Abstract
Metastatic cancer is extremely difficult to treat, and the presence of metastases greatly reduces a cancer patient’s likelihood of long-term survival. The ZEB1 transcriptional repressor promotes metastasis through downregulation of microRNAs (miRs) that are strong inducers of epithelial differentiation and inhibitors of stem cell factors. Given that each miR can target multiple genes with diverse functions, we posited that the prometastatic network controlled by ZEB1 extends beyond these processes. We tested this hypothesis using a mouse model of human lung adenocarcinoma metastasis driven by ZEB1, human lung carcinoma cells, and human breast carcinoma cells. Transcriptional profiling studies revealed that ZEB1 controls the expression of numerous oncogenic and tumor-suppressive miRs, including miR-34a. Ectopic expression of miR-34a decreased tumor cell invasion and metastasis, inhibited the formation of promigratory cytoskeletal structures, suppressed activation of the RHO GTPase family, and regulated a gene expression signature enriched in cytoskeletal functions and predictive of outcome in human lung adenocarcinomas. We identified several miR-34a target genes, including Arhgap1, which encodes a RHO GTPase activating protein that was required for tumor cell invasion. These findings demonstrate that ZEB1 drives prometastatic actin cytoskeletal remodeling by downregulating miR-34a expression and provide a compelling rationale to develop miR-34a as a therapeutic agent in lung cancer patients.
Introduction
Metastasis currently represents a tipping point in a cancer patient’s likelihood of achieving long-term survival. Metastatic deposits cannot be eradicated with any standard treatment options and are the most common cause of death from epithelial malignancies (1). Understanding the basic biological processes that govern metastasis represents a critical barrier to attaining long-term survival for patients afflicted with epithelial cancer.
In one working hypothesis, metastasis is initiated by a population of tumor cells that undergo epithelial-to-mesenchymal transition (EMT) in response to extracellular cues, leading to loss of polarized features, detachment from neighboring cells, increased motility, and invasion into surrounding matrix (2). Context-dependent cellular plasticity has been identified in side populations of established human cancer cell lines and in cell lines derived from mouse models of human epithelial cancers; these cells are marked by increased expression of aldehyde dehydrogenase or prominin-1 (CD133) (3, 4). The unique biological features of these cells and their dearth within primary tumors has led to the belief that they originate from rare populations of pluripotent cells or have uncommon gain-of-function somatic mutations (5).
EMT is driven by several families of transcriptional repressors (ZEB, SNAIL, and basic helix-loop-helix factors) (6). ZEB factors contain 2 widely separated clusters of zinc fingers that bind to paired CAGGTA/G E-box–like promoter elements. They induce EMT by downregulating the expression of epithelial genes, including E-cadherin (7, 8). During development, ZEB expression is upregulated in cells that undergo EMT and migrate by extracellular signals such as TGF-β and Notch ligands (9, 10). Beyond its physiological roles, ZEB1 is overexpressed in many human cancers (e.g., prostate, colon, breast, and pancreatic) and has been implicated in metastasis and cellular events thought to precede it, including reduced expression of basement membrane components and induction of EMT (9). Increased ZEB1 levels correlate with poor prognosis in a variety of epithelial tumor types (11). In tumor cells, ZEB1 represses the expression of certain microRNAs (miRs) — including miR-183, miR-203, and miR-200 family members (miR-200a, miR-200b, miR-200c, miR-141, and miR-429) — that function not only as strong inducers of epithelial differentiation, but also as inhibitors of stem cell properties through repression of the stem cell factors SOX2, BMI1, and KLF4 (9). Reciprocally, miR-200 family members directly target the ZEB1 3′–untranslated region (3′-UTR); hence, ZEB1 and miR-200 are interconnected through a double-negative feedback loop (12–15). The relevance of these findings to metastasis is supported by findings in a mouse model of human lung adenocarcinoma driven by expression of Trp53R172HΔG and KrasG12D alleles (KP mice) that recapitulates features of poor-prognosis human lung adenocarcinomas, including overlapping oncogenic driver mutations, distribution of metastases, and gene expression signatures (16, 17). Metastatic tumor cell lines derived from these mice (KP cells) have high basal ZEB1 expression, form polarized epithelial spheres in 3D cultures, and undergo ZEB1-dependent EMT in response to specific extracellular cues that can be reversed by ectopic expression of the miR-200b/a/429 cluster, whereas nonmetastatic KP cells have low basal ZEB1 expression and do not form spheres or undergo EMT (17). Thus, ZEB1 plays a key role in determining the metastatic fate of epithelial cancers.
Given that each miR downregulated by ZEB1 has the capacity to target multiple genes with diverse functions, we here posited that the scope of prometastatic biological processes controlled by ZEB1 extends beyond EMT and stem-ness, testing this hypothesis in KP cells and human lung and breast carcinoma cells. Our findings showed that ZEB1 controlled an unexpectedly large miR network implicated in diverse cellular functions, activated the RHO family of GTPases, and enhanced the formation of promigratory cytoskeletal structures by downregulating miR-34a. We elucidated a gene expression signature regulated by miR-34a that was enriched in cytoskeletal functions and prognostic in human lung adenocarcinomas and revealed several miR-34a target genes, including Arhgap1, a RHO GTPase-activating protein (RHOGAP). Together with the known therapeutic effects of miR-34a on prostate tumors (18), these findings provide a strong rationale for developing miR-34a as a therapeutic agent to inhibit tumor growth and metastasis in lung cancer patients.
Results
ZEB1 regulates the expression of diverse miRs.
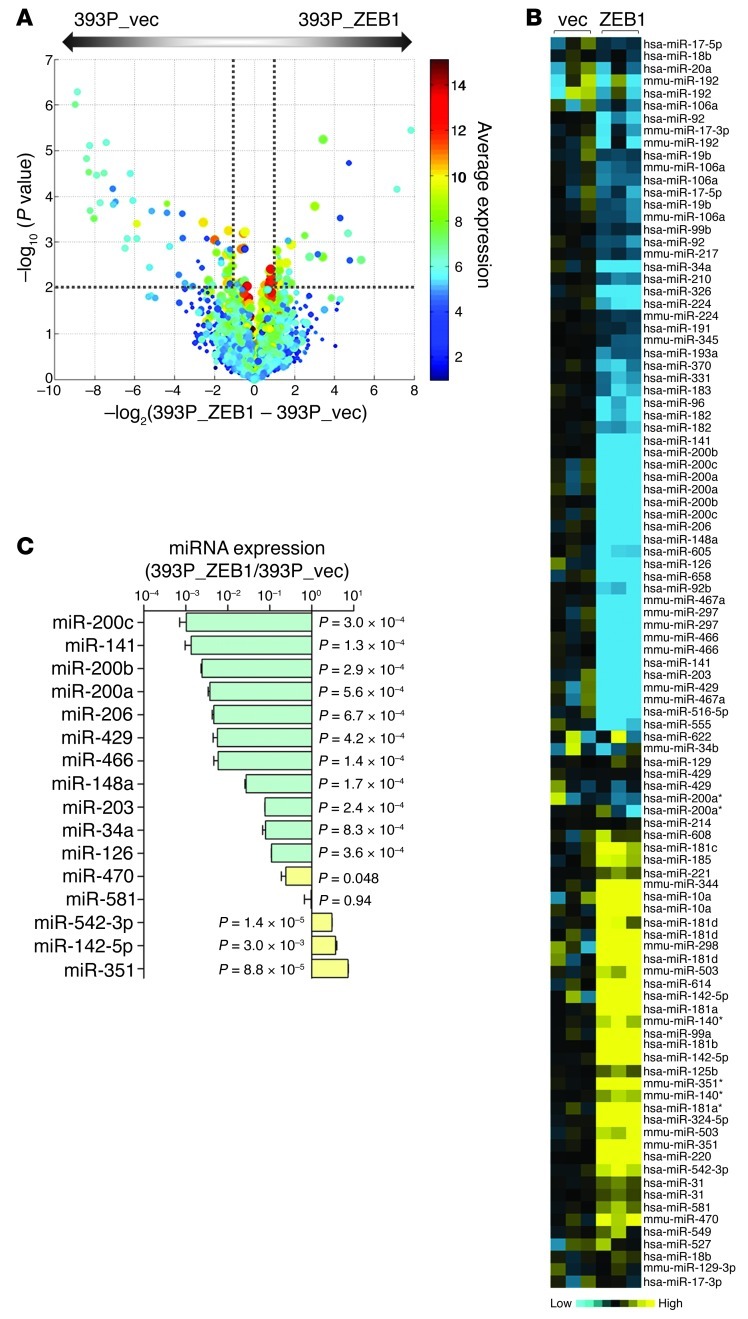
Microarray-based interrogation of global miR expression was carried out on a nonmetastatic KP cell line (393P) that undergoes EMT and gains invasive and metastatic capabilities after ectopic ZEB1 expression (referred to herein as 393P_ZEB1 cells; GEO accession no. GSE38386). Zeb1 levels in 393P_ZEB1 cells were 4-fold higher than in 393P_vector cells and were similar to endogenous ZEB1 levels in human lung cancer cell lines (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI63608DS1). Using 393P_vector as the reference, we found that 46 miRs were differentially expressed in 393P_ZEB1 cells: 27 downregulated (fold change less than 0.5, P < 0.01) and 19 upregulated (fold change greater than 2.0, P < 0.01; Table 1 and Figure 1, A and B). Of the differentially expressed miRs, 19 were clustered within 7 genomic loci that are transcribed and processed together (Table 1). Quantitative RT-PCR (Q-PCR) analysis confirmed up- or downregulation of 14 of 16 miRs sampled, including 6 known direct transcriptional targets of ZEB1 (miR-200a, miR-200b, miR-200c, miR-141, miR-429, and miR-203) and 8 other miRs not known to be regulated by ZEB1 (Figure 1C).
Table 1.
miRs regulated by ZEB1

Figure 1. ZEB1 regulates the expression of numerous miRs.
(A) Volcano plot depiction of findings from microarray analysis showing the miRs differentially expressed in 393P_vector and 393P_ZEB1 cells. The –log10 of P values (y axis) is plotted against the log2 of fold change between 2 groups (x axis). The size of the circle for each probe is proportional to the miR detection rate for the entire experiment. Each symbol is color coded according to average expression of the probe across the 2 groups (scale at right). Dotted lines delineate the cutoffs for miRs significantly downregulated (left) or upregulated (right) in 393P_ZEB1 cells. (B) Heat map depiction of miRs differentially expressed in 393P_ZEB1 cells (ZEB1), using 393P_vector cells (vec) as reference. (C) Taqman microRNA assays (Q-PCR) to confirm miRs differentially expressed by microarray. Data are mean ± SD (n = 3 samples). P values are indicated (2-tailed Student’s t test).
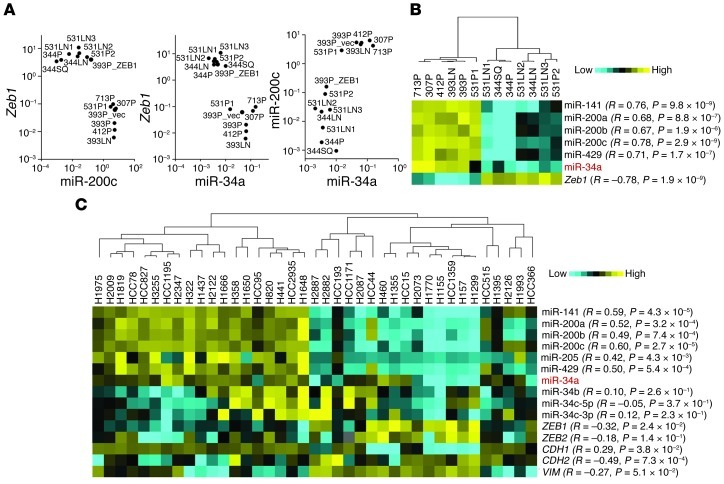
Several of the miRs regulated by ZEB1 function as oncogenes (e.g., miR-181b, miR-181d, and miR-10a) or tumor suppressors (e.g., miR-34a, miR-210, miR-326, miR-193a, miR-370, miR-206, miR-126, and miR-203), or lack reported roles in cancer development (e.g., miR-331, miR-605, miR-470, miR-581, and miR-351) (19–27). miR-34a was of particular interest as a candidate downstream mediator of ZEB1, given its tumor-suppressing activity in various models (18, 27) and prominent downregulation by ZEB1 (0.0015-fold; Table 1). In a panel of KP cell lines that have different basal Zeb1 levels, miR-34a levels correlated negatively with Zeb1 (R = –0.78; P = 1.9 × 10–9, 1-tailed Spearman rank correlation test) and positively with miR-200c (R = 0.78; P = 2.9 × 10–9; Figure 2, A and B); similar findings were observed in a panel of 39 human lung cancer cell lines (Figure 2C).
Figure 2. miR-34a levels correlate tightly with the ZEB1/miR-200 axis.
(A and B) Expression analysis of a panel of KP cell lines. (A) Log-scale cluster plots of normalized miR-200c and Zeb1 mRNA levels in 13 KP cell lines. Data are mean ± SD (n = 3 samples). 393P_ZEB1 and 393P_vector cells are included as controls. (B) Heat map representation of gene expression in A. (C) Expression analysis of a panel of human lung cancer cell lines. EMT marker expression in these 39 human cell lines was reported previously (17). Correlation (R and P, 1-tailed Spearman’s rank correlation test) is indicated for each gene relative to miR-34a.
Although located on a separate chromosome from miR-34a, expression of the miR-34b/c cluster is frequently coregulated with miR-34a (27, 28). However, TaqMan PCR assays confirmed the evidence from our microarray studies (Figure 1) that ZEB1 downregulated the expression of miR-34a, but not that of miR-34b or miR-34c (Supplemental Figure 2). Furthermore, miR-34a levels did not correlate with miR-34b or miR-34c in the panel of human lung cancer cell lines (Figure 2C). Collectively, these findings suggest that miR-34a expression is regulated through mechanisms different from those controlling the miR-34b/c cluster in the human and murine lung adenocarcinoma cells examined in this study.
ZEB1 downregulates miR-34a through ΔNp63.
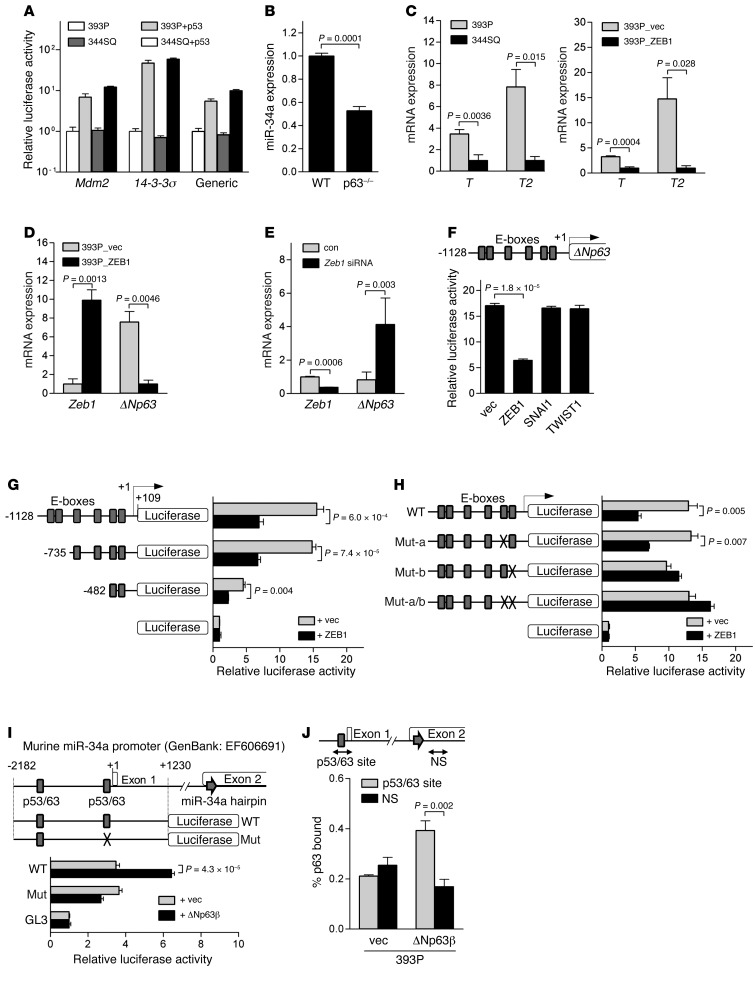
Although miR-34a is a known transcriptional target of p53 (27, 29), KP cells have the same germline Trp53 mutation, and the activity of p53 reporter constructs was not different between 2 KP cell lines with high and low miR-34a levels (Figure 3A), which indicates that differential p53 activity does not contribute to relative miR-34a levels in KP cells. As an alternative intermediary, we turned our attention to ΔN isoforms of p63 (ΔNp63), which lack the transactivation domain and are transcriptional targets of ZEB1 in 393P cells (Supplemental Figure 3 and ref. 30). ΔNp63 acts in a dominant-negative fashion against p53, TAp63, and TAp73, but has been shown to function as a transcriptional activator of specific genes such as T and T2 (31). Murine embryonic fibroblasts (MEFs) deficient in all p63 isoforms had reduced miR-34a levels (Figure 3B), which suggests that p63 positively regulates miR-34a expression. On the basis of these findings in MEFs, we sought to determine whether ΔNp63 serves as an intermediate in ZEB1-induced miR-34a repression in KP cells. Basal levels of ΔNp63 transcriptional target genes T and T2 were higher in low-ZEB1 cell line 393P than they were in high-ZEB1 cell line 344SQ, and introduction of ZEB1 into 393P cells decreased T and T2 levels (Figure 3C), which demonstrated that ZEB1 repressed ΔNp63 transcriptional activity. Ectopic ZEB1 expression downregulated ΔNp63 mRNA levels in 393P cells (Figure 3D), and siRNA-mediated knockdown of ZEB1 upregulated ΔNp63 mRNA levels in 344SQ cells (Figure 3E). The activity of a ΔNp63 promoter fragment (–1,128 to +109) was repressed by exogenous ZEB1, but not SNAI1 or TWIST1 (Figure 3F). Promoter deletion studies demonstrated that ZEB1 repressed the activity of a minimal ΔNp63 promoter fragment (–482 to +109) containing 2 putative ZEB1-binding sites (E-boxes; Figure 3G), and site-directed mutagenesis of the most proximal E-box element abrogated ZEB1-induced repression of the ΔNp63 promoter (Figure 3H).
Figure 3. ZEB1 downregulates miR-34a levels through ΔNp63.
(A) Luciferase reporter assays on KP cells cotransfected with p53 or empty expression vectors and reporters containing promoters from Mdm2, 14-3-3σ, or consensus p53-binding sites (Generic). Results are expressed relative to empty vector transfectants, set at 1.0. (B) Q-PCR analysis of miR-34a in WT or Trp63-null (p63–/–) MEFs. (C) Q-PCR values relative to 344SQ cells (left) and 393P_ZEB1 cells (right), set at 1.0. (D and E) Q-PCR values relative to 393P_vector cells (D) and control siRNA-transfected 344SQ cells (E), set at 1.0. (F) Location of putative E-boxes in the ΔNp63 promoter are shown above ΔNp63 promoter luciferase assays on cotransfected 393P cells. (G) ΔNp63 promoter reporter constructs are shown above luciferase activities in cotransfected 393P cells, expressed relative to basal reporter activity (set at 1.0). (H) ΔNp63 promoter constructs are WT or lack 1 (Mut-a and Mut-b) or 2 (Mut-a/b) proximal E-boxes. Luciferase activities in cotransfected 393P cells are also shown, expressed relative to basal reporter activity (set at 1.0). (I) Luciferase reporter constructs that do (Mut) or do not (WT) contain a site-directed mutation in a p53-binding site. Luciferase activities in cotransfected 393P cells are also shown, expressed relative to basal reporter (GL3) activity (set at 1.0). (J) DNA amount at specific (p53/p63) and nonspecific (NS) sites of the miR-34a promoter precipitated from transiently transfected 393P cells by anti-Myc antibody, corrected for nonspecific binding activity by IgG precipitation and expressed as percent p63 bound. P values were determined by 2-tailed Student’s t test. Data are mean ± SD (n = 3).
We next examined whether ΔNp63 regulates miR-34a expression and acts directly on the miR-34a gene promoter in KP cells. Ectopic ΔNp63β expression in 344SQ cells upregulated miR-34a, but not miR-34b or miR-34c (Supplemental Figure 4), and increased the activity of a WT promoter, but not a mutant miR-34a promoter lacking a p53-binding site (Figure 3I). Binding of endogenous p63 to the miR-34a promoter was examined by performing ChIP assays using an antibody that recognizes all p63 isoforms. In 344SQ cells, total p63 binding roughly reflected ΔNp63 binding, because ΔN isoforms were 67.0-fold more abundantly expressed than were TA isoforms (Supplemental Figure 5A). Murine keratinocytes were included as a positive control. The percentage of total p63 that bound to the p53/p63 site and to a nonspecific site in the miR-34a promoter was 0.19% and 0.06%, respectively (P = 0.08; Supplemental Figure 5B). To more specifically examine ΔNp63 binding to that site, ChIP assays were performed on 393P cells stably transfected with Myc-tagged ΔNp63. Using an anti-Myc antibody, we found the percentage of ectopic ΔNp63 that bound to the p53/p63 site and to a nonspecific site in the miR-34a promoter to be 0.39% and 0.17%, respectively (P = 0.002; Figure 3J). We concluded that ΔNp63 serves as an intermediate in ZEB1-induced miR-34a repression in KP cells.
miR-34a abrogates tumor cell invasion and metastasis and induces transcriptional changes that are prognostic in human lung adenocarcinomas.
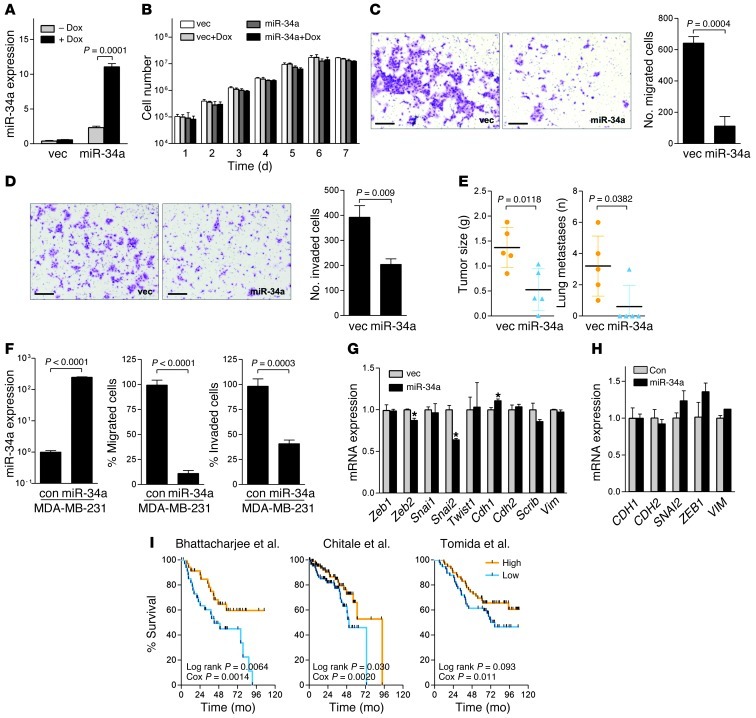
To examine the biological role of miR-34a repression, miR-34a was exogenously expressed under the control of a doxycycline-inducible promoter in 344SQ cells, which have high ZEB1 and low miR-34a expression (Figure 4A). Grown in monolayer, 344SQ_miR-34a cells proliferated at a rate similar to that of 344SQ_vector cells and demonstrated no biochemical evidence of apoptosis (Figure 4B and Supplemental Figure 6A). However, when grown in suspension, 344SQ_miR34a cells did not proliferate and demonstrated evidence of apoptosis (Supplemental Figure 6, B and C), which suggests that miR-34a enhanced the susceptibility of 344SQ cells to anoikis. 344SQ_miR34a cells exhibited reduced migration and invasion in Boyden chambers (Figure 4, C and D) and generated flank tumors in syngeneic mice that were smaller and metastasized to the lung less frequently (Figure 4E). Conversely, transfection of miR-34a hairpin inhibitor into 393P cells, which have low ZEB1 and high miR-34a expression, induced a 84% decrease in endogenous miR-34a levels and 1.6- and 1.5-fold increases in cell migration and invasion, respectively (Supplemental Figure 7, A–C). In MDA-MB-231 human breast cancer cells and H1299 human lung cancer cells, which have high basal ZEB1 expression and undergo EMT in response to TGF-β (13, 32), overexpression of miR-34a antagonized migration and invasion (Figure 4F and Supplemental Figure 8, A–C), which was not a consequence of apoptosis or reduced proliferation (Supplemental Figure 8, D and E). There was no evidence of EMT reversal on the basis of expression of epithelial and mesenchymal markers in 344SQ cells and MDA-MB-231 cells (Figure 4, G and H). Thus, miR-34a downregulation was required for ZEB1-induced metastatic properties and apparently mediated these actions through EMT-independent mechanisms.
Figure 4. miR-34a regulates multiple biological properties of tumor cells.
(A–D) 344SQ_miR-34a cells and 344SQ_vector cells were cultured in the presence or absence of doxycycline (Dox). (A) Q-PCR analysis of miR-34a levels. (B) Cell numbers in monolayer. Migrating (C) and invading (D) cells in Boyden chambers were photographed and counted. Scale bars: 100 μm. (E) Primary tumor weight and total lung metastases from flank tumors in syngeneic mice (mean ± SD, n = 5). P values were determined by 2-tailed Student’s t test. (F) MDA-MB-231 cells were transiently transfected with a random sequence miR precursor molecule control or with pre–miR-34a precursor. Shown are Q-PCR analysis of miR-34a levels, expressed relative to control transfectants (set at 1.0), and migration and invasion assays in Boyden chambers. (G and H) Q-PCR analysis of epithelial (Cdh1 and Scrib) and mesenchymal (Cdh2 and Vim) markers and their transcriptional regulators (Zeb1, Zeb2, Snai1, Snai2, and Twist1) in 344SQ_vector and 344SQ_miR-34a cells (G) and in MDA-MB-231 cells transiently transfected with pre-miR control or pre–miR-34a precursor (H). Results are expressed relative to control transfectants (set at 1.0). Data are mean ± SD (n = 3). *P < 0.01. (I) Kaplan-Meier analysis of 3 independent cohorts of lung cancer patients (33–35), comparing the differences in risk between tumors with high (>0) or low (<0) scores (36), reflecting the presence or absence, respectively, of overlap with the murine miR-34a signature. P values from log-rank (differences between arms) and univariate Cox (gene signature score as a continuous variable) tests are shown.
To gain insight into the biologic processes induced by miR-34a repression, microarray-based interrogation was carried out on RNA from tumor samples (344SQ_miR-34a and 344SQ_vector), which revealed a total of 805 genes that were differentially expressed (fold change greater than 1.5, P < 0.01; Supplemental Figure 9; GEO accession no. GSE38341). Q-PCR analysis of the same RNA samples confirmed differential expression of 22 of 24 genes sampled (Supplemental Figure 10). The 512 downregulated genes were enriched in, among other Gene Ontology terms mitosis (P = 3.80 × 10–24, Fisher exact test), cell cycle (P = 3.40 × 10–23), mRNA processing (P = 4.72 × 10–10), and cytoskeleton (P = 3.04 × 10–7), whereas the 293 upregulated genes were enriched in protein dimerization activity (P = 7.55 × 10–4) and glutathione transferase activity (P = 1.12 × 10–3). Human lung adenocarcinomas for which both gene expression and clinical outcome data are publicly available (33–35) were scored based on the presence or absence of this 805-gene signature (high or low t score, respectively), as described previously (36); the absence of this expression signature in primary lung tumors correlated with poor prognosis in 3 independent cohorts of lung cancer patients (Figure 4I). We conclude that the genes regulated by miR-34a are functionally diverse and have prognostic value in lung adenocarcinoma patients.
miR-34a inhibits promigratory cytoskeletal processes and attenuates RHO GTPase activity.
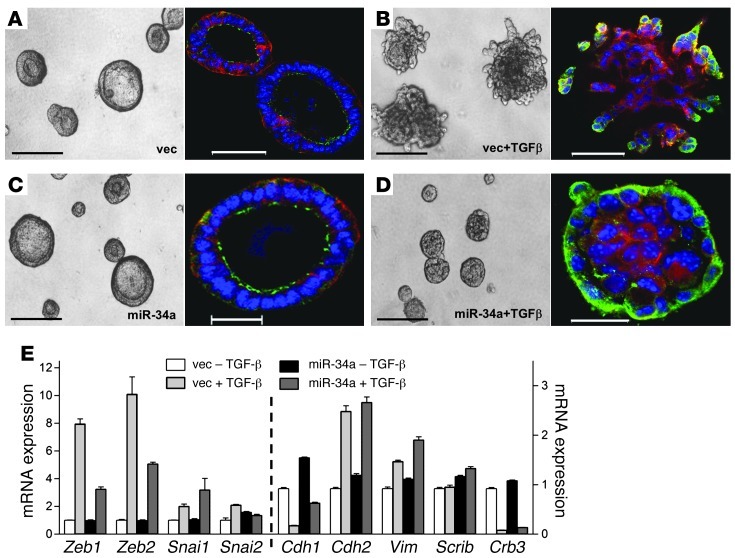
Given the prominent effect of miR-34a on cell migration and invasion in Boyden chambers and the enrichment of the miR-34a transcriptome in cytoskeletal functions, we next carried out studies in 3D cultures to visualize tumor cell polarization and invasion. In Matrigel, 344SQ cells form hollow, polarized spheres with apical localization of ZO-1 and basolateral localization of α6 integrin; these structures lose apical-basal polarity, become hyperproliferative, and form invasive membrane protrusions after TGF-β treatment (17). 344SQ_miR-34a and 344SQ_vector cells formed hollow polarized spheres in the absence of TGF-β (Figure 5, A and C) and became aberrantly polarized and filled in their central cores after TGF-β treatment (Figure 5, B and D). However, TGF-β–treated 344SQ_vector structures formed invasive projections (Figure 5B), whereas 344SQ_miR-34a structures remained spherical in morphology (Figure 5D), despite upregulation of mesenchymal markers in both (Figure 5E). Thus, miR-34a abrogated TGF-β–induced formation of invasive cellular protrusions in spite of biochemical evidence of EMT.
Figure 5. miR-34a blocks invasion, but does not reverse EMT.
(A–D) miR-34a repressed TGF-β–induced invasion in 3D Matrigel cultures. 344SQ_vector cells formed polarized epithelial spheres (A) that became hyperproliferative and invasive in the presence of TGF-β (B). 344SQ_miR-34a cells formed polarized epithelial spheres (C) that did not become invasive in the presence of TGF-β (D). Shown are light (left) and fluorescent (right) microscopic images of structures formed after 10 days in Matrigel containing doxycycline in the presence or absence of TGF-β (10 ng/ml). Blue, Topro-3; red, anti–α6 integrin; green, anti–ZO-1. Scale bars: 200 μm (light); 50 μm (fluorescent). (E) miR-34a did not abrogate TGF-β–induced EMT. Q-PCR analysis of epithelial markers (Cdh1, Scrib, and Crb3) and mesenchymal markers (Cdh2 and Vim) and their transcriptional regulators (Zeb1, Zeb2, Snai1, and Snai2) in 344SQ_vector and 344SQ_miR-34a cells after 10 days in Matrigel cultures containing doxycycline in the presence or absence of TGF-β. Results are expressed relative to empty vector transfectants treated without TGF-β (set at 1.0). Data are mean ± SD (n = 3).
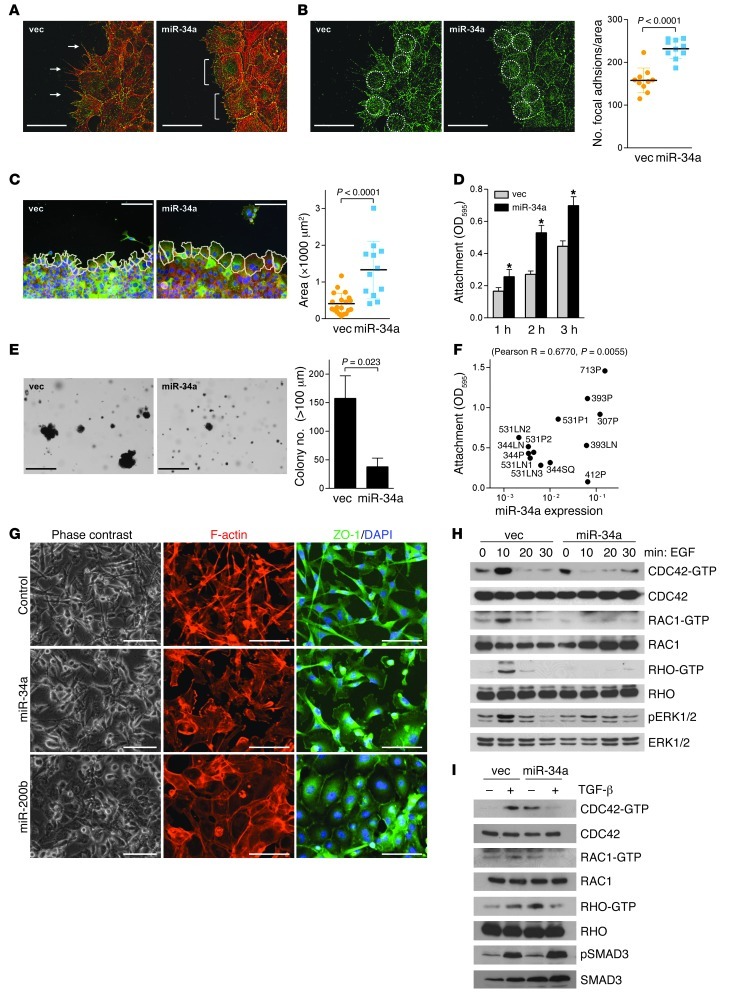
We next carried out scratch injury assays to examine whether miR-34a regulates the formation of focal adhesions and promigratory membranous protrusions. At the leading edge, 344SQ_vector cells formed lamellipodia with prominent filopodial extensions, whereas 344SQ_miR-34a cells formed no filopodia (Figure 6A), generated more focal adhesions per surface area (mean, 232 versus 158; Figure 6B), and exhibited increased cell cross-sectional area (1,330 μm2 versus 410 μm2; Figure 6C). Consistent with these findings, 344SQ_miR-34a cells anchored more avidly to plastic (Figure 6D) and formed fewer colonies in soft agar (Figure 6E). In the larger panel of KP cell lines, anchorage avidity correlated positively with endogenous miR-34a levels (R = 0.6770; P = 0.0055, 1-tailed Pearson correlation test; Figure 6F). In MDA-MB-231 cells, exogenous miR-34a decreased cortical F-actin–containing filopodial extensions and increased cell size without reversing mesenchymal features on the basis of ZO-1 localization, which was predominantly cytoplasmic (Figure 6G). In contrast, exogenous miR-200b in MDA-MB-231 cells decreased filopodial extensions and shifted ZO-1 to a membranous distribution typical of a mesenchymal-to-epithelial transition (Figure 6G). Given the central role of RHO GTPases in actin cytoskeletal remodeling (37, 38), we quantified activated (GTP-bound) RHO family members after treatment with promigratory cytokines, which revealed that ectopic miR-34a expression in 344SQ cells attenuated EGF- or TGF-β–induced increases in GTP-bound CDC42, RAC1, and RHO (Figure 6, H and I). Thus, miR-34a increased cellular adhesions, attenuated cytokine-induced RHO GTPase activation, and inhibited formation of membrane protrusions and migration in response to external stimuli.
Figure 6. miR-34a regulates actin cytoskeletal remodeling and RHO family GTPase activity.
(A–C) Cells imaged at leading edge of scratch-wounded confluent cultures. (A) Filopodia (arrows) formed in 344SQ_vector, but not 344SQ_miR-34a, cells. Brackets denote lamellipodia. (B) Focal adhesions in anti-vinculin–stained cultures, counted per defined surface area of confluent cells (circles) using ImageJ. Data are mean ± SD (n = 10). Red, phalloidin; green, vinculin. (C) Cells were outlined (white lines), and their surface areas were measured using ImageJ. Data are mean ± SD (n = 20 [344SQ_vector]; 12 [344SQ_miR-34a]). (D) Attached cells were quantified 1, 2, or 3 hours after seeding by optical densitometry (595 nm) of cells stained with crystal violet. Data are mean ± SD (n = 3). *P < 0.01. (E) Cells seeded in soft agar were stained with nitrotetrazolium blue 3 weeks after seeding, and colonies larger than 100 μm in diameter were counted. Data are mean ± SD (n = 3). (F) Attached cells were quantified by optical densitometry 3 hours after seeding and expressed relative to miR-34a levels from Figure 2A. Correlation (R and P, 1-tailed Pearson’s correlation test) is indicated. (G) MDA-MB-231 cells transiently transfected with control, miR-34a, or miR-200b precursors and imaged under fluorescence (blue, DAPI; red, phalloidin; green, anti–ZO-1). As a comparison, miR-200b–transfected cells demonstrated mesenchymal-to-epithelial transition. (H and I) Western blot analysis of GTP-bound (CDC42-GTP) and total (CDC42) RHO family GTPases. Phospho-ERK1/2 (H) and phospho-SMAD3 (I) were included as positive controls for EGF- and TGF-β–induced signaling, respectively. Scale bars: 50 μm (A, B, and G); 100 μm (C); 500 μm (E). See complete unedited blots in the supplemental material.
Arhgap1 is a miR-34a target gene required for the regulation of RHO GTPase activity and tumor cell invasion.
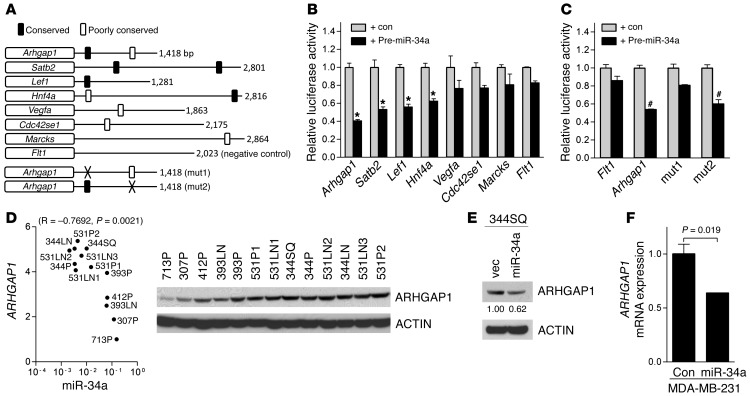
We next examined whether miR-34a inhibits RHO GTPases at the level of GTP loading or upstream at the level of focal adhesion kinase (FAK), which is autophosphorylated at Tyr397 and initiates RHO GTPase activation after recruitment to ligand-bound EGFR (39). Arguing against the latter possibility, EGF-induced FAK-Tyr397 phosphorylation was unchanged by exogenous miR-34a, as determined by Western blot analysis (data not shown). At the level of GTP loading, RHO GTPases cycle from inactive (GDP-bound) to active (GTP-bound) states by binding to guanine nucleotide exchange factors (GEFs), and in the reverse direction by binding to GAPs (37). To examine whether miR-34a directly targets GEFs or GAPs, we used a prediction algorithm (TargetScan; http://www.targetscan.org) to scan the genome for putative miR-34a binding sites and discovered sites in the 3′-UTR of a RHOGAP (Arhgap1) and a CDC42 downstream effector (Cdc42se1) (Figure 7A). Reporter assays were carried out to determine whether miR-34a binds directly to these 3′-UTRs as well as those of other predicted miR-34a target genes identified in our analysis that are not directly involved in RHO GTPase regulation, but are potentially important in tumorigenesis (Figure 7B). Relative to its effect on a negative control 3′-UTR (Flt1), cotransfection of miR-34a precursors repressed 3′-UTRs of Arhgap1 by 59%, Satb2 (a regulator of chromatin structure; ref. 40) by 47%, Lef1 (a regulator of Wnt signaling; ref. 41) by 44%, and Hnf4a (a known miR-34a target; ref. 42) by 37%, but did not affect other reporters (Figure 7B). Site-directed mutagenesis of 2 predicted binding sites in the Arhgap1 3′-UTR with differing PCt values (0.71 and <0.1) revealed that miR-34a suppressed Arhgap1 3′-UTR reporter activity through the conserved binding site, but not the nonconserved site (Figure 7C).
Figure 7. Arhgap1 is a miR-34a target gene.
(A) 3′-UTRs analyzed. Shown are size (right) and location of predicted miR-34a binding sites. Poorly conserved sites were defined on the basis of criteria established by TargetScan. mut1 and mut2, mutations generated in 2 putative miR-34a binding sites in the Arhgap1 3′-UTR. Flt1 3′-UTR is a negative control. (B and C) miR-34a–induced repression of 3′-UTR reporters. Luciferase assays on 344SQ cells transiently cotransfected with the indicated 3′-UTR reporters and control or miR-34a precursors. Data are mean ± SD (n = 3). *P < 0.001; #P < 0.01. (D) Correlation of miR-34a levels in KP cell lines (from Figure 2A), with densitometric analysis of ARHGAP1 protein expression by Western analysis. ACTIN was used as a loading control. Correlation (R and P, 1-tailed Spearman rank correlation test) is indicated. (E) miR-34a downregulated ARHGAP1. Western blot analysis of ARHGAP1 in 344SQ_vector and 344SQ_miR-34a cells. Densitometric analysis (numbers below ARHGAP1 gel) is shown relative to 344SQ_vector cells (set at 1.0). (F) Q-PCR analysis of ARHGAP1 mRNA levels in MDA-MB-231 cells 72 hours after transient transfection with pre-miR negative control or pre–miR-34a precursors. Data are mean ± SD (n = 3). P values were determined by 2-tailed Student’s t test. See complete unedited blots in the supplemental material.
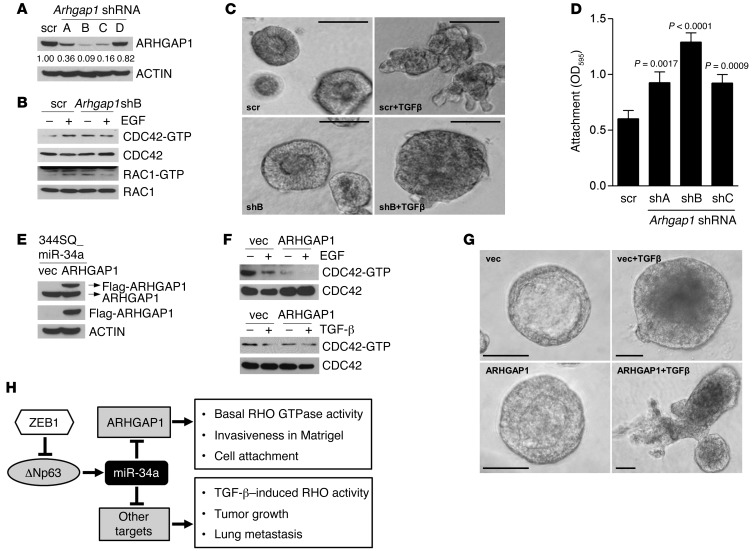
In the KP cell line panel, ARHGAP1 and miR-34a levels correlated negatively (R = –0.7692; P = 0.0021, 1-tailed Spearman rank correlation test; Figure 7D), and exogenous miR-34a decreased ARHGAP1 levels in 344SQ cells (Figure 7E). Conversely, transfection of miR-34a hairpin inhibitor into 393P cells induced a 2.0-fold increase in ARHGAP1 expression (Supplemental Figure 11A). In MDA-MB-231 and H1299 cells, ectopic miR-34a downregulated ARHGAP1 (Figure 7F and Supplemental Figure 11B). To determine whether ARHGAP1 downregulation recapitulates the effects of miR-34a, ARHGAP1 was depleted by introduction of shRNAs (Figure 8A), which increased basal levels of GTP-bound CDC42 and RAC1 (Figure 8B), attenuated EGF-induced levels of GTP-bound CDC42 and RAC1 (Figure 8B), reduced invasive projections on tumor spheres after TGF-β treatment (Figure 8C), and increased cell adherence (Figure 8D).
Figure 8. ARHGAP1 mediates specific phenotypic effects of miR-34a.
(A) Western blot analysis of ARHGAP1 in 344SQ cells stably transfected with scrambled shRNA (scr) or 1 of 4 Arhgap1 shRNAs (shA, shB, shC, or shD). ACTIN was used as loading control. (B) Western blot analysis of GTP-bound (-GTP) and total RHO family GTPases. (C) Spheres formed by scrambled shRNA– and Arhgap1 shB–transfected 344SQ cells after 10 days in 3D Matrigel cultures in the presence or absence of TGF-β (10 ng/ml). Scale bars: 100 μm. (D) Attached cells were quantified by optical densitometry 3 hours after seeding. Data are mean ± SD (n = 3). P values were determined by 2-tailed Student’s t test. (E) Western blot analysis of 344SQ_miR-34a cells stably transfected with Flag-tagged ARHGAP1 cDNA or empty vector using antibodies against ARHGAP1 (top), Flag (middle), or ACTIN as a loading control (bottom). Arrows at top denote locations of endogenous (ARHGAP1) and ectopic (Flag-ARHGAP1) ARHGAP1. (F) Western blot analysis of GTP-bound and total CDC42 in 344SQ_miR34a/vector and 344SQ_miR34a/ARHGAP1 cells treated for 10 minutes with or without EGF or TGF-β. (G) 344SQ_miR34a/vector and 344SQ_miR34a/ARHGAP1 cells were cultured for 10 days in Matrigel in the presence or absence of TGF-β and photographed under phase-contrast microscopy. TGF-β induced loss of lumen formation in both transfectants, but invasive projections formed only in 344SQ_miR34a/ARHGAP1 cells. Scale bars: 50 μm. (H) Proposed model illustrating the ARHGAP1-dependent and -independent phenotypic effects of miR-34a. See complete unedited blots in the supplemental material.
The anti-invasive effect of Arhgap1 shRNA was paradoxical, given that RHO is required for proinvasive actin cytoskeletal remodeling. We performed experiments to confirm this finding and to determine whether ARHGAP1 is required for miR-34a–induced phenotypic features. ARHGAP1 expression was reconstituted in 344SQ_miR-34a cells through stable transfection of an Arhgap1 cDNA expression vector (Figure 8E), and the 344SQ_miR-34a/ARHGAP1 double transfectants were compared with 344SQ_miR-34a/vector cells from the standpoint of basal and cytokine-induced RHO activity and invasive activity. ARHGAP1 reconstitution repressed basal GTP-bound CDC42 and rescued TGF-β–induced sphere invasion in Matrigel (Figure 8, F and G), which suggests that miR-34a represses TGF-β–induced tumor cell invasion by downregulating ARHGAP1. However, ARHGAP1 reconstitution did not restore RHO activation in response to EGF or TGF-β treatment (Figure 8F) or rescue tumor growth or metastasis (data not shown), which suggests that these phenotypic effects of miR-34a are mediated through other target genes.
Discussion
Studies using experimental tumor models have established a strong link between high levels of EMT activators and loss of cell polarity, reduced expression of basement membrane components, and increased propensity for metastasis (43–47). The discovery that EMT activators endow epithelial tumor cells with pluripotency led to the current belief that metastatic propensity is directly related to plasticity in response to extracellular cues (12, 48–50). Here, positing that the scope of prometastatic biological processes controlled by ZEB1 extends beyond EMT and stem-ness, we discovered that ZEB1 drove promigratory cytoskeletal processes and metastasis by downregulating the expression of miR-34a. Exogenous miR-34a decreased tumor cell invasion and metastasis, inhibited the formation of promigratory cytoskeletal structures, suppressed activation of the RHO GTPase family, and regulated a gene expression signature that was enriched in cytoskeletal functions and prognostic in human lung adenocarcinomas. Biological reprogramming of this magnitude supports a central role for miR-34a in metastasis regulation by ZEB1.
RHO family members play key parts in the regulation of actin cytoskeletal remodeling and tumorigenesis. RAC1 is required for the development of primary lung adenocarcinomas in mice that express mutant KRAS (51). The activities of RHO, RAC1, and CDC42 are coordinated to regulate membrane protrusions and cell-matrix adhesions at the leading edge of migrating cells to control forward movement (52). Effectors of RHO GTPases include RHO-associated protein kinase, focal adhesions, and membrane protrusions, which together mediate cell adhesion to extracellular matrix, link matrix attachments to intracellular signaling pathways, and drive actomyosin contractility and cell locomotion (52). Beyond these roles, a large body of evidence implicates RAC1 in the assembly, disassembly, and maintenance of adherens junctions and tight junctions, which play a central role in the regulation of apical-basal polarity (52). Tightly regulating these processes is a miR network that targets RHO GTPases and their associated GEFs and GAPs (53). Examples include RHOA (miR-31, miR-133, and miR-155), RHOC (miR-138 and miR-10b), CDC42 (miR-29), TIAM1 (miR-10b), and ARHGDIA (miR-151) (53). Here we showed that miR-34a inhibited cytokine-induced RHO family GTPase activation and discovered that a RHOGAP, Arhgap1, was a miR-34a target gene. ARHGAP1 reconstitution in miR-34a–overexpressing cells did not rescue RHO activation in response to EGF or TGF-β treatment, which was expected, given that RHOGAPs inhibit RHO GTPase activity. However, TGF-β–induced invasion was abrogated in metastasis-prone tumor cells by ARHGAP1 depletion and was rescued in miR-34a–overexpressing cells by ARHGAP1 reconstitution. The proinvasive effect of ARHGAP1 was paradoxical, given that RHO GTPase activity stimulates the formation of actin cytoskeletal structures that drive cell migration. Although the mechanism is unclear, ARHGAP1 binds to a number of proteins other than RHO family GTPases — including BNIP2, SRC, UBC, and PIK3R1 (BioGRID; http://thebiogrid.org) — that regulate diverse biological processes and may have contributed to the proinvasive effect of ARHGAP1 through RHO GTPase–independent mechanisms. Collectively, these findings suggest that ARHGAP1 mediates some, but not all, of the biological effects of miR-34a (Figure 8H).
We discovered that ZEB1 regulated a larger number of miRs than had previously been reported (12, 13, 15). This multiplicity was due in part to 19 miRs clustered within 7 genomic loci that are transcribed and processed together. ZEB1 downregulated certain miRs and upregulated others, which could be related either to the capacity of ZEB1 to function as a transcriptional repressor or activator (54–56) or to indirect regulation of miRs by ZEB1. In support of the latter possibility, we found that ZEB1 indirectly repressed miR-34a through ΔNp63. The reported biological functions of the 46 miRs were diverse, encompassing hypoxic response (miR-210), cell differentiation (miR-326), proliferation (e.g., miR-224, miR-206, miR-542-3p, and miR-126), apoptosis (miR-96, miR-193a, and miR-181a), and migration (miR-206, miR-503, and miR-181b), among other functions (Supplemental Table 1), which indicates that ZEB1 might control a number of biological processes by regulating the expression of these miRs.
The p63 transcription factor family plays a central role in the regulation of embryonic development, normal adult tissue homeostasis, and malignancy (57). The tumor-suppressive properties of TAp63 are exerted through the upregulation of a wide variety of miRs, including let-7, miR-15/16a, miR-145, miR-129, miR-26, miR-30, and miR-146a (57). Senescence in keratinocytes is activated through ΔNp63-induced downregulation of miR-138, miR-181a, miR-181b, and miR-130b (58). TAp63 is also a transcriptional activator of Dicer, an endoribonuclease required for miR biogenesis (29). The findings presented here build on this growing body of evidence that miRs are central mediators of the diverse biological actions of p63 by showing that miR-34a was upregulated by ΔNp63 and was a potent tumor suppressor in a Kras/Trp53-driven lung adenocarcinoma model. Furthermore, our finding that ΔNp63 served as a downstream mediator of ZEB1 completes a feedback circuit initiated by p63, which transcriptionally activates the miR-200b/a/429 cluster (59) and, in turn, directly targets ZEB1 (9, 13, 14), thereby relieving the ZEB1-induced repression of ΔNp63 shown here. There are numerous other p63/miR circuits, including one involving miR-193-5p, which targets p63 and is directly repressed by p63 (60). Thus, miR homeostasis is tightly regulated through multiple mechanisms involving p63 and ZEB1.
The evidence presented here that miR-34a is a potent repressor of tumor growth and metastasis in a mouse model of human lung cancer bolsters evidence from other mouse models that miR-34a is a promising therapeutic agent. Delivery of miR-34a oligomers systemically by tail vein inhibits tumor growth in mice bearing lung adenocarcinomas, suppresses metastasis to the lung and other organs, and prolongs the survival of mice bearing orthotopic human prostate carcinomas (61). The mechanisms by which miR-34a exerts its therapeutic effects are tumor cell type specific. For example, in the lung adenocarcinoma metastasis model shown here, miR-34a downregulation enhanced promigratory cytoskeletal processes, but was not required for stem cell features, based on formation of polarized epithelial spheres, whereas it targets the stem cell marker CD44 in prostate cancer cells and represses stem-ness in prostate, glioblastoma, pancreatic, and gastric cancer cells (18, 49). The distinct mechanisms by which miRs exert tumor suppressor functions in a given tumor type might be leveraged to create combinatorial treatment approaches. In metastatic KP cells, the miR-200 family members and miR-34a are all sharply downregulated, and ectopic expression of the miR-200b/a/429 cluster locks KP cells into an epithelial state and abrogates metastasis (17). Thus, combined delivery of miR-34a and miR-200 family members might be complementary in these cells. Safe and efficient approaches using lipid-based nanoparticles (neutral or charged) have been developed that deliver miRs locally or systemically to the tumor tissue where they regulate their target genes (62, 63). Physical and chemical moieties of the particles that facilitate the targeted distribution and the controlled and sustained release of miRs are under clinical investigation (64). External moieties, such as aptamers and ligands that enhance miR uptake by cancer cells, are being developed to direct the particles to a particular tissue (65, 66). Moreover, efforts are underway to initiate clinical trials that deliver miRs into patients with advanced cancer.
Methods
Antibodies and plasmid constructs.
Antibodies against ERK, phospho-ERK, SMAD3, phospho-SMAD3, PARP, cleaved caspase-3 (Cell Signaling Technology), ACTIN (Sigma-Aldrich), p63 (Abcam), and ARHGAP1 were purchased (Santa Cruz Biotechnologies). Doxycycline (Sigma-Aldrich), EGF (Invitrogen), and TGF-β (Calbiochem) were purchased. Human SNAI1 cDNA (catalog no. 16218), murine Twist1 cDNA (catalog no. 1783; Addgene), murine Arhgap1 cDNA, and murine Arhgap1 shRNA (Origene) were purchased. To construct the miR-34a overexpression vector, a tet operator–H1 promoter fusion (tH1) was cloned into the XmnI and BamHI sites of pENTR2B (Invitrogen). A 487-nt fragment containing miR-34a (∼200 nt either side of the mature miR) was amplified from human cDNA by PCR and directionally cloned into the BamHI and EcoRI sites of pENTR2B-tH1. The pLV711G lentiviral expression construct contains a Gateway cassette (Invitrogen) and the regulatory protein (T-REx) under the control of the EF-1a promoter (67). Gateway technology (Invitrogen) was used for the transfer of miR-34a into the pLV711G vector to create a single lentiviral vector enabling doxycycline-responsive expression of miR-34a.
Cell culture studies.
Murine (307P, 344LN, 344P, 344SQ, 393LN, 393P, 412P, 531LN1, 531LN2, 531LN3, 531P1, 531P2, and 713P) and human lung cancer cells (H2009 and H1299) were cultured in RPMI 1640 (Mediatech) with 10% FBS (Sigma-Aldrich) in the presence or absence of 1 μg/ml doxycycline (Sigma-Aldrich). WT and p63–/– murine embryonic fibroblasts (MEFs) were maintained in DMEM (Mediatech) with 10% FBS. MDA-MB-231 cells were cultured in Leibovitz’s L-15 (Mediatech) with 10% FBS. Cells were transfected using Dharmafect-DUO (Dharmacon). For soft agar assays, 5 × 104 cells (in 0.3% agar) were seeded into 6-well plates layered with 0.8% agar, and colonies were stained with 0.5 mg/ml nitrotetrazolium blue (Sigma-Aldrich) 21 days later. For migration and invasion assays, 1 × 105 cells were cultured in the upper wells of Transwell and Matrigel chambers, respectively (BD Biosciences), and allowed to migrate toward 10% FBS in the bottom wells. After 16 hours of incubation, migrating or invading cells were stained with 0.1% crystal violet, photographed, and counted. Cellular proliferation was measured in anchorage-dependent and -independent conditions by counting cells seeded onto high- or low-adherence plates, respectively (Greiner Bio-One), using the Countess automated cell counter (Invitrogen). For immunocytochemistry, cells were cultured on collagen-coated coverslips and then stained with DAPI (Sigma-Aldrich), Alexa Fluor 568–conjugated phalloidin (Invitrogen), and anti-vinculin (Millipore) antibody. Cells were cultured in 3D Matrigel cultures (BD Biosciences) and stained with immunofluorescently tagged antibodies, as described previously (17). A Zeiss LSM 510 confocal microscope was used to capture fluorescent images of Matrigel cultures. Fluorescence-stained slides depicting filopodia, lamellipodia, and focal adhesions were imaged on a Nikon Ti microscope equipped with a CoolSnap HQ2 camera (Photometrics). Image stacks of 100-nm sections were then deconvoled with Autoquant (Media Cybernetics). For attachment assays, 1 × 105 cells were seeded on 24-well plates and incubated for 1–3 hours. After washing with PBS twice, attached cells were stained with 0.1% crystal violet, and optical density was measured at 595 nm.
miR expression profiling.
Total RNA was isolated from 393P_vector and 393P_ZEB1 cells, profiled using the DiscovArray platform by Asuragen, and analyzed as described previously (17). The platform included all but 91 of the 1,086 probes in the Sanger miRBase version 9.2 covering humans, rats, and mice and an additional 12,894 exploratory probes covering multiple other species. Transcriptomic data sets were deposited in GEO (accession no. GSE38386). Heat maps were generated using Cluster and TreeView software ( http://rana.lbl.gov/EisenSoftware.htm).
Quantitative RT-PCR (Q-PCR).
Total RNA was isolated from the cells using TRIzol (Invitrogen) according to the manufacturer’s protocol. To analyze mRNA levels, Q-PCR assays were performed after reverse transcription with Superscript III reverse transcriptase (Invitrogen) using a SYBR-Green–based system (Applied Biosystems). mRNA levels were normalized on the basis of mRNA for ribosomal protein L32 (Rpl32). See Supplemental Table 2 for primer sequences. miR levels were quantified using Taqman microRNA assays (Applied Biosystems) according to the manufacturer’s protocol and normalized on the basis of snoRNA-135.
Luciferase reporter assays.
Cells were seeded on 24-well plates (1 × 105 cells/well) 1 day before transfection, and then transfected with 500 ng luciferase reporter plasmids and 50 ng hRL-control vector. After 48 hours, luciferase activity was measured using Dual-Luciferase Reporter Assay System (Promega). A murine miR-34a promoter region was isolated by PCR from TC-1 murine ES cell genomic DNA and ligated into the pGL3-basic vector (Promega). p53-reporter plasmids (Mdm2, 14-3-3σ, and generic promoters) were gifts from M.-H. Lee (University of Texas MD Anderson Cancer Center, Houston, Texas, USA), and the ΔNp63 and TAp63 promoter reporters were gifts from I. Shachar (Weizmann Institute of Science, Rehovot, Israel). For the 3′-UTR assay, murine 3′-UTRs were amplified by PCR from genomic DNA and ligated into pCI-neo-hRL vector (13). 3′-UTR reporters (500 ng) and pGL3-control (50 ng; Promega) were cotransfected into 344SQ cells seeded on 24-well plates (1 × 105 cells/well) in the presence or absence of pre–miR-34a precursor (5 nM; Ambion). A PCR-based site-directed mutagenesis strategy was carried out to generate mutant constructs.
ChIP assays.
393P cells were transiently transfected with Myc-ΔNp63β or empty vector. Cells were cross-linked with 1% formaldehyde and then incubated in lysis buffer (50 mM Tris-HCl, pH 8.1; 1% SDS; 10 mM EDTA; and protease inhibitor cocktail) on ice for 10 min. After sonication (Cole-Parmer GEX-130 sonicator; 50% power, pulse on for 10 s, pulse off for 10 s, 20 cycles), samples were immunoprecipitated with anti-Myc tag antibody (Millipore) or anti-mouse IgG (Santa Cruz). DNA was eluted and purified with PCR purification kit (Qiagen), and quantitative PCR was carried out with specific primers to amplify the p53/63-binding region of the miR-34a promoter (forward, 5′-CAGCCTGGAGGAGGATCGA-3′; reverse, 5′-TCCCAAAGCCCCCAATCT-3′) or a nonspecific region within exon 2 as a negative control (forward, 5′-AAGCGGGTTTCAAGTGCATCTCAG-3′; reverse, 5′-TCAGGCTACTAAACCAGTTGCCCT-3′). To detect binding of endogenous p63 to miR-34a promoter elements, cellular proteins from 393P cells and murine primary keratinocytes were cross-linked to DNA using 1% formaldehyde, and chromatin was prepared as described earlier (68). p63-DNA complexes were diluted 10-fold in ChIP dilution buffer and incubated overnight at 4°C with 2 μg anti–pan-p63 antibody (4A4; Abcam) or 2 μg IgG. Resulting chromatin was resuspended in 300 μl double-distilled H2O. The percent DNA bound was calculated as (2ΔCt × 2.5)p63 antibody – (2ΔCt × 2.5)IgG, where ΔCt is Ctinput chromatin – Ctsample.
Western blotting.
Cells were lysed in 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and protease/phosphatase inhibitors (Sigma-Aldrich). Cell lysates were separated by SDS-PAGE, transferred onto PVDF membrane, and then incubated with primary antibodies and HRP-conjugated secondary antibodies. Protein bands were visualized with Pierce ECL Western Blotting substrate (Thermo). RAC1, CDC42, and RHO activation assay kits (Upstate) were used for GTPase assays.
Animal husbandry.
Syngeneic (129/Sv) mice (n = 5 per group) were injected subcutaneously in the right flank with 344SQ cells (1 × 106 per mouse) that had been stably transfected with miR-34a or empty vectors. Mice were treated with doxycycline (2 mg/ml) in drinking water (2% sucrose), monitored daily for tumor growth, sacrificed at 6 weeks, and necropsied to isolate primary tumors and sites of metastasis, which were confirmed histologically by analysis of hematoxylin and eosin–stained, formalin-fixed tissues.
Affymetrix gene expression profiling.
Total RNA was extracted from primary tumors from mice injected with 344SQ_vector and 344SQ_miR-34a cells using RiboPure kit (Ambion), and then hybridized to Affymetrix GeneChip Mouse Genome 430 2.0 array (Asuragen). Data processing and determination of differentially expressed genes were carried out essentially as described previously (69). Transcriptomic data sets were deposited in GEO (accession no. GSE38341).
Statistics.
With the exception of mRNA and miR profiling, data were analyzed using 2-tailed Student’s t test and Spearman rank correlation test. A P value less than 0.05 was considered significant.
Study approval.
All mouse studies were approved by the IACUC at the University of Texas MD Anderson Cancer Center. Mice received standards of care and were euthanized according to the standards set forth by the IACUC.
Supplementary Material
Acknowledgments
This work was supported by R01 CA157450 (to J.M. Kurie). J.M. Kurie is the Elza and Ina A. Shackelford Endowed Professor in Lung Cancer Research. D.L. Gibbons was supported by NCI K08 CA151651, an International Association for the Study of Lung Cancer Fellow Grant, and received financial support from Dr. Waun Ki Hong (MD Anderson Cancer Center). C.J. Creighton was supported by P30 CA125123. D. Chakravarti was funded by a CPRIT training grant (RP101502). Z.H. Rizvi was supported by HHMI-Medical Research Fellows Program. A. Pertsemlidis was funded by R01 CA129632 and P50 CA70907 (the UT Southwestern/MD Anderson Cancer Center Lung Specialized Program of Research Excellence). E.R. Flores was funded by R01CA134796 and is a Leukemia and Lymphoma of America Scholar. G.J. Goodall was supported by NHMRC project grant 1008327. We thank Suraya Roslan for technical assistance.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(9):3170–3183. doi:10.1172/JCI63608.
References
- 1.Mehlen P, Puisieux A. Metastasis: a question of life or death. Nat Rev Cancer. 2006;6(6):449–458. doi: 10.1038/nrc1886. [DOI] [PubMed] [Google Scholar]
- 2.Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5(9):744–749. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- 3.Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 4.Ginestier C, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: an evolving concept. Nat Rev Cancer. 2012;12(2):133–143. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- 6.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanchez-Tillo E, et al. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene. 2010;29(24):3490–3500. doi: 10.1038/onc.2010.102. [DOI] [PubMed] [Google Scholar]
- 8.Aghdassi A, et al. Recruitment of histone deacetylases HDAC1 and HDAC2 by the transcriptional repressor ZEB1 downregulates E-cadherin expression in pancreatic cancer. Gut. 2012;61(3):439–448. doi: 10.1136/gutjnl-2011-300060. [DOI] [PubMed] [Google Scholar]
- 9.Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop—a motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11(9):670–677. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brabletz S, et al. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO J. 2011;30(4):770–782. doi: 10.1038/emboj.2010.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Browne G, Sayan AE, Tulchinsky E. ZEB proteins link cell motility with cell cycle control and cell survival in cancer. Cell Cycle. 2010;9(5):886–891. doi: 10.4161/cc.9.5.10839. [DOI] [PubMed] [Google Scholar]
- 12.Wellner U, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11(12):1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 13.Gregory PA, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 14.Burk U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9(6):582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bracken CP, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68(19):7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 16.Zheng S, El-Naggar AK, Kim ES, Kurie JM, Lozano G. A genetic mouse model for metastatic lung cancer with gender differences in survival. Oncogene. 2007;26(48):6896–6904. doi: 10.1038/sj.onc.1210493. [DOI] [PubMed] [Google Scholar]
- 17.Gibbons DL, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23(18):2140–2151. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu C, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17(2):211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang B, et al. TGFbeta-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene. 2010;29(12):1787–1797. doi: 10.1038/onc.2009.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiss FU, et al. Retinoic acid receptor antagonists inhibit miR-10a expression and block metastatic behavior of pancreatic cancer. Gastroenterology. 2009;137(6):2136–2145. doi: 10.1053/j.gastro.2009.08.065. [DOI] [PubMed] [Google Scholar]
- 21.Meng F, Wehbe-Janek H, Henson R, Smith H, Patel T. Epigenetic regulation of microRNA-370 by interleukin-6 in malignant human cholangiocytes. Oncogene. 2008;27(3):378–386. doi: 10.1038/sj.onc.1210648. [DOI] [PubMed] [Google Scholar]
- 22.Huang X, et al. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell. 2009;35(6):856–867. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferretti E, et al. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J. 2008;27(19):2616–2627. doi: 10.1038/emboj.2008.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao XN, et al. MicroRNA-193a represses c-kit expression and functions as a methylation-silenced tumor suppressor in acute myeloid leukemia. Oncogene. 2011;30(31):3416–3428. doi: 10.1038/onc.2011.62. [DOI] [PubMed] [Google Scholar]
- 25.Yan D, et al. MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. J Biol Chem. 2009;284(43):29596–29604. doi: 10.1074/jbc.M109.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miko E, et al. miR-126 inhibits proliferation of small cell lung cancer cells by targeting SLC7A5. FEBS Lett. 2011;585(8):1191–1196. doi: 10.1016/j.febslet.2011.03.039. [DOI] [PubMed] [Google Scholar]
- 27.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siemens H, et al. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10(24):4256–4271. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- 29.Su X, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467(7318):986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fontemaggi G, et al. deltaEF1 repressor controls selectively p53 family members during differentiation. Oncogene. 2005;24(49):7273–7280. doi: 10.1038/sj.onc.1208891. [DOI] [PubMed] [Google Scholar]
- 31.Cho MS, Chan IL, Flores ER. DeltaNp63 transcriptionally regulates brachyury, a gene with diverse roles in limb development, tumorigenesis and metastasis. Cell Cycle. 2010;9(12):2434–2441. doi: 10.4161/cc.9.12.12051. [DOI] [PubMed] [Google Scholar]
- 32.Shintani Y, Maeda M, Chaika N, Johnson KR, Wheelock MJ. Collagen I promotes epithelial-to-mesenchymal transition in lung cancer cells via transforming growth factor-beta signaling. Am J Respir Cell Mol Biol. 2008;38(1):95–104. doi: 10.1165/rcmb.2007-0071OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhattacharjee A, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98(24):13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chitale D, et al. An integrated genomic analysis of lung cancer reveals loss of DUSP4 in EGFR-mutant tumors. Oncogene. 2009;28(31):2773–2783. doi: 10.1038/onc.2009.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomida S, et al. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. J Clin Oncol. 2009;27(17):2793–2799. doi: 10.1200/JCO.2008.19.7053. [DOI] [PubMed] [Google Scholar]
- 36.Gibbons DL, et al. Expression signatures of metastatic capacity in a genetic mouse model of lung adenocarcinoma. PLoS One. 2009;4(4):e5401. doi: 10.1371/journal.pone.0005401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420(6916):629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 38.Gimona M, Buccione R, Courtneidge SA, Linder S. Assembly and biological role of podosomes and invadopodia. Curr Opin Cell Biol. 2008;20(2):235–241. doi: 10.1016/j.ceb.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 39.Tomar A, Schlaepfer DD. Focal adhesion kinase: switching between GAPs and GEFs in the regulation of cell motility. Curr Opin Cell Biol. 2009;21(5):676–683. doi: 10.1016/j.ceb.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Savarese F, et al. Satb1 and Satb2 regulate embryonic stem cell differentiation and Nanog expression. Genes Dev. 2009;23(22):2625–2638. doi: 10.1101/gad.1815709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim NH, et al. p53 and microRNA-34 are suppressors of canonical Wnt signaling. Sci Signal. 2011;4(178):ra71. doi: 10.1126/scisignal.2001744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takagi S, Nakajima M, Kida K, Yamaura Y, Fukami T, Yokoi T. MicroRNAs regulate human hepatocyte nuclear factor 4alpha, modulating the expression of metabolic enzymes and cell cycle. J Biol Chem. 2010;285(7):4415–4422. doi: 10.1074/jbc.M109.085431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spaderna S, et al. A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology. 2006;131(3):830–840. doi: 10.1053/j.gastro.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 44.Spaderna S, Brabletz T, Opitz OG. The miR-200 family: central player for gain and loss of the epithelial phenotype. Gastroenterology. 2009;136(5):1835–1837. doi: 10.1053/j.gastro.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 45.Yang J, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117(7):927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 46.Mani SA, et al. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci U S A. 2007;104(24):10069–10074. doi: 10.1073/pnas.0703900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7(6):415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 48.Yang MH, et al. Bmi1 is essential in Twist1-induced epithelial-mesenchymal transition. Nat Cell Biol. 2010;12(10):982–992. doi: 10.1038/ncb2099. [DOI] [PubMed] [Google Scholar]
- 49.Shimono Y, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138(3):592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Proia TA, et al. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell. 2011;8(2):149–163. doi: 10.1016/j.stem.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kissil JL, et al. Requirement for Rac1 in a K-ras induced lung cancer in the mouse. Cancer Res. 2007;67(17):8089–8094. doi: 10.1158/0008-5472.CAN-07-2300. [DOI] [PubMed] [Google Scholar]
- 52.Mack NA, Whalley HJ, Castillo-Lluva S, Malliri A. The diverse roles of Rac signaling in tumorigenesis. Cell Cycle. 2011;10(10):1571–1581. doi: 10.4161/cc.10.10.15612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valastyan S, Weinberg RA. Roles for microRNAs in the regulation of cell adhesion molecules. J Cell Sci. 2011;124(pt 7):999–1006. doi: 10.1242/jcs.081513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Postigo AA, Depp JL, Taylor JJ, Kroll KL. Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. EMBO J. 2003;22(10):2453–2462. doi: 10.1093/emboj/cdg226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nishimura G, et al. DeltaEF1 mediates TGF-beta signaling in vascular smooth muscle cell differentiation. Dev Cell. 2006;11(1):93–104. doi: 10.1016/j.devcel.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 56.Eger A, et al. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24(14):2375–2385. doi: 10.1038/sj.onc.1208429. [DOI] [PubMed] [Google Scholar]
- 57.Boominathan L. The guardians of the genome (p53, TA-p73, and TA-p63) are regulators of tumor suppressor miRNAs network. Cancer Metastasis Rev. 2010;29(4):613–639. doi: 10.1007/s10555-010-9257-9. [DOI] [PubMed] [Google Scholar]
- 58.Rivetti d V, et al. p63-microRNA feedback in keratinocyte senescence. Proc Natl Acad Sci U S A. 2012;109(4):1133–1138. doi: 10.1073/pnas.1112257109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Knouf EC, et al. An integrative genomic approach identifies p73 and p63 as activators of miR-200 microRNA family transcription. Nucleic Acids Res. 2012;40(2):499–510. doi: 10.1093/nar/gkr731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ory B, et al. A microRNA-dependent program controls p53-independent survival and chemosensitivity in human and murine squamous cell carcinoma. J Clin Invest. 2011;121(2):809–820. doi: 10.1172/JCI43897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11(12):849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trang P, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther. 2011;19(6):1116–1122. doi: 10.1038/mt.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wiggins JF, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70(14):5923–5930. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Duncan R. The dawning era of polymer therapeutics. Nat Rev Drug Discov. 2003;2(5):347–360. doi: 10.1038/nrd1088. [DOI] [PubMed] [Google Scholar]
- 65.Estevez MC, et al. Nanoparticle–aptamer conjugates for cancer cell targeting and detection. Methods Mol Biol. 2010;624:235–248. doi: 10.1007/978-1-60761-609-2_16. [DOI] [PubMed] [Google Scholar]
- 66.Chen Y, Zhu X, Zhang X, Liu B, Huang L. Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy. Mol Ther. 2010;18(9):1650–1656. doi: 10.1038/mt.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brown CY, et al. Robust, reversible gene knockdown using a single lentiviral short hairpin RNA vector. Hum Gene Ther. 2010;21(8):1005–1017. doi: 10.1089/hum.2009.107. [DOI] [PubMed] [Google Scholar]
- 68.Flores ER, et al. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416(6880):560–564. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- 69.Creighton CJ, et al. Insulin-like growth factor-I activates gene transcription programs strongly associated with poor breast cancer prognosis. J Clin Oncol. 2008;26(25):4078–4085. doi: 10.1200/JCO.2007.13.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.