Abstract
Identifying the molecular and cellular basis of complex neuropsychiatric disorders (cNPDs) has been limited by the inaccessibility of central neurons, variability within broad diagnostic classifications, and the interplay of genetic and environmental factors. Recent work utilizing neuronally differentiated human induced pluripotent stem cells (hiPSCs) from Mendelian and polygenic cNPDs is beginning to illuminate neuritic, synaptic or cell body variations accompanied by specific gene or protein expression alterations largely mimicking known pathology. In some cases, phenotypes have only emerged after application of cellular stress or long duration of differentiation. Pathological and cellular expression features are fully or partially responsive to pharmacological treatment highlighting the potential utility of differentiated hiPSCs for discovery of personalized therapeutics and for identifying pathogenetically relevant targets in subgroups of patients within a broad syndromic classification. Because of the inherent variability in developing and differentiating hiPSC lines and the multiple comparisons implicit in ‘omics technologies, rigorous algorithms for assuring statistical significance and independent confirmation of results, will be required for robust modeling of cNPDs.
Introduction
The major neuropsychiatric disorders share several features that encumber disease modeling and drug discovery. As in most complex disorders, symptomatology in neuropsychiatric disease results from a complicated interplay between nature and nurture. The diagnostic categories, as determined by a consensus of authorities in the field (e.g. the American Psychiatric Association‘s Diagnostic and Statistical Manual of Mental Disorders [DSM]), are often composed of multiple endophenotypes that have distinct but also overlapping etiologies. Moreover, living brain tissue is largely inaccessible for laboratory studies. Finally, it is difficult to extrapolate from non-primate animal behavioral models to specific human diseases, and likely insufficient to address higher order human neural processing [1].
Recent cell culture advances have allowed the routine conversion of patient-specific fibroblasts (or other ostensibly committed somatic cells) to cells that emulate primordial pluripotent human embryonic stem cells (hESCs) including the capacity to give rise to diverse tissue- and organ-specific cell types [1a,1b]. These reprogrammed cells have been termed “human induced pluripotent stem cells (hiPSCs)”. Other emerging technologies have demonstrated the ability to convert fibroblasts directly to neurons without an intervening stem or progenitor cell stage (so called “induced neurons [iNs]”) [2] to change fibroblasts directly to neural stem/progenitor cells [3] or to render neural progenitor cells indefinitely but conditionally self-renewing (“induced conditional self-renewing progenitor [ICSP] cells”) [4]. All of these techniques offer the potential for modeling cNPDs in vitro given that the resulting neural cells have been derived directly from the affected patient with the disorder in question and presumably harbor all of that individual‘s genetic predispositions. For the purpose of this review, we will focus solely on the potential of hiPSCs to model neuropsychiatric disorders given that the other techniques have been less well-studied to date.
hiPSCs derived from patients with a variety of neurologic conditions have been differentiated in vitro towards electrophysiologically active neurons with characteristics consistent with forebrain, dopaminergic, glutamatergic, or motoneuronal phenotypes, among others [5]. Neurons differentiated from hiPSCs have been shown to engraft and project axons after transplantation in rodent brains at least in the case of a Parkinson‘s disease (PD) model [6].
With regard to modeling “diseases-in-a-dish”, two major questions can in principle be asked. First, can a cellular phenotype be discerned within hiPSC-derived neural cultures that is representative and predictive of the pathophysiology underlying the disease-of-interest and can it be altered in vitro such that a potential therapy for actual patients might emerge? Second, and of greater complexity, how can the environmental stimuli that contribute to the emergence of cNPDs be modeled, and the effects understood, and possibly reversed or slowed in cultures of hiPSCs undergoing neuronal and glial differentiation and maturation in vitro? Though this field is still in its infancy, some progress has been made on both fronts. While this progress has been greatest in Mendelian disorders of the brain, secrets regarding aspects of environmentally-influenced complex disorders are also starting to be revealed.
