grateful to Irene Kuhn for valuable discussion and critical reading of the manuscript and Alexander Borowski for help with pathological evaluation. We also thank all other members of the Bissell laboratory for comments and discussions. The work from M.J. Bissell’s laboratory was supported by grants from the US Department of Energy, Office of Biological and Environmental Research and Low Dose Radiation Program (contract no. DE-AC02-05CH1123); by the National Cancer Institute (awards R37CA064786, U54CA126552, U01CA143233, U54CA112970, and U54CA143836 — Bay Area Physical Sciences–Oncology Center, University of California, Berkeley, California, USA); and by a US Department of Defense Innovator Award (W81XWH0810736). R. Meier was supported by the Swiss National Science Foundation (PBZHB-121019 and PASMP3-127673/1) and by the Claudia von Schilling Foundation for Breast Cancer Research.
Footnotes
Conflict of interest: Roland Meier is an employee of and holds restricted stock in Genentech.
Citation for this article: J Clin Invest. 2012;122(9):3211–3220. doi:10.1172/JCI60498.
See the related Commentary beginning on page 3048.
References
- 1.Rimawi MF, et al. Epidermal growth factor receptor expression in breast cancer association with biologic phenotype and clinical outcomes. Cancer. 2010;116(5):1234–1242. doi: 10.1002/cncr.24816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358(11):1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 3.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 4.Paez JG, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 5.Mueller KL, Hunter LA, Ethier SP, Boerner JL. Met and c-Src cooperate to compensate for loss of epidermal growth factor receptor kinase activity in breast cancer cells. Cancer Res. 2008;68(9):3314–3322. doi: 10.1158/0008-5472.CAN-08-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhargava R, et al. EGFR gene amplification in breast cancer: correlation with epidermal growth factor receptor mRNA and protein expression and HER-2 status and absence of EGFR-activating mutations. Mod Pathol. 2005;123(8):889–895. doi: 10.1038/modpathol.3800438. [DOI] [PubMed] [Google Scholar]
- 7.Ferrer-Soler L, Vazquez-Martin A, Brunet J, Menendez JA, De Llorens R, Colomer R. An update of the mechanisms of resistance to EGFR-tyrosine kinase inhibitors in breast cancer: Gefitinib (Iressa) -induced changes in the expression and nucleo-cytoplasmic trafficking of HER-ligands. Int J Mol Med. 2007;20(1):3–10. [PubMed] [Google Scholar]
- 8.Weaver VM, et al. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137(1):231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Briand P, Nielsen KV, Madsen MW, Petersen OW. Trisomy 7p and malignant transformation of human breast epithelial cells following epidermal growth factor withdrawal. Cancer Res. 1996;56(9):2039–2044. [PubMed] [Google Scholar]
- 10.Briand P, Petersen OW, Van Deurs B. A new diploid nontumorigenic human breast epithelial cell line isolated and propagated in chemically defined medium. In Vitro Cell Dev Biol. 1987;23(3):181–188. doi: 10.1007/BF02623578. [DOI] [PubMed] [Google Scholar]
- 11.Petersen OW, Ronnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci U S A. 1992;89(19):9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang F, et al. Reciprocal interactions between beta1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: a different perspective in epithelial biology. Proc Natl Acad Sci U S A. 1998;95(25):14821–14826. doi: 10.1073/pnas.95.25.14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Itoh M, Nelson CM, Myers CA, Bissell MJ. Rap1 integrates tissue polarity, lumen formation, and tumorigenic potential in human breast epithelial cells. Cancer Res. 2007;67(10):4759–4766. doi: 10.1158/0008-5472.CAN-06-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu H, Radisky DC, Wang F, Bissell MJ. Polarity and proliferation are controlled by distinct signaling pathways downstream of PI3-kinase in breast epithelial tumor cells. J Cell Biol. 2004;164(4):603–612. doi: 10.1083/jcb.200306090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bissell MJ, Kenny PA, Radisky DC. Microenvironmental regulators of tissue structure and function also regulate tumor induction and progression: the role of extracellular matrix and its degrading enzymes. Cold Spring Harb Symp Quant Biol. 2005;70:343–356. doi: 10.1101/sqb.2005.70.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, et al. BJ-TSA-9, a novel human tumor-specific gene, has potential as a biomarker of lung cancer. Neoplasia. 