Abstract
Interleukin-12 (IL12) enhances anti-tumor immunity when delivered to the tumor microenvironment. However, local immunoregulatory elements dampen the efficacy of IL12. The identity of these local mechanisms used by tumors to suppress immunosurveillance represents a key knowledge gap for improving tumor immunotherapy. From a systems perspective, local suppression of anti-tumor immunity is a closed-loop system - where system response is determined by an unknown combination of external inputs and local cellular cross-talk. Here, we recreated this closed-loop system in vitro and combined quantitative high content assays, in silico model-based inference, and a proteomic workflow to identify the biochemical cues responsible for immunosuppression. Following an induction period, the B16 melanoma cell model, a transplantable model for spontaneous malignant melanoma, inhibited the response of a T helper cell model to IL12. This paracrine effect was not explained by induction of apoptosis or creation of a cytokine sink, despite both mechanisms present within the co-culture assay. Tumor-derived Wnt-inducible signaling protein-1 (WISP-1) was identified to exert paracrine action on immune cells by inhibiting their response to IL12. Moreover, WISP-1 was expressed in vivo following intradermal challenge with B16F10 cells and was inferred to be expressed at the tumor periphery. Collectively, the data suggest that (1) biochemical cues associated with epithelial-to-mesenchymal transition can shape anti-tumor immunity through paracrine action and (2) remnants of the immunoselective pressure associated with evolution in cancer include both sculpting of tumor antigens and expression of proteins that proactively shape anti-tumor immunity.
Introduction
Monoclonal antibodies form one of the largest classes of molecular targeted therapies for cancer.1 Therapies that target molecules relevant in the pathogenesis of cancer promise efficacy in defined patient groups with minimal side effects. Breast cancer is a prime example where a monoclonal antibody - trastuzumab - has remarkable efficacy in patients that overexpress the corresponding epidermal growth factor (EGF) receptor.2,3 While pre-clinical development of trastuzumab focused on tumor-intrinsic effects,4,5 clinical response correlates with the induction of anti-tumor immunity that includes antibody-dependent cell-mediated cytotoxicity.6,7 In contrast to the fixed epitope recognized by a monoclonal antibody, using the patient’s own immune system to provide a similarly selective but also adaptive therapy has intrigued immunologists and cancer biologists for decades.8 One of the key cytokines that promotes cell-mediated cytotoxic immunity to tumor-associated antigens is Interleukin-12.
Interleukin-12 (IL12) is an important cytokine that links innate to adaptive immunity and signals via a canonical Janus kinase (JAK) and signal transducer and activator of transcription (STAT) signaling pathway.9 Sufficient and sustained IL12 signaling polarizes naïve CD4+ T cells into a T helper type 1 (TH1) phenotype.10 In CD8+ Cytotoxic T Lymphocytes (CTLs), IL12 provides a critical third signal that, in conjunction with antigen-specific stimulation via the T cell receptor and co-stimulation, produces strong CTL effector function, survival, and memory formation.11 Originally called Natural Killer (NK) Cell Stimulating Factor, IL12 also enhances the ability of NK cells to lyse target cells, a mechanism exploited for tumor immunotherapy. As an adjuvant, IL12 promotes NK-cell mediated killing of breast cancer cells in patients treated with trastuzumab.12 Toxicity restricts the systemic delivery of IL12 as an adjuvant for tumor immunotherapy.13 However, CTLs engineered to express IL12 in the tumor microenvironment eradicate established B16 melanoma tumors in animals preconditioned to reduce the number of intratumoral CD4+ Foxp3+ T regulatory cells.14 Local delivery of IL12 increased the retention of CTLs and NK cells within the tumor microenvironment. This finding is important as it suggests that local delivery of IL12 into the tumor microenvironment is important for tumor regression and the presence of immunosuppressive elements within the tumor microenvironment limit the effectiveness of local delivery. Yet, how these immunosuppressive elements limit the local action of IL12 remains unclear.
Various hypotheses have been proposed regarding how tumor cells promote immunoescape. Within the tumor microenvironment, immunosuppressive cells are thought to catabolize cytokines and thereby deprive immune cells that enter the tumor microenvironment of biochemical cues necessary for sustained activity, a concept called a “cytokine sink.”15 In vitro, CD4+ Foxp3+ T regulatory cells function as “sinks” for key cytokines that regulate immune cell activity.16 Tumors induce the apoptosis of immune cells that enter the tumor microenvironment by expressing ligands for members of the tumor necrosis factor superfamily, such as Fas ligand (FasL/CD95L)17,18 or tumor necrosis factor (TNF)-related apoptosis-inducing lig- and (TRAIL).19,20 Indirectly, tumors also promote tolerance by producing biochemical cues that suppress immune function, including transforming growth factor β (TGF-β), IL6, and IL10.21,22 The objective of this study was to evaluate these competing hypotheses regarding how cells derived from an immunosuppressive cellular model for melanoma (e.g., the B16 cell model23) suppress the action of IL12. An in vitro co-culture model was used to evaluate three competing hypotheses that included formation of a cytokine sink for IL12, tumor-induced apoptosis, and secretion of biochemical cues that cross-regulate IL12 signaling.
Results
B16F0 cells suppress the cellular response to IL12 following an induction period
We created an in vitro model of local tumor-derived immunosuppression by co-culturing the B16F0 cell line with the 2D6 cell line, a model of TH1 effector cells that responds to IL12, and developed a high content assay to characterize the dynamic response of the 2D6 cells to IL12. A 2×2 factorial experimental design was used to parse the cellular response due to the direct effect of IL12 stimulation from potential immunosuppressive effects of B16F0 co-culture. In total, the resulting data set contained 567 data points that included measures of cell fate and key proteins associated with the IL12 signaling pathway. These measures were obtained at 6 data points, under four experimental conditions, and in technical triplicate. Flow cytometry was used to quantify the nature and dynamics of the cellular response of 2D6 cells to IL12. Viable 2D6 cell events were identified by cell morphology and confirmed by back-gating on membrane integrity, as indicated by 7-AAD staining (Figures 1A and B), and CD45 expression (Figure 1C). The specific measures of the 2D6 response to IL12 included expression of IL12Rβ2 (Figure 1D) and phosphorylation of STAT4 (pY693 - pSTAT4 - Figure 1E), integrative measures of IL12-dependent (IFN-γ and IL-10) and IL12-independent cytokine production (TNF-α), and an independent measure of IL12p70 stimulation. As expected, STAT4 became phosphorylated in response to IL12 (Figure 1B). However for subsequent analysis, the activation state of STAT4 was limited to live cells as fluorescence associated with pSTAT4 was decreased in cells that lost membrane integrity, as illustrated by 2D6 cells cultured in cRPMI plus IL12 for 30 hours (see right panel in Figure 1B). More importantly, the probability distribution functions for IL12Rβ2 and pSTAT4 exhibited unimodal distributions, whereby median values can be used as appropriate summary statistics.
Figure 1.