To date, abnormalities have been discerned in hiPSC-derived neurons from four Mendelian disorders: morphological and electrophysiological aberrations have been noted in spinal muscular atrophy (SMA) and Rett syndrome (RS) hiPSC-derived neurons; stress sensitivity in dopaminergic neurons derived from LRRK-associated Parkinson‘s Disease (PD) hIPSCs; and reduced neuron-generating potential in hIPSCs derived from cases of familial dysautonomia. Work in these models has begun to pave a path toward understanding how drug discovery could be accomplished using patient-specific hiPSCs. In contrast, disorders such as schizophrenia and sporadic PD reinforce the difficulty of establishing informative cell models without a clear starting point for investigation, whether by specific genetic or environmental signals or even a definitive, pathologically involved cell type.
Advances in modeling of Mendelian neuropsychiatric disorders with hiPSCs
As noted above, one of the first Mendelian disorders modeled using hiPSCs was SMA, a motor neuron defect caused by decreased levels of the protein called “survival motor neuron protein 1” (SMN-1). Differentiation of hiPSCs from such patients displayed a selective and progressive reduction in motor neuron number with reduced motor neuron size as well as abnormalities in markers of mature synapses in long duration cultures. The SMN protein levels and quantities of nuclear aggregates, termed “gems”, associated with SMA could be increased by tobramycin or valproic acid, compounds previously reported to affect protein levels. Disappointingly, these increases in SMN protein alone were not sufficient for restoring motor neuron numbers [7], perhaps reflecting exposure too late in differentiation or the action of as-yet-unknown pathological factors. Nonetheless, this study demonstrated that hiPSC-derived neurons in culture may mirror some of the pathophysiological hallmarks of a neurological disease, and that a drug effect, however incomplete in the present case, can be measured. Converting such a study into a drug screening program seems a challenging yet conceivable next step.
A study employing hiPSCs generated from patients with familial IKBKAP-linked dysautonomia introduced the use of mRNA transcriptome profiling to compare “diseased” hiPSC-derived neurons to control neurons, an unbiased approach that broadens the potential of hiPSC derivatives to uncover features not apparent by morphology or any one particular phenotype. The study found that expression of ~100 genes was altered as a consequence of the IKBKAP point mutation, many highly relevant to neural crest differentiation, such as decreases of ASCL1 or Mash1, and of several genes expressed selectively in neural crest cells. Indeed, a defect was observed in the number and migration of neural crest cells that emerged from hiPSC lines consistent with the expression deficits.
As a proof-of-concept that this in vitro model might be used to select pharmacological interventions, the authors found, after testing a number of candidate compounds, that kinetin partially suppressed defective splicing and specifically increased IKBKAP levels in hiPSC-derived neural crest precursor cells from familial dysautonomia but not unaffected patients [8]. Importantly, incubation with kinetin during the initial differentiation of the pluripotent cells, but not the hiPSC-derived neural crest precursors, also promoted partial restoration of neural crest cell number (though not improved migration). These early attempts at restoration of normal function with drug therapy appeared to suggest a critical period during which drug exposure pre-empted development of the abnormal phenotype. Whether or not medicines that operate this early in differentiation can be used safely for such a purpose remains an open question. As a cautionary note, this study also revealed defects in endodermal differentiation found only by rigorously tracking the differentiation of each germ layer. Should these findings prove robust, it is raises concern that such an unintended drug action would have been missed if direct conversion of fibroblasts to neurons via the iN method [2] were used instead of iPSCs.