2005;7(12):1073–1080. doi: 10.1593/neo.05406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferraro E, Peluso D, Via A, Ausiello G, Helmer-Citterich M. SH3-Hunter: discovery of SH3 domain interaction sites in proteins. Nucleic Acids Res. 2007;35(web server issue):W451–W454. doi: 10.1093/nar/gkm296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayer BJ. SH3 domains: complexity in moderation. J Cell Sci. 2001;114(pt 7):1253–1263. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- 19.Hawthorn L, Luce J, Stein L, Rothschild J. Integration of transcript expression, copy number and LOH analysis of infiltrating ductal carcinoma of the breast. BMC cancer. 2010;10:460. doi: 10.1186/1471-2407-10-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hegde PS, et al. Delineation of molecular mechanisms of sensitivity to lapatinib in breast cancer cell lines using global gene expression profiles. Mol Cancer Ther. 2007;6(5):1629–1640. doi: 10.1158/1535-7163.MCT-05-0399. [DOI] [PubMed] [Google Scholar]
- 21.Konecny GE, et al. Activity of the dual kinase inhibitor lapatinib (GW572016) against HER-2-overexpressing and trastuzumab-treated breast cancer cells. Cancer Res. 2006;66(3):1630–1639. doi: 10.1158/0008-5472.CAN-05-1182. [DOI] [PubMed] [Google Scholar]
- 22.Banasiak D, Barnetson AR, Odell RA, Mameghan H, Russell PJ. Comparison between the clonogenic, MTT, and SRB assays for determining radiosensitivity in a panel of human bladder cancer cell lines and a ureteral cell line. Radiat Oncol Investig. 1999;7(2):77–85. doi: 10.1002/(SICI)1520-6823(1999)7:2<77::AID-ROI3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 23.Carmichael J, DeGraff WG, Gazdar AF, Minna JD, Mitchell JB. Evaluation of a tetrazolium-based semiautomated colorimetric assay: assessment of chemosensitivity testing. Cancer Res. 1987;47(4):936–942. [PubMed] [Google Scholar]
- 24.Kakunaga T. A quantitative system for assay of malignant transformation by chemical carcinogens using a clone derived from BALB-3T3. Int J Cancer. 1973;12(2):463–473. doi: 10.1002/ijc.2910120217. [DOI] [PubMed] [Google Scholar]
- 25.Shih C, Padhy LC, Murray M, Weinberg RA. Transforming genes of carcinomas and neuroblastomas introduced into mouse fibroblasts. Nature. 1981;290(5803):261–264. doi: 10.1038/290261a0. [DOI] [PubMed] [Google Scholar]
- 26.Rizki A, Mott JD, Bissell MJ. Polo-like kinase 1 is involved in invasion through extracellular matrix. Cancer Res. 2007;67(23):11106–11110. doi: 10.1158/0008-5472.CAN-07-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petrelli F, Cabiddu M, Cazzaniga M, Cremonesi M, Barni S. Targeted therapies for the treatment of breast cancer in the post-trastuzumab era. Oncologist. 2008;13(4):373–381. doi: 10.1634/theoncologist.2007-0173. [DOI] [PubMed] [Google Scholar]
- 28.Muhsin M, Graham J, Kirkpatrick P. Gefitinib. Nat Rev Drug Discov. 2003;2(7):515–516. doi: 10.1038/nrd1136. [DOI] [PubMed] [Google Scholar]
- 29.Rizki A, et al. A human breast cell model of preinvasive to invasive transition. Cancer Res. 2008;68(5):1378–1387. doi: 10.1158/0008-5472.CAN-07-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kenny PA, et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol. 2007;1(1):84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giricz O, Calvo V, Pero SC, Krag DN, Sparano JA, Kenny PA. GRB7 is required for triple-negative breast cancer cell invasion and survival. Breast Cancer Res Treat. 2012;133(2):607–615. doi: 10.1007/s10549-011-1822-6. [DOI] [PubMed] [Google Scholar]
- 32.Pawitan Y, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7(6):R953–R964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haverty PM, et al. High-resolution genomic and expression analyses of copy number alterations in breast tumors. Genes Chromosomes Cancer. 2008;47(6):530–542. doi: 10.1002/gcc.20558. [DOI] [PubMed] [Google Scholar]
- 34.Micke P, et al. Gene copy number aberrations are associated with survival in histologic subgroups of non-small cell lung cancer. J Thorac Oncol. 2011;6(11):1833–1840. doi: 10.1097/JTO.0b013e3182295917. [DOI] [PubMed] [Google Scholar]
- 35.Coldren CD, et al. Baseline gene expression predicts sensitivity to gefitinib in non-small cell lung cancer cell lines. Mol Cancer Res. 2006;4(8):521–528. doi: 10.1158/1541-7786.MCR-06-0095. [DOI] [PubMed] [Google Scholar]
- 36.Bjorge J, Chan T, Antczak M, Kung H, Fujita D. Activated type I phosphatidylinositol kinase is associated with the epidermal growth factor (EGF) receptor following EGF stimulation. Proc Natl Acad Sci U S A. 1990;87(10):3816–3820. doi: 10.1073/pnas.87.10.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang F, et al. Phenotypic reversion or death of cancer cells by altering signaling pathways in three-dimensional contexts. J Natl Cancer Inst. 2002;94(19):1494–1503. doi: 10.1093/jnci/94.19.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolch W, Heidecker G, Lloyd P, Rapp UR. Raf-1 protein kinase is required for growth of induced NIH/3T3 cells. Nature. 1991;349(6308):426–428. doi: 10.1038/349426a0. [DOI] [PubMed] [Google Scholar]
- 39.Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nat Rev Mol Cell Biol. 2004;5(11):875–885. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- 40.Castellano E, Downward J. RAS interaction with PI3K: more than just another effector pathway. Genes Cancer. 2011;2(3):261–274. doi: 10.1177/1947601911408079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moodie SA, Wolfman A. The 3Rs of life: Ras, Raf and growth regulation. Trends Genet. 1994;10(2):44–48. doi: 10.1016/0168-9525(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 42.Thamilselvan V, Craig DH, Basson MD. FAK association with multiple signal proteins mediates pressure-induced colon cancer cell adhesion via a Src-dependent PI3K/Akt pathway. FASEB J. 2007;21(8):1730–1741. doi: 10.1096/fj.06-6545com. [DOI] [PubMed] [Google Scholar]
- 43.Xie D, et al. DAB2IP coordinates both PI3K-Akt and ASK1 pathways for cell survival and apoptosis. Proc Natl Acad Sci U S A. 2009;106(47):19878–19883. doi: 10.1073/pnas.0908458106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng CF, Guan KL. Properties of MEKs, the kinases that phosphorylate and activate the extracellular signal-regulated kinases. J Biol Chem. 1993;268(32):23933–23939. [PubMed] [Google Scholar]
- 45.Sholl LM, et al. EGFR mutation is a better predictor of response to tyrosine kinase inhibitors in non-small cell lung carcinoma than FISH, CISH, and immunohistochemistry. Am J Clin Pathol. 2010;133(6):922–934. doi: 10.1309/AJCPST1CTHZS3PSZ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsao MS, et al. Erlotinib in lung cancer - molecular and clinical predictors of outcome. N Engl J Med. 2005;353(2):133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 47.Generali D, et al. EGFR mutations in exons 18-21 in sporadic breast cancer. Ann Oncol. 2007;18(1):203–205. doi: 10.1093/annonc/mdl322. [DOI] [PubMed] [Google Scholar]
- 48.Baselga J, et al. Phase II and tumor pharmacodynamic study of gefitinib in patients with advanced breast cancer. J Clin Oncol. 2005;23(23):5323–5333. doi: 10.1200/JCO.2005.08.326. [DOI] [PubMed] [Google Scholar]
- 49.Green MD, et al. Gefitinib treatment in hormone-resistant and hormone receptor-negative advanced breast cancer. Ann Oncol. 2009;20(11):1813–1817. doi: 10.1093/annonc/mdp202. [DOI] [PubMed] [Google Scholar]
- 50.Irwin ME, Mueller KL, Bohin N, Ge Y, Boerner JL. Lipid raft localization of EGFR alters the response of cancer cells to the EGFR tyrosine kinase inhibitor gefitinib. J Cell Physiol. 2011;226(9):2316–2328. doi: 10.1002/jcp.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cipriano R, et al. FAM83B mediates EGFR- and RAS-driven oncogenic transformation. J Clin Invest. 2012;122(9):3197–3210. doi: 10.1172/JCI60517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arcaro A, et al. Critical role for lipid raft-associated Src kinases in activation of PI3K-Akt signalling. Cell Signal. 2007;19(5):1081–1092. doi: 10.1016/j.cellsig.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 53.Hekman M, et al. Associations of B- and C-Raf with cholesterol, phosphatidylserine, and lipid second messengers: preferential binding of Raf to artificial lipid rafts. J Biol Chem. 2002;277(27):24090–24102. doi: 10.1074/jbc.M200576200. [DOI] [PubMed] [Google Scholar]
- 54.Koh EY, Chen T, Daley GQ. Novel retroviral vectors to facilitate expression screens in mammalian cells. Nucleic Acids Res. 2002;30(24):e142. doi: 10.1093/nar/gnf142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lochter A, Srebrow A, Sympson CJ, Terracio N, Werb Z, Bissell MJ. Misregulation of stromelysin-1 expression in mouse mammary tumor cells accompanies acquisition of stromelysin-1-dependent invasive properties. J Biol Chem. 1997;272(8):5007–5015. doi: 10.1074/jbc.272.8.5007. [DOI] [PubMed] [Google Scholar]
- 56.Ella KM, Meier GP, Bradshaw CD, Huffman KM, Spivey EC, Meier KE. A fluorescent assay for agonist-activated phospholipase D in mammalian cell extracts. Anal Biochem. 1994;218(1):136–142. doi: 10.1006/abio.1994.1152. [DOI] [PubMed] [Google Scholar]
- 57.Gril B, et al. Effect of lapatinib on the outgrowth of metastatic breast cancer cells to the brain. J Natl Cancer Inst. 2008;100(15):1092–1103. doi: 10.1093/jnci/djn216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miller LD, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102(38):13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu G, et al. NetAffx: Affymetrix probesets and annotations. Nucleic Acids Res. 2003;31(1):82–86. doi: 10.1093/nar/gkg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.