A high content assay characterized the immunosuppressive action of B16F0 cells on the cellular response to IL12. (A) Cytometry events were parsed into cellular events (dashed gate) and live cells (red oval) based upon a data-driven gate of the forward- (FSC-A) and side-scattering (SSC-A) properties. Representative results for three time points - 0 hours (left panel), 12 hours (center panels), and 30 hours (right panels) - and two stimulation conditions - cultured in cPRMI alone (top row) and in cRPMI plus IL12 (bottom row) - are shown. Dead or dying cells within the dashed gate were indicated by back-gating on 7-AAD staining (yellow shading). (B) Positive 7-AAD staining was identified using a data-driven threshold (dotted horizontal red line) such that 95% of the 2D6 cells cultured for 12 hours with IL12 exhibited a MFI below the threshold - center panel. 2D6 cells cultured for 12 hours in cRPMI alone (left panel) and 2D6 cells cultured in cRPMI plus IL12 for 30 hours (right panel) are shown for comparison. (C) CD45 was used to identify 2D6 cells in B16F0-2D6 co-culture experiments (unstained 2D6 - gray shaded, unstained B16F0 - red dashed line, CD45-stained B16F0 - blue line, and CD45-stained 2D6 - black line). Changes in IL12 receptor β2 (Panel D) and phosphorylated STAT4 (Panel E) were used to quantify intracellular changes in response to IL12 stimulation in 2D6 cells. The results from three 2D6 cell populations are shown: unstained cells (gray shaded), cells cultured in cRPMI for 12 hours (black solid line), and cells cultured with cRPMI plus IL12 for 12 hours (red dashed line). In panel B, the solid vertical red line indicates the data-driven threshold for background pSTAT4 (i.e., 95% of unstained cells exhibited a MFI below the threshold). In panels C – E, the dotted vertical line indicates the data-driven threshold for the upper limit of protein expression or activity for 95% of the unstained (Panel C - CD45, Panel D - IL12Rβ2) or unstimulated (Panel E - pSTAT4) 2D6 cells. Results representative of three technical replicates obtained for each of two biological replicates.
In analyzing the dynamics of the in vitro system, the addition of IL12 increased and co-culture of 2D6 cells with B16F0 cells decreased the survival of 2D6 cells in culture (Figure 2A and Supplemental Table S1, p < 001). Expression of components of the IL12 receptor were not significantly influenced by either IL12 stimulation or B16F0 co-culture (Supplemental Figure S1). The concentration of IL12 declined as a function of time (Figure 2B and Supplemental Figure S2) but remained above the IL12 EC50 of 0.2 pM (Supplemental Figure S3). As the 2D6 cell line is maintained in media supplemented with IL12, there was a basal level of STAT4 phosphorylation that declined with time in cells cultured in cRPMI alone (Figure 2C). As expected, STAT4 became phosphorylated in response to IL12 and remained phosphorylated for the duration of the experiment with 2D6 cells alone. However, the STAT4 response in 2D6 cells co-cultured with B16F0 cells was initially similar to 2D6 cells cultured alone but exhibited a significant decrease in STAT4 phosphorylation at the 24 and 30 hour time points. The functional response of 2D6 cells to IL12 stimulation was also suppressed by B16F0 co-culture, as indicated by the concentrations of IL-10 and IFN-γ in the conditioned media were unchanged at the 24 and 30 hour time points compared to the 12 hour time point (Figures 2D and 2E). Independent of IL12 stimulation, 2D6 cells also make TNFα that was suppressed at the 24 and 30 hour time points following B16F0 co-culture (Figure 2F). The first 12 hours of the high content assay served as an internal control and indicates that there is no acute effect on 2D6 cells upon co-culture with B16F0 cells.
Figure 2.

B16F0 cells suppressed the cellular response to IL12. Changes in cell viability (A) and STAT4 activation (C) were quantified as a function of time by flow cytometry for the four experimental conditions (B16F0 + 2D6 co-culture - dotted lines, x’s and squares; 2D6 alone - solid lines, o’s and triangles; unstimulated - blue x’s and o’s; IL12 stimulated - red squares and triangles). The changes in cellular IL12Rβ2 and pSTAT4 were represented by median values of the unimodal distribution in MFI. Changes in IL12p70 (B) and enrichment of IL10 (D), IFNγ (E), and TNFγ (F) in the conditioned media were quantified as a function of time using cytometric bead array (mean ± SD, N = 3). Mean response at each time point and condition were used to create trend lines (solid and dotted lines). A two-sided Student’s t test was used to assess statistical significance, where * indicates p < 0.015. Results from three technical replicates are shown and are representative of two biological replicates. In panels D, E, and F, the cytometric bead results for 2D6 alone and 2D6 + IL12 are also reported in Klinke et al.33
One possible explanation of the observed suppression of the response to IL12 is that the addition of B16F0 into the in vitro culture locally deplete IL12 (i.e., B16F0 cells create a cytokine sink). We used model-based inference to estimate the rate of IL12 catabolism (Figure 3 and Supplemental Figure S6). Despite adding the same amount of IL12 to the same tissue culture volume, the initial concentrations in cultures containing B16F0 cells were approximately 30 pM lower than cultures with 2D6 cells alone. The drop in initial concentration could be due to an increase in the number of IL12 binding sites contained within the system. Copy numbers of IL12 receptor β1 and IL12 receptor beta2 expressed by B16F0, 2D6, and B16F10 cells were measured by flow cytometry (Figure 3). Given that B16F0 were observed to express 1.8 × 105 copies of IL12 receptor β2, the drop in IL12 in the media corresponds to an increase in fractional occupancy of IL12 receptor β2 on B16F0 cells of 0.2. A more important factor that could influence the observed dynamics is the ratio of the reaction rate of IL12 (i.e., the catabolism rate of IL12) to the rate of diffusion of IL12 within the system. The posterior distribution in the initial reaction rate for 2D6-B16F0 co-culture was narrower than 2D6 culture alone and shifted to higher values (Figure 3C). Using an initial reaction rate of 10−3 pM/sec, the Mears Criterion was estimated to be 1 × 10−6, which implies that external diffusion does not play a role in regulating 2D6 cell response to IL12 (see IL12 Sink Model in Supplemental File 1 for more detail). Collectively, the results of the high content co-culture assay suggest that the presence of B16F0 suppresses the response of 2D6 cells to IL12 following an induction period that is independent of a local reduction in IL12 (i.e., a cytokine sink effect) and of a reduction in cell viability. While both of these phenomena act within the co-culture assay, they are insufficient to explain the observed cessation in 2D6 cytokine production. An alternative hypothesis for the observed induction period is that B16F0 secrete soluble biochemical cues that accumulate within the system and act in a paracrine fashion to suppress the cellular response to IL12.
Figure 3.
B16F0 cells create a cytokine sink for IL12. Model-based inference was used to interpret changes in IL12 (panel A) in 2D6 culture (triangles and red lines) versus 2D6-B16F0 co-culture (squares and blue lines) due to differences in initial concentrations (panel B) and initial reaction rates (panel C). IL12 receptor β1 and β2 were observed to exhibit different patterns of expression in the B16 cells (B16F0 - Panel D, B16F10 - Panel E) compared to 2D6 cells (Panel F). Receptor copy number was quantified by flow cytometry and using Quantum Simply Cellular anti-mouse IgG-conjugated microspheres. In panel A, the solid lines correspond to the maximum expectation value of the model predictions and the dotted lines enclose 95% of the posterior distribution in the model predictions. One data point at the 30 hour time point and two data points at the 6 hour time point were determined to be outliers, as indicated by *.
B16 melanoma model secretes biochemical cues
To discover whether such a paracrine mechanism exists, we used a proteomics approach, based upon two-dimensional gel electrophoresis and MALDI-TOF mass spectrometry, to characterize the secretome of B16F0 cells. Proteins secreted from the B16 cells after 24h were resolved on a 2D-PAGE (Figure 4A and Supplemental Figure S7) and identified using peptide mass fingerprinting. Forty five proteins were identified, out of a total of 63 spots selected for identification, and 39 were unique (see Table 1 and Supplemental File 2). The secretome proteins represent diverse classes such as matricellular proteins (secreted protein, acidic and rich in cysteine (SPARC); Wnt-inducible signaling protein-1 (WISP-1)), metabolic enzymes (alpha enolase, pyruvate kinase), cytoskeletal components (actin), heat shock/chaperone proteins (Hsp90aa1, Hspa8, Calr, Pdia3) and annexins (annexin A4, A5, A6). These identified proteins were interpreted in the form of protein interaction network (Figure 4B) using IPA to identify their interaction partners and possibly under represented proteins. This protein interaction network had functions associated with ”post-translational modification” and ”protein folding” (p < 0:0001). Three proteins from the data set were involved with the MHC Class I complex: Hsp90b1, Pdia3 and Calr. The most significant pathways enriched in the secretome as a result of the association with the MHC Class I molecules were ”Lipid antigen presentation by CD1” (p < 0:001) and ”Antigen presentation pathway” (p < 0:005) with 26s proteasome being central to the network. High levels of MHC I molecules were previously shown to be present in exosomes that were secreted by dendritic cells.24 Upon investigating the ExoCarta exosome database,25 we found that out of the 39 identified proteins, 20 proteins associate with exosomes in humans and/or mice. Using a Fisher’s Exact Test, we found this functional annotation to be significant (p < 1e−10 - see Supplemental Table S3 for 2×2 Contingency table). In the protein interaction network, 13 out of 28 nodes were proteins associated with exosomes, with the cytoplasmic compartment of the interaction network more dominant with exosomal proteins. The enrichment of WISP-1 and alpha-enolase within the B16F0 conditioned media, as observed by western blot, were used to confirm the peptide mass fingerprinting results (Supplemental Figure S8A). WISP-1 was also observed to be expressed by the B16F10 cell line and to exhibit time-dependent enrichment in B16F0-conditioned media (Supplemental Figure S8C). GAPDH, a protein commonly associated with exosomes, was also observed to be enriched in B16F0 conditioned media and used to validate the exosome prediction (Supplemental Figure S8A). Of the proteins contained within the protein interaction network, SPARC and WISP-1 have not been associated with exosomes and were associated with the extracellular space. Collectively, the proteins identified in the B16F0 secretome suggest three potential paracrine biochemical actors within the co-culture model: exosomes, SPARC, and WISP-1.
Figure 4.

The secretome of B16F0 cells was characterized using a 2D-GE/MALDI-TOF MS proteomic workflow. (A) Medium conditioned by B16F0 cells was concentrated and resolved in the first dimension on IPG strip 4–7 pH, 7cm. The second dimension is a 12% SDS-PAGE spanning molecular weight region 10–100kDa, stained with Coomassie blue and scanned using Typhoon 9400 scanner. The spot numbers on the gel indicate the 45 identified proteins, shown in Table 1. (B) Inferred IPA protein network structure. Proteins identified in the conditioned media were overlaid onto a global molecular network developed from information contained in the Ingenuity Knowledgebase and arranged according to their subcellular localization. Genes or gene products are represented as nodes, and the biological relationship between two nodes is represented as an edge (line). Solid lines indicate a direct relationship and dashed lines indicate an indirect relationship between nodes. Nodes shaded gray are the focus molecules present in the data set and unshaded nodes are the inferred nodes from the Ingenuity Knowledgebase. Post-translational modification; protein folding; and DNA replication, recombination and repair were functional annotations associated with the network (p < 0.0001). The proteins highlighted by a solid box are associated with exosomes. PSMD10 mRNA has been associated with exosomes (dotted box).
Table 1.
List of proteins that were secreted by B16F0 cell line after 24 hours in serum free media, identified using 2D-GE/MALDI-TOF MS workflow, and formed the data set for IPA analysis. Proteins that were used as focus proteins in the protein interaction network (bolded) and that are known to be associated with exosomes (‡) are highlighted. The identification summary of these proteins including the peptide hits and sequence coverage are listed in Supplemental File 2.
| SPOT ID |
Protein Name | GENE SYMBOL |
Theoretical | |
|---|---|---|---|---|
| MW | pI | |||
| 1 | Endoplasmin | Hsp90b1‡ | 90 | 4.7 |
| 2 | Heat shock protein HSP 90-alpha | Hsp90aa1‡ | 85 | 4.9 |
| 4 | Calreticulin | Calr‡ | 46 | 4.3 |
| 5 | Rab GTPase-binding effector protein 2 | Rabep2 | 62 | 4.9 |
| 6 | Protein disulfide-isomerase | P4hb‡ | 55 | 4.8 |
| 8,35 | ATP synthase subunit beta, mitochondrial | Atp5b | 52 | 5 |
| 9 | SPARC | Sparc | 33 | 4.7 |
| 10 | Annexin A5 | Anxa5‡ | 36 | 4.8 |
| 14 | ATP synthase subunit d, mitochondrial | Atp5h | 19 | 5.5 |
| 15 | 26S proteasome non-ATPase regulatory subunit 10 | Psmd10 | 25 | 5.7 |
| 16 | N-acetyltransferase 9 | Nat9 | 28 | 5.2 |
| 17 | Prohibitin | Phb‡ | 30 | 5.5 |
| 18 | TIP41-like protein | Tiprl | 31 | 5.4 |
| 19 | Nuclear distribution protein nudE homolog 1 | Nde1 | 36 | 5.3 |
| 20 | Annexin A4 | Anxa4‡ | 36 | 5.4 |
| 25,28,29 | T-complex protein 1 subunit theta | Cct8‡ | 59 | 5.4 |
| 30 | RuvB-like 2 | Ruvbl2‡ | 51 | 5.5 |
| 32 | Tubulin alpha-1C chain | Tuba1c‡ | 50 | 5 |
| 33 | Na(+)/H(+) exchange regulatory cofactor NHE-RF3 | Pdzk1‡ | 56 | 5.3 |
| 36,37,38 | Actin, cytoplasmic 2 | Actg1‡ | 42 | 5.3 |
| 40 | Homer protein homolog 1 | Homer1 | 41 | 5.4 |
| 41 | Activator of 90 kDa heat shock protein ATPase homolog 1 | Ahsa1‡ | 38 | 5.4 |
| 42 | Guanine nucleotide-binding protein subunit alpha-11 | Gna11‡ | 42 | 5.7 |
| 43,45 | Protein disulfide-isomerase A3 | Pdia3‡ | 54 | 5.7 |
| 44 | Baculoviral IAP repeat-containing protein 4 | Xiap | 56 | 5.7 |
| 47 | Annexin A6 | Anxa6‡ | 76 | 5.3 |
| 48 | RNA polymerase I-specific transcription initiation factor | Rrn3 | 75 | 5.3 |
| 49 | Collagen type IV alpha-3-binding protein | Col4a3bp | 71 | 5.3 |
| 50 | Calpain-9 | Capn9 | 79 | 5.1 |
| 51 | Heat shock cognate 71 kDa protein | Hspa8‡ | 71 | 5.4 |
| 52 | Cytochrome P450 2D11 | Cyp2d11 | 57 | 6.2 |
| 53 | Pyruvate kinase isozymes M1/M2 | Pkm2‡ | 58 | 6.7 |
| 54 | 5-azacytidine-induced protein 2 | Azi2 | 46 | 6.2 |
| 56 | Ornithine aminotransferase, mitochondrial | Oat | 46 | 5.7 |
| 57 | Transcobalamin-2 | Tcn2‡ | 46 | 5.9 |
| 58 | WNT1-inducible-signaling pathway protein 1 | Wisp1 | 43 | 6 |
| 60 | Alpha-enolase | Eno1‡ | 47 | 6.4 |
| 62 | Cytosolic 5’-nucleotidase 3 | Nt5c3 | 37 | 6.2 |
| 63 | Ankyrin repeat domain-containing protein 16 | Ankrd16 | 40 | 6.5 |
WISP-1 suppresses 2D6 response to IL12
Given the emerging evidence of cross talk between the Wnt signaling pathway and immunity, 26 we hypothesized that WISP-1 plays a role in suppressing the cellular response to IL12. To test this hypothesis, we co-cultured 2D6 cells with B16F0 cells in the presence of a neutralizing antibody against WISP-1 (αWISP-1). Based upon the high content assay results, we selected the 32 hour time point to assay whether neutralization of WISP-1 would restore IL12 dependent activation of STAT4. A 2×2×2 factorial experimental design was used where IL12 stimulation, B16F0 co-culture, and addition of a neutralizing antibody against WISP-1 (αWISP-1) were the three independent variables. As expected, IL12 stimulation increased the activity of STAT4 in 2D6 cells cultured alone and co-culture with B16F0 cells decreased the activity of STAT4 (Figures 5A and 5C). The addition of αWISP-1 did not alter the activity of STAT4 in 2D6 cells cultured alone (Figures 5B and 5C). However, neutralization of WISP-1 did restore the activity of STAT4 in IL12 stimulated 2D6 cells when cultured in the presence of B16F0 cells (Figures 5B and 5C). The difference in pSTAT4 fluorescence intensity in IL12 stimulated 2D6 cells co-cultured with B16F0 cells plus αWISP-1 mAb was identified as significant using a Student’s t-test (p < 0:005) relative to B16F0 co-culture alone (negative control) and co-culture with an antibody isotype control (IC). As the viability of 2D6 cells was decreased at 32 hours (see Figure 2A) and cell viability had a confounding influence on STAT4 activity (see Figure 1B), we shifted the rmWISP-1 experiment to an earlier time point to increase basal viability. As the signal-to-noise properties of a biological assay influences inference,27 increasing doses of rmWISP-1 were used to demonstrate a causal relationship between WISP-1 and STAT4 activity and were used to compensate for a decrease in signal associated with an earlier time point (i.e., a decrease in the observed difference in STAT4 activity between control and co-cultured conditions as shown in Figure 2C). In IL-12 stimulated 2D6 cells, rmWISP-1 also inhibited STAT4 phosphorylation in a dose-dependent manner (Figure 5D - Pr(Effect < 0|data, M) > 0.9999), where model-based inference was used to assess the significance of WISP-1 inhibition (see Supplemental File 1). In addition, neutralization of WISP-1 in 2D6-B16F0 co-culture or addition of rmWISP-1 to 2D6 cells did not change cell viability relative to controls. Functionally, cytokines produced by 2D6 cells at 32 hours also reflected the changes observed in STAT4 activity upon addition of αWISP-1 (Figure 5E). Collectively, these data suggest that WISP-1 contributes to the observed decrease in response to IL12 upon co-culture with B16F0 cells and that the inhibition by WISP-1 is independent of changes in cell viability.
Figure 5.

WISP-1 inhibited cellular response to IL12. Neutralization of WISP-1 restored STAT4 activity (Panels A – C) and cytokine response to IL12 (Panel E), while rmWISP-1 dose-dependently inhibited STAT4 phosphorylation (Panels D). IL12Rβ2 and pSTAT4 were assayed by flow cytometry in 2D6 cells that were cultured for 32 hours under different conditions: +/− IL12 stimulation and +/− B16F0 co-culture. The assay was performed in the absence (Panel A) and the presence of an αWISP-1 antibody (Panel B). The median fluorescence intensity associated with STAT4 phosphorylation is summarized for all conditions (Panel C - mean ± SD, N ≥ 3). A significant difference in pSTAT4 fluorescence intensity in response to αWISP-1 mAb conditioning is indicated by ** (p < 0.001) relative to negative and antibody isotype control (IC) indicated by *. STAT4 phosphorylation was assayed in 2D6 cells that were stimulated for 24 hours with IL-12 and increasing concentrations of rmWISP-1 (Panel D). Model-based inference was used to quantify the extent and significance of rmWISP-1 inhibition of STAT4 activation (see Supplemental File 1). The solid line corresponds to the maximum expectation value of the model predictions and the dotted lines enclose 95% of the posterior distribution in the model predictions. One control replicate (indicated by *) was considered an outlier based upon an estimate of the experimental variance and excluded from the analysis. Using the indicated experimental conditions, TNFα, IFNγ, and IL10 were quantified in the conditioned media at 32 hours using cytometric bead array (Panel E: mean ± SD, N = 3). In the scatter plots, the horizontal dotted and vertical solid red lines indicate the data-driven thresholds for background fluorescence associated with pSTAT4 and IL12Rβ2 staining, respectively.
WISP-1 is expressed at the periphery of melanoma tumors
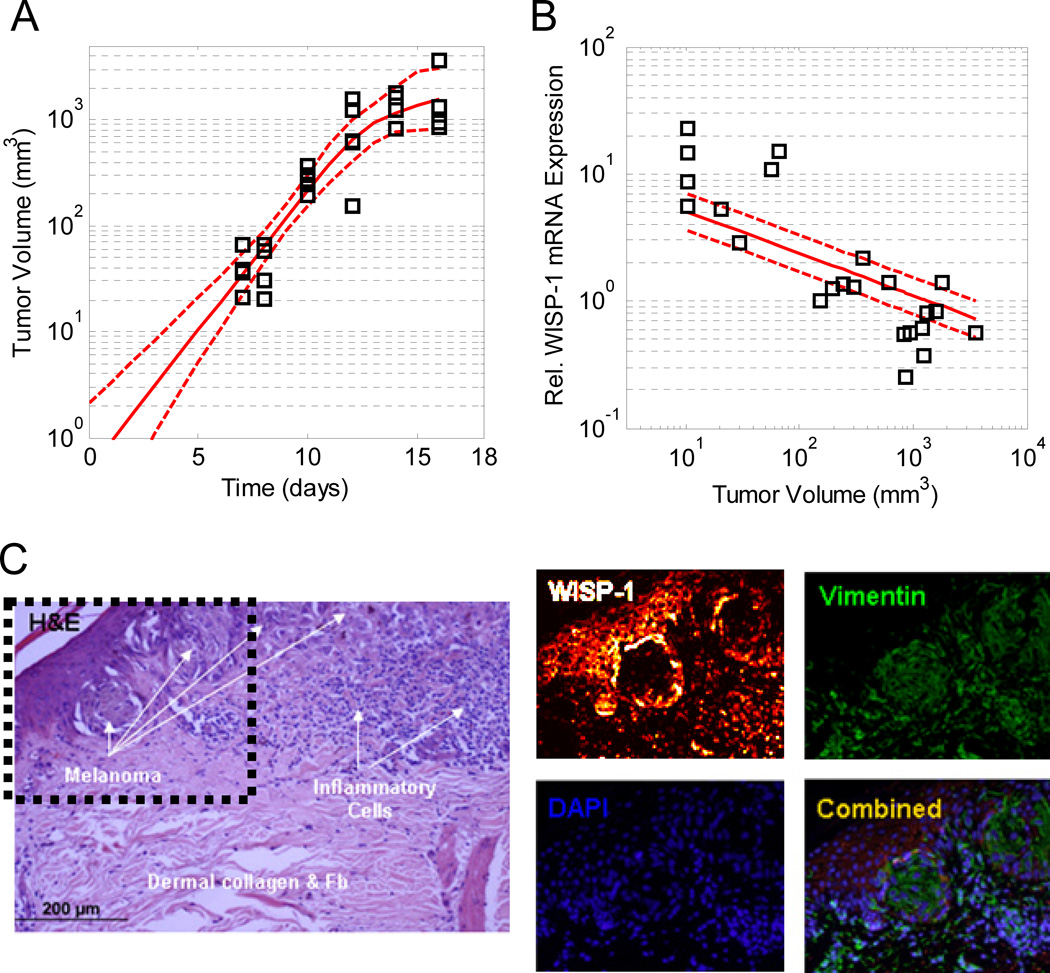
To ascertain whether WISP-1 expression may contribute to the nonimmunogenicity of the B16 cell lines, we assayed WISP-1 expression in B16F10-derived tumors following intradermal challenge in syngeneic hosts as an initial effort to translate our in vitro findings to more clinically relevant systems. Similar to B16F0 cells, WISP-1 was observed in B16F10-conditioned media (Supplemental Figure S8B). The injected B16F10 formed tumors that were palpable at day 5 and quantifiable at day 7 (Figure 6A). The growth rate was initially exponential and gradually decreased at day 12. Tumors were excised at various time points, homogenized, and assayed for WISP-1 mRNA by qRT-PCR (Figure 6B). Relative to GAPDH mRNA, WISP-1 expression was inversely proportional to tumor volume. To interpret this trend, we postulated three competing quantitative hypotheses regarding the observed trend: WISP-1 is expressed in the center of the expanding tumor (Model 1), all cells constitutively express WISP-1 (Model 2), and WISP-1 is expressed at the periphery of the expanding tumor (Model 3 - see Supplemental File 1 for more detail). Using in silico model-based inference, we found that the evidence favors Model 3 (i.e., Pr(Model 3|data) > 0.95), where the posterior distribution in the predictions for Model 3 is also shown in Figure 6B. The observed WISP-1 expression deviates from the best model predictions at the extremes, such that WISP-1 expression is greater than predicted in smaller tumors and less than predicted in larger tumors. One possible explanation for this deviation is that depth of the peripheral region that expresses WISP-1 may shrink as the tumor grows in size. Consistent with the prediction that WISP-1 is expressed in the periphery of B16F10 tumors, a similar pattern of WISP-1 expression is observed in human skin melanoma tissue (Figure 6C).
Figure 6.
The pattern of WISP-1 mRNA expression in B16F10-derived tumors in vivo is consistent with expression at the tumor periphery. The volume of B16F10-derived tumors increased with time following intradermal challenge with 105 B16F10 cells (Panel A). Model-based inference was used to extrapolate the observed tumor growth data. Normalized WISP-1 mRNA expression decreased with increasing tumor volume (Panel B). The lines correspond to the posterior distribution in the model predictions obtained from quantitative hypothesis 3: tumor cells in outer shell of the tumor express WISP-1 (see Supplemental File 1). The solid lines correspond to the maximum expectation value of the model predictions and the dotted lines enclose 95% of the posterior distribution in the model predictions. WISP-1 is expressed at the periphery of human melanoma nests (Panel C). As reported in46, sequential sections of human melanoma tissue samples were immunostained with hematoxylin and eosin (H&E) and antibodies against vimentin and WISP-1. Relative to Shao et al.46, the 8-bit image that represents WISP-1 staining was recolored using a black-red-orange-white color map. Nuclei were identified using DAPI. The dotted box in the H&E stained panel highlights region of interest around the melanoma nests.
Discussion
Identifying the cross talk between tumor and immune cells that occurs within the tumor microenvironment is becoming recognized as a critical knowledge gap for improving clinical efficiency of tumor immunotherapies.28 One of the challenges with understanding this cellular cross talk is that it occurs within the tumor microenvironment and that samples obtained from the periphery may not directly inform the identify of the biochemical cues responsible for tumor immunoescape29. This is not surprising as, from a systems perspective,30 the tumor and immune cells within the tumor microenvironment comprise a closed-loop system.9 In a closed-loop system, changes in system response (e.g., cytokine production by immune cells) cannot be predicted solely from system input (e.g., cytokine stimulation) but also reflect the cumulative effects of cross talk among components of the system (e.g., cytokine catabolism and secretion of counter-regulatory biochemical cues), as demonstrated by the cessation of cytokine production by the 2D6 model after 12 hour in vitro co-culture with the B16 melanoma cell model. While gene expression can be studied in cells isolated from excised tumors, increased gene expression is necessary but not sufficient to infer that the resulting gene product is released into the extracellular space and is able to exert a paracrine biological effect. At the protein level, targeted experimental studies, such as those using antibody microarrays, can be used to identify proteins that may facilitate intercellular communication. However, prior knowledge has a significant influence on the conclusions derived from such targeted studies as the experimental design presumes that the relevant proteins are contained within the set of probed antibodies. In summary, identifying the particular causal suppression mechanisms at work in a cancer from observations of biological state (e.g., immunohistology to reveal the frequency of infiltrating T cells and cytokine expression) is one of the most pervasive problems in the analysis of physiological systems.31
In designing this experimental study, we tried to minimize the influence of prior knowledge by creating an in vitro model of tumor-mediated immunosuppression that is amenable to proteomic characterization. Proteomics is an emerging tool for quantifying biochemical mechanisms for cell-to-cell communication.32 Previously, we have shown that the response of the 2D6 T helper cell model to IL12 can be described as an open-loop system.33 Here, we developed a high content assay to show that B16F0 cells, a nonimmunogenic tumor cell line, suppressed the response of the 2D6 T helper cell model to IL12 - a characteristic of a closed-loop system. We found that competing hypotheses, namely induction of apoptosis and creation of a cytokine sink, were insufficient to explain the observed phenomenon, despite the fact that they are acting within the system. To minimize the influence of bias, we used a proteomic workflow to identify secreted biochemical cues that were enriched in media conditioned by the B16F0 cell line. The proteomic analysis suggested the presence of SPARC, exosomes, and WISP-1 in the conditioned media. Finally, we confirmed that tumor-derived WISP-1 inhibited the immune cell response to IL12 and is upregulated in vivo.
WISP-1 is a member of the family of connective tissue growth factors that participate in stem cell differentiation and tumorigenesis.34 In syngeneic animal transplant models, WISP-1 gene expression is upregulated in pulmonary metastases derived from D122 Lewis lung carcinoma and B16F10 melanoma cells.35 In a mouse model of pulmonary fibrosis, WISP-1 induces epithelial-to-mesenchymal transition.36 Rat fibroblasts treated with recombinant or overexpressed WISP-1 showed increased proliferation and morphological transformation and injecting these cells in nude mice led to tumorigenesis.37 In humans, there are conflicting reports regarding the correlation of WISP-1 expression to clinical outcome.38,39 As evidenced by the in vivo dependence of WISP-1 expression to tumor size, sampling bias may explain these conflicting reports. While these prior studies highlight the direct role that WISP-1 plays in tumorigenesis, the data presented here suggest a paracrine role for WISP-1 in regulating anti-tumor immunity by modulating the response to IL-12. Given the presented data, we have not ruled out either SPARC or exosome-derived mRNA/miRNA as potential additional mechanisms that modulate different aspects of the system.
Conclusion
In summary, the biochemical cues secreted by the B16 tumor model in conjunction with the tumor-induced cytokine sink for IL-12 present some intriguing new targets for future in vivo validation. More importantly, this co-culture model illustrates that immunosuppressive tumors may use multiple mechanisms to suppress anti-tumor immunity. This is consistent with a view that cancer is a evolutionary process.40 Carcinogens and radiation introduce genetic variation - some of the mutations are beneficial while other mutations are detrimental to tumor cell survival. Evolutionary theory suggests that the resulting cancer cells are alternative solutions, encoded in genetic and epigenetic modifications, to a multivariate optimization problem where the tumor microenvironment and immune system provide the selective landscape. Prior work has shown that the immune system sculpts tumor antigens.41 Here, we observed that the B16 model used multiple mechanisms to suppress the local action of IL12, namely creating a sink for IL12 through the upregulation of IL12Rβ2 and secreting WISP-1. Given that IL12 is not required for in vitro culture of the B16 cell lines and that the B16 model is considered a spontaneous transplantable animal model for malignant melanoma, the data suggest that the immune system selects for tumor cells within this heterogeneous tumor cell population that exhibit an intrinsic ability to suppress antitumor immunity. Thus, the profile of antigens expressed by and, as shown here, the proteins expressed by tumor cells are remnants of that selective process. Interestingly, redundancy is a valuable strategy in engineering systems to perform robustly in the presence of external perturbations. It seems that redundancy is also an optimal strategy in oncogenesis. The presence of multiple inhibitory mechanisms that may differ among tumor cells in how they act upon common immunosurveillance pathways, like IL12, may frustrate translational research. However, this general quantitative systems approach may identify likely mechanisms in similar models for cancer and thereby inform therapeutic strategies designed to counteract these multiple escape mechanisms and to enhance the effectiveness of monotherapies, like anti-CTLA4.
Experimental
Antibodies and reagents
PE-conjugated mouse anti-mouse CD212 (IL12Rβ1), purified hamster anti-mouse IL12 receptor β2, FITC-conjugated mouse anti-armenian and syrian hamster IgG1 (G94-56), Alexa Fluor 647-conjugated mouse anti-STAT4 (pY693), PerCP-Cy5.5-conjugated Mouse Anti-Mouse CD45.2, BD Phosflow lyse/fix buffer 5X, BD Phosflow Perm Buffer III and BD Cytometric Bead Array Mouse Inflammation Kit were purchased from BD Biosciences (San Diego, CA). ChromPure mouse IgG, rat IgG, and sheep IgG (whole molecule) were purchased from Jackson Immuno Research (West Grove, PA). Recombinant mouse IL12p70 was purchased from eBioscience (San Diego, CA). Recombinant mouse WISP-1 (rmWISP-1 1680WS), anti-WISP1 (rat MAB1680 and sheep AF1680), anti-sheep IgG-HRP (HAF016) and an anti-rabbit IgG-HRP (HAF008) were purchased from R&D Systems (Minneapolis, MN). Quantum Simply Cellular uniform microspheres conjugated to anti-mouse IgG were purchased from Bangs Laboratories (Fishers, IN). Anti-biotin and anti-enolase-1 antibodies were purchased from Cell Signaling Technology (Danvers, MA) and anti-GAPDH was purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Trypsin-Versene Mixture was purchased from BioWhittaker (Walkersville, MD). DPBS 1X and RPMI-1640 were purchased from Mediatech (Manassas, VA). Sodium Azide (Na Azide) was purchased from United States Biochemical Corp (Cleveland, OH). All cell cultures were maintained at 37°C in 5% CO2 in RPMI 1640 plus supplements (referred to as complete RPMI or cRPMI). For every 500 ml of RMPI 1640, the RPMI supplements were 10% heat-inactivated Fetal Bovine Serum (FBS) (Hyclone, Logan, UT), 5ml of 100 I.U./ml Penicillin-Streptomycin (Biowhittaker, Walkersville, MD), 5ml of 200 mM L-Glutamine (Mediatech, Manassas, VA), 5ml of 100X Non-Essential Amino Acids (Chemicon International, Temecula, CA), 5ml of 100 mM Sodium Pyruvate (MP Biomedicals, Solon, OH), 5ml of 1M HEPES (Fisher Bioreagents, Pittsburgh, PA), and 2µl β-mercaptoethanol (Sigma Chemical, St. Louis, MO).
Mice, cell culture and stimulation
Female C57BL/6 mice were purchased from Charles River Breeding Laboratory (Wilmington, MA). All of our investigations have been approved by the McMaster Animal Research Ethics Board. An IL12-dependent cell line, 2D6, was kindly provided by Dr. Grusby (Harvard University, Cambridge, MA) and maintained as described in42. The B16F0 and B16F10 cell lines were purchased from American Tissue Culture Collection (ATCC, Bethesda, MD). For the proteomics experiments, B16F0 and B16F10 cells were grown in 75 cm2 tissue culture flask with 15ml cRPMI, which was replaced every 48 hours. Cells were cultured at 37°C and 5% CO2. For in vivo validation of WISP-1 expression, mice were challenged intradermally with 105 B16F10 cells. Tumor growth was monitored daily and measured with calipers every other day. Tumor volume was calculated as width × length × depth.
To assess the impact of B16F0-2D6 co-culture on the cellular response to IL12, 2×2 factorial experimental design was used with B16F0 co-culture and IL12 stimulation as the two factors. A saturation concentration of IL12 (150 pM) was selected for the IL12 stimulation conditions. This concentration of IL12 was greater than the effective concentration necessary to elicit a 50% of maximal response in STAT4 at 2 hours (EC50 = 0.2 pM - Figure S3). 2D6 cells were washed and plated at 6 × 104 cells per well in 96-well U-bottom plates for 12 hours with cRPMI to reduce background STAT4 activation (i.e., preconditioning). The volume in each well was 120 µl. After preconditioning, 6 × 104 B16F0 cells per well were added to the B16F0-2D6 co-culture groups, rmIL12p70 (150 pM) was added to the IL12 treatment groups, and DPBS was added into negative controls. Cells were subsequently cultured at 37°C in 5% CO2 for the indicated time period. At each time point, the plate was centrifuged 1250 RPM for 5 minutes at 4°C. The supernatant from each well was collected and stored at −20°C for subsequent cytometric bead analysis. To assess the contribution of a paracrine WISP-1 feedback loop in regulating 2D6 response to IL12, a 3×2 factorial experimental design was used at a single 32 hour time point. Cells were exposed to combinations of three factors: B16F0 co-culture, rmIL12p70 (150 pM), and a neutralizing antibody against WISP-1 (500 µg/ml). In a separate experiment, 2D6 cells alone were exposed to rmIL12p70 (150 pM) and increasing concentrations of rmWISP-1 for 24 hours. At least two biological replicates were performed for each experiment and three technical replicates were obtained for each biological replicate.
Flow cytometry and cytometric bead array
Stimulated 2D6 cells were fixed, permeabilized, and stained with fluorophore-conjugated antibodies specific for CD45, IL12Rβ1, IL12Rβ2, and phosphorylated STAT4 (pSTAT4), as described previously.42 The cells were analyzed using a FACSAria flow cytometer and FACSDiva Version 6.1.1 software (BD Biosciences). No stain controls were used as negative flow cytometry controls. Single stain controls were used to establish fluorescent compensation parameters. Cellular events were identified by forward and side scatter characteristics. On average, 2 × 104 events were analyzed at each timepoint. Concentrations of biochemical cues (IL12p70, IFN-γ, IL-10, and TNFα) contained within the cell culture media were measured using a cytometric bead array (BD Bioscience) and analyzed on a FACSCaliber with CellQuest Pro software. To determine the concentration of cytokines in each sample, BD cytometric bead array software was used. Flow cytometry data was exported as FCS3.0 files and analyzed using R/Bioconductor.43 The statistical difference between two treatment conditions was assessed using a two sample t-Test that assumes unequal variance, where a p-value of less than 0.05 was considered statistically significant.
RNA extraction from solid tumors and quantitative real-time PCR
Tumors were excised, snap-frozen in liquid nitrogen and stored at −80°C. Tumors were homogenized in Trizol (Invitrogen) using a Polytron PT 1200C (Kinematica) and total RNA was extracted according to the manufacturer’s specifications. RNA samples were further purified using an RNeasy mini kit (Qiagen) and treated with Ambion’s DNA-free kit. Reverse transcription was performed with Superscript III First-Strand (Invitrogen) according to the manufacturer’s instructions. Quantitative PCR was carried out on an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems) using Perfecta SYBR Green SuperMix, ROX (Quanta Biosciences). Reaction efficiency was determined for individual primer sets using a minimum of 5 serial dilutions to ensure similar efficiency between target and endogenous control reactions. Data were analyzed via the delta/delta CT method using the Sequence Detector Software version 2.2 (Applied Biosystems). Primer sequences were as follows: WISP-1 (FWD ACACATCAAGGCAGGGAAGA; REV GCAGCTCAGGTTGCAGAAG) and GAPDH (FWD AGGAGCGAGACCCCACTAAC; REV GGTTCACACCCATCACAAAC). GAPDH was used as an endogenous control. An empirical Bayesian approach was used to establish a level of significance associated three competing hypothesis regarding the dependence of normalized WISP-1 with tumor size.
Proteomics
Sample preparation for 2-DE and MALDI-TOF MS workow B16F0 cells were cultured in 175 cm2 flasks in the presence of Dulbecco’s modified eagle medium (DMEM) (Mediatech, Manassas, VA) medium supplemented with 10% (v/v) heat inactivated Fetal Bovine Serum (FBS) (Hyclone, Logan, UT), 100 I.U./ml Penicillin-Streptomycin (1% v/v) (Biowhittaker, Walkersville, MD). After 2 days, or when the cells were about 70% confluent, the medium was discarded, the cells were rinsed thrice with serum free DMEM. Cells were then incubated briefly in the serum free media (approximately 2 minutes) and this was collected as the 0 h secretome to screen for background proteins. Cells were further incubated in the serum free media for 24 h and this conditioned medium (CM) was collected as the 24 h secretome. The CM was spun down at 1200 rpm for 10 minutes at 4°C to remove cellular debris and stored at −80°C until further use. 500 mL CM was collected for both, 0h and 24h secretome, consisting of 10 biological replicates, and concentrated 100-fold using Centriprep centrifugal filters (Millipore, Carrigtwohill, Co.Cork, Ireland) with a molecular weight cutoff of 3-kDa and cleaned up using a 2-D Clean-Up Kit (GE Healthcare, Piscataway, NJ). The proteins were identified using a proteomic workflow based upon 2-D electrophoresis and MALDI-TOF mass spectrometry (see Supplemental File 1 for additional details regarding the proteomic workflow). Differentially regulated proteins identified by 2DE and peptide mass fingerprints were analyzed using Ingenuity Pathway Analysis (IPA; Ingenuity Systems, Mountain View, CA), as described previously.44 The proteomics results were subsequently validated using western blot, as described in Kulkarni et al.44, using primary antibodies specific for either WISP-1, GAPDH, or enolase-1. Recombinant mouse WISP-1 was used as a positive control.
Models and inference
In silico model-based inference was used to determine the significance associated with dynamic changes in the behavior of the observed system. Here we used mathematical models to interpret dynamic changes in cell viability in 2D6 cells upon in vitro culture, in the concentration of IL12 in the conditioned media, in the size of B16F10-derived tumors in vivo, and in WISP-1 expression within samples obtained from homogenized B16F10-derived in vivo tumors. The mathematical models were described using either algebraic relationships or ordinary differential equations. An empirical Bayesian approach was used to establish a level of confidence in the model predictions, given the available data and the uncertainty in the model parameters.45 Details are supplied in the Supplemental File 1.
Supplementary Material
Insight Statement.
Identifying local mechanisms for immunosuppression is a key knowledge gap for improving the efficacy of immunotherapies for cancer. Leveraging concepts from the analysis of physiological systems, an integrated in vitro-in silico-in vivo approach was developed to evaluate competing hypotheses regarding tumor-mediated suppression of immune response to Interleukin-12, a key cytokine that helps shape anti-tumor immunity. Collectively, the data suggest that (1) biochemical cues associated with epithelialto- mesenchymal transition can shape anti-tumor immunity through paracrine action and (2) remnants of the immunoselective pressure associated with evolution in cancer include both sculpting of tumor antigens and expression of proteins that proactively shape anti-tumor immunity.
Acknowledgements
We thank N. Cheng for assistance with some of the high content assays, J. Kaiser for performing one of the western blots, and J. Barnett for access to laboratory facilities. This work was supported by grants from the Terry Fox Foundation (JLB), the National Science Foundation (CAREER 1053490 to DJK), the National Cancer Institute (R15CA132124 to DJK), the National Institute of Allergy and Infectious Diseases (R56AI076221 to DJK), and National Institute of Health (RR016440 and RR020866). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Science Foundation, the National Cancer Institute, the National Institute of Allergy and Infectious Diseases, or the National Institutes of Health.
Footnotes
Author contributions: DJK conceived and coordinated the study; YK and JW performed the proteomics experiments; EC performed the high content assay experiments; AJRM and JB provided in vivo measurement of WISP-1 expression; YK and DJK analyzed the data; DJK performed the in silico model-based inference; DJK drafted the manuscript; and all of the authors discussed the results and approved the final manuscript.
Competing interests The authors declare that they have no competing financial interest.
References
- 1.Weiner LM, Dhodapkar MV, Ferrone S. Lancet. 2009;373:1033–1040. doi: 10.1016/S0140-6736(09)60251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlotter CM, Vogt U, Allgayer H, Brandt B. Breast Cancer Res. 2008;10:211. doi: 10.1186/bcr2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Widakowich C, de Azambuja E, Gil T, Cardoso F, Dinh P, Awada A, Piccart-Gebhart M. Int J Biochem Cell Biol. 2007;39:1375–1387. doi: 10.1016/j.biocel.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 4.Yarden Y. Oncology. 2001;61:1–13. doi: 10.1159/000055396. [DOI] [PubMed] [Google Scholar]
- 5.Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Jr WD, Leahy DJ. Nature. 2003;421:756–760. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- 6.Herpen CMV, van der Laak JA, de Vries IJM, van Krieken JH, de Wilde PC, Balvers MG, Adema GJ, Mulder PHD. Clin Cancer Res. 2005;11:1899–1909. doi: 10.1158/1078-0432.CCR-04-1524. [DOI] [PubMed] [Google Scholar]
- 7.Bekaii-Saab TS, Roda JM, Guenterberg KD, Ramaswamy B, Young DC, Ferketich AK, Lamb TA, Grever MR, Shapiro CL, Carson IWE. Mol Cancer Ther. 2009;8:2983–2991. doi: 10.1158/1535-7163.MCT-09-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunn GP, Old LJ, Schreiber RD. Annu Rev Immunol. 2004;22:329–360. doi: 10.1146/annurev.immunol.22.012703.104803. [DOI] [PubMed] [Google Scholar]
- 9.Klinke DJ. Mol. Cancer. 2010;9:242. doi: 10.1186/1476-4598-9-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Athie-Morales V, Smits HH, Cantrell DA, Hilkens CMU. J Immunol. 2004;172:61–69. doi: 10.4049/jimmunol.172.1.61. [DOI] [PubMed] [Google Scholar]
- 11.Curtsinger JM, Mescher MF. Curr Opin Immunol. 2010;22:333–340. doi: 10.1016/j.coi.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parihar R, Nadella P, Lewis A, Jensen R, De HC, Dierksheide JE, Van-Buskirk AM, Magro CM, Young DC, Shapiro CL, Carson IWE. Clin Cancer Res. 2004;10:5027–5037. doi: 10.1158/1078-0432.CCR-04-0265. [DOI] [PubMed] [Google Scholar]
- 13.Colombo MP, Trinchieri G. Cytokine Growth Factor Rev. 2002;13:155–168. doi: 10.1016/s1359-6101(01)00032-6. [DOI] [PubMed] [Google Scholar]
- 14.Kerkar SP, Muranski P, Boni A, Kaiser A, Boni A, Sanchez-Perez L, Yu ZY, Palmer D, Reger R, Borman ZA, Zhang L, Morgan RA, Gattinoni L, Rosenberg SA, Trinchieri G, Restifo NP. Cancer Res. 2010;70:6725–6734. doi: 10.1158/0008-5472.CAN-10-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gattinoni L, Finkelstein SE, Klebanoff CA, Antony PA, Palmer DC, Spiess PJ, Hwang LN, Yu ZY, Wrzesinski C, Heimann DM, Surh CD, Rosenberg SA, Restifo NP. J Exp Med. 2005;202:907–912. doi: 10.1084/jem.20050732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feinerman O, Jentsch G, Tkach KE, Coward JW, Hathorn MM, Sneddon MW, Emonet T, Smith KA, Altan-Bonnet G. Mol Syst Biol. 2010;6:437. doi: 10.1038/msb.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Connell J, O’Sullivan GC, Collins JK, Shanahan F. J Exp Med. 1996;184:1075–1082. doi: 10.1084/jem.184.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryan AE, Shanahan F, O’Connell J, Houston AM. Cancer Res. 2005;65:9817–9823. doi: 10.1158/0008-5472.CAN-05-1462. [DOI] [PubMed] [Google Scholar]
- 19.Giovarelli M, Musiani P, Garotta G, Ebner R, Di CE, Kim Y, Cappello P, Rigamonti L, Bernabei P, Novelli F, Modesti A, Coletti A, Ferrie AK, Lollini PL, Ruben S, Salcedo T, Forni G. J Immunol. 1999;163:4886–4893. [PubMed] [Google Scholar]
- 20.McLornan DP, Barrett HL, Cummins R, McDermott U, McDowell C, Conlon SJ, Coyle VM, Van SS, Wilson R, Kay EW, Longley DB, Johnston PG. Clin Cancer Res. 2010;16:3442–3451. doi: 10.1158/1078-0432.CCR-10-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicolini A, Carpi A, Rossi G. Cytokine Growth Factor Rev. 2006;17:325–337. doi: 10.1016/j.cytogfr.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Ben-Baruch A. Breast Cancer Res. 2003;5:31–36. doi: 10.1186/bcr554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fidler IJ, Nicolson GL. J Natl Cancer Inst. 1976;57:1199–1202. doi: 10.1093/jnci/57.5.1199. [DOI] [PubMed] [Google Scholar]
- 24.Wiley RD, Gummuluru S. Proc Natl Acad Sci U. S. A. 2006;103:738–743. doi: 10.1073/pnas.0507995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathivanan S, Simpson RJ. Proteomics. 2009;9:4997–5000. doi: 10.1002/pmic.200900351. [DOI] [PubMed] [Google Scholar]
- 26.Gattinoni L, Ji Y, Restifo NP. Clin Cancer Res. 2010;16:4695–4701. doi: 10.1158/1078-0432.CCR-10-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klinke DJ, Ustyugova IV, Brundage KM, Barnett JB. Biophys. J. 2008;94:4249–4259. doi: 10.1529/biophysj.107.120451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fox BA, Schendel DJ, Butterfield LH, Aamdal S, Allison JP, Ascierto PA, Atkins MB, Bartunkova J, Bergmann L, Berinstein N, Bonorino CC, Borden E, Bramson JL, Britten CM, Cao X, Carson WE, Chang AE, Characiejus D, Choudhury AR, Coukos G, de Gruijl T, Dillman RO, Dolstra H, Dranoff G, Durrant LG, Finke JH, Galon J, Gollob JA, Gouttefangeas C, Grizzi F, Guida M, Hakansson L, Hege K, Herberman RB, Hodi FS, Hoos A, Huber C, Hwu P, Imai K, Jaffee EM, Janetzki S, June CH, Kalinski P, Kaufman HL, Kawakami K, Kawakami Y, Keilholtz U, Khleif SN, Kiessling R, Kotlan B, Kroemer G, Lapointe R, Levitsky HI, Lotze MT, Maccalli C, Maio M, Marschner JP, Mastrangelo MJ, Masucci G, Melero I, Nelief C, Murphy WJ, Nelson B, Nicolini A, Nishimura MI, Odunsi K, Ohashi PS, O’Donnell-Tormey J, Old LJ, Ottensmeier C, Papamichail M, Parmiani G, Pawelec G, Proietti E, Qin S, Rees R, Ribas A, Ridolfi R, Ritter G, Rivoltini L, Romero PJ, Salem ML, Scheper RJ, Seliger B, Sharma P, Shiku H, Singh-Jasuja H, Song W, Straten PT, Tahara H, Tian Z, van Der Burg SH, von Hoegen P, Wang E, Welters MJ, Winter H, Withington T, Wolchok JD, Xiao W, Zitvogel L, Zwierzina H, Marincola FM, Gajewski TF, Wigginton JM, Disis ML. J Transl Med. 2011;9:214. doi: 10.1186/1479-5876-9-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gajewski TF, Fuertes M, Spaapen R, Zheng Y, Kline J. Curr Opin Immunol. 2011;23:286–292. doi: 10.1016/j.coi.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mullard A. Nature Rev Drug Disc. 2011;10:894. [Google Scholar]
- 31.Khoo MCK. Physiological control systems: analysis, simulation, and estimation. IEEE Press; 2000. [Google Scholar]
- 32.Jørgensen C, Sherman A, Chen GI, Pasculescu A, Poliakov A, Hsiung M, Larsen B, Wilkinson DG, Linding R, Pawson T. Science. 2009;326:1502–1509. doi: 10.1126/science.1176615. [DOI] [PubMed] [Google Scholar]
- 33.Klinke DJ, Cheng N, Chambers E. Sci Signal. 2011 doi: 10.1126/scisignal.2002657. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuo GW, Kohls CD, He BC, Chen L, Zhang W, Shi Q, Zhang BQ, Kang Q, Luo J, Luo X, Wagner ER, Kim SH, Restegar F, Haydon RC, Deng ZL, Luu HH, He TC, Luo Q. Histol Histopathol. 2010;25:795–806. doi: 10.14670/hh-25.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Margalit O, Eisenbach L, Amariglio N, Kaminski N, Harmelin A, Pfeffer R, Shohat M, Rechavi G, Berger R. Br J Cancer. 2003;89:314–319. doi: 10.1038/sj.bjc.6600977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konigshoff M, Kramer M, Balsara N, Wilhelm J, Amarie OV, Jahn A, Rose F, Fink L, Seeger W, Schaefer L, Gunther A, Eickelberg O. J Clin Invest. 2009;119:772–787. doi: 10.1172/JCI33950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu L, Corcoran RB, Welsh JW, Pennica D, Levine AJ. Genes Dev. 2000;14:585–595. [PMC free article] [PubMed] [Google Scholar]
- 38.Davies SR, Davies ML, Sanders A, Parr C, Torkington J, Jiang WG. Int J Oncol. 2010;36:1129–1136. doi: 10.3892/ijo_00000595. [DOI] [PubMed] [Google Scholar]
- 39.Davies SR, Watkins G, Mansel RE, Jiang WG. Ann Surg Oncol. 2007;14:1909–1918. doi: 10.1245/s10434-007-9376-x. [DOI] [PubMed] [Google Scholar]
- 40.Greaves M, Maley CC. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, Schreiber RD. Nature. 2001;410:1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 42.Finley SD, Gupta D, Cheng N, Klinke DJ. Immunol Cell Biol. 2011;89:100–110. doi: 10.1038/icb.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klinke DJ, Brundage KM. Cytometry A. 2009;75:699–706. doi: 10.1002/cyto.a.20746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kulkarni YM, Suarez V, Klinke DJ. BMC. Cancer. 2010;10:291. doi: 10.1186/1471-2407-10-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klinke DJ. BMC Bioinform. 2009;10:371. doi: 10.1186/1471-2105-10-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shao H, Cai L, Grichnik JM, Livingstone AS, Velazquez OC, Liu ZJ. Oncogene. 2011;30:4316–4326. doi: 10.1038/onc.2011.142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.