For Rett syndrome (RS), an X-linked neurodevelopmental disorder largely associated with mutations in the DNA-methyl-binding protein MecP2, the study of hiPSCs from affected patients both confirmed known aspects of the disease‘s pathophysiology and offered some new insights. Retrotransposon DNA target sequences of MecP2 are thought to be preferentially expressed in neurons, suggesting the importance of examining MecP2 function in differentiated neural cells rather than in pluripotent cells or non-neural clinically accessible tissues [9]. RS hiPSC-derived neurons possess abnormal dendritic spine density and neurite formation similar to those observed in brains of murine RS models and post-mortem brain samples. Recordings from these neurons reveal altered electrophysiological potentials reminiscent of those recorded in mouse models. Despite similar cell cycle populations, the cell body sizes of RS-derived neurons are smaller suggesting a link to the brain volumes reductions observed in RS patients. RS hiPSC-derived neurons exhibited improved morphological parameters in the presence of insulin-like-growth factor (IGF-1) which had been previously demonstrated to improve the viability of mouse RS models [10].
Extending the hiPSC approach to a multifactorial disorder, models of PD have been generated from sporadic cases [11] as well as from early-onset PD patients with mutations of either LRRK2 or PINK1. hiPSC neurons derived from a single LRRK2 patient showed an increase in alpha-synuclein protein, a finding commonly observed post-mortem in PD, and increased susceptibility to peroxide-induced death. In addition, oxidative stress also produced alterations in gene expression in LRRK2 mutant neurons [12]. Caspase 3 immunofluorescence was increased in tyrosine hydroxylase (TH) immunoreactive neurons derived from LRRK2 mutant Parkinson‘s hiPSCs compared to TH-negative and control hiPSC-derived neurons after exposure to each of three compounds: 6-hydroxydopamine, a proteasome inhibitor MG-132, or hydrogen peroxide. (Other measures of apoptosis, unfortunately, were not made). Neuronal cultures derived from PINK-1-associated PD hiPSCs lacked differentiation or morphological differences from control hiPSC-derived neurons, but both Parkin translocation and mitochondrial copy number in response to mitochondrial depolarization were abnormal, indicating the requirement for PINK1 in Parkin function and mitochondrial autophagy in response to stress [13]. To date, no abnormalities have been identified in hiPSC neurons from sporadic cases of PD [11], although these lines still need to be evaluated in all the models established for the genetic cases. Given that sporadic and PINK-1 cultures did not exhibit changes in synuclein, how the common pathophysiological feature of a synucleinopathy arises remains an open question.
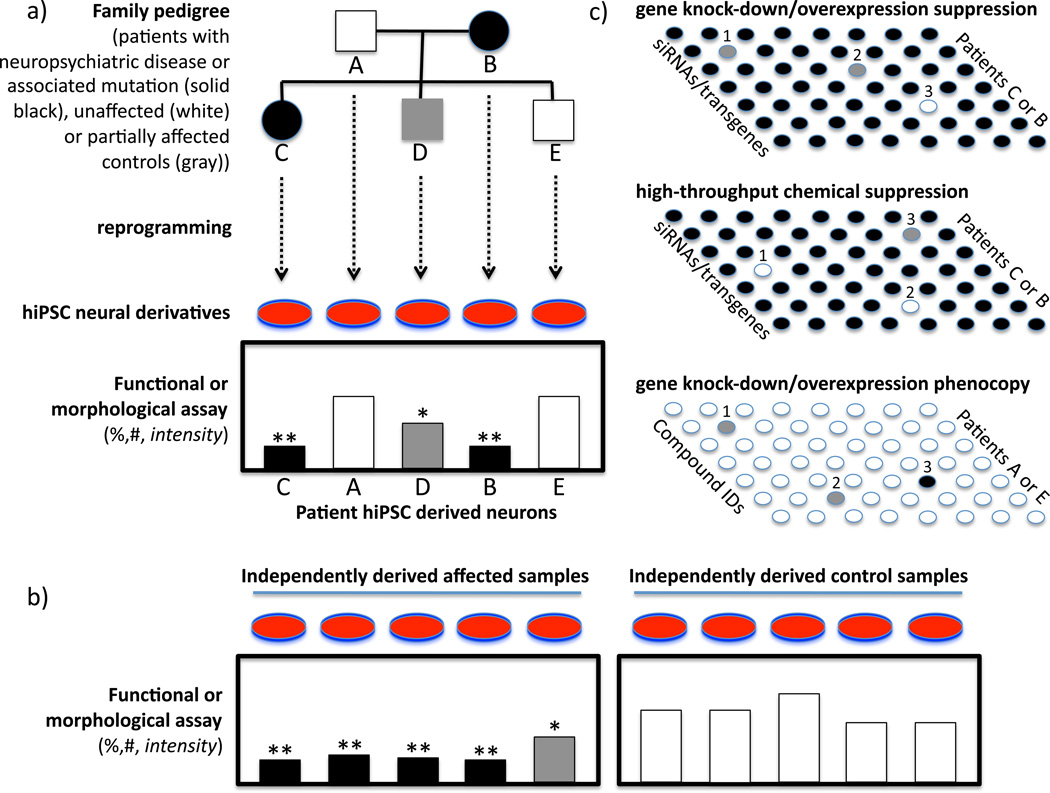
These results are promising, and, taken together, reveal both the opportunity and complexity offered by hiPSC models of a disease. For most of the diseases discussed above, the technically-challenging process of establishing hiPSC lines and neurons needs to be reproduced from larger groups of patients to confirm the original findings typically gleaned from a single case or a small number of cases (See Figure 1a,b). In the case of familial clusters, lines should be established and findings reproduced and tracked in affected and unaffected family members to conclude that any observations are attributable to the genetic defect in question. Moreover, not all carriers of a genetic trait, e.g., LRRK2, are affected, offering the opportunity to study unaffected gene carriers and the possible differences in susceptibility to environmental stressors or therapeutic interventions. This approach could advance the effort to subdivide broad syndromic categories into individual etiologies and propel drug discovery for personalized therapeutics.
Figure 1. Modeling complex disease and drug discovery with hiPSCs.
A. Hypothetical pedigree analysis showing co-segregation of clinical features and measurable hiPSC phenotypes in a family cluster
B. Confirmation of original hiPSC phenotype in an independent sample of hiPSC lines from multiple affected and unaffected individuals to validate specificity of hiPSC findings characteristic of the original family
C. Three high-throughput screening approaches to utilizing hiPSC cultures: chemical (top) or genetic suppression (middle) to renormalize phenotypic findings observed in affected cases, and genetic screens (bottom) to find candidates that reproduce in normal hiPSC cells the phenotypes of affected individuals. Validation of genetic or chemical intervention in independent sets of patients is required to confirm findings and to understand how broadly in the population the personalized therapeutic approach can be applied.
Modeling a complex disorder of unclear etiology: Schizophrenia
The potential power of hiPSCs for possibly modeling diseases lies in the following situations: (a) disorders that are poorly modeled by animals; (b) disorders for which underlying pathophysiological mechanisms are poorly understood because of the complexity of the multiple cells and genes involved and/or the cells and genes are unknown; (c) disorders where the cells and genes may be heavily influenced by extrinsic cell-non-autonomous forces (often multiple) that are difficult to isolate, control, and measure or the identity of such influences is uncertain; (d) conditions that affect multiple organs or an abnormal protein is present in all tissues yet the pathology is manifest predominantly in only one organ; (e) studies that require large numbers of uniform, controllable cells; (f) the disease-to-be-studied requires cell types that are difficult to obtain from living patients (e.g., from CNS tissue).
Neuropsychiatric disorders – among the most challenging of all diseases to unravel – would appear to fit these guidelines. For these diseases, where pathogenesis and endophenotypes are so poorly defined, the need is tremendous to delineate subtypes of these complex disorders that are based on mechanistic criteria and not solely on symptomatology which can be non-specific and even misleading because of its vulnerability to confounding factors.
hiPSCs have been generated from patients with sporadic schizophrenia [14] as well those carrying mutant DISC1 (“Disrupted in schizophrenia 1”), a neurodevelopmental regulator associated with psychiatric illness [15]. DISC1 was originally found in a family with multiple neuropsychiatric conditions including schizophrenia, bipolar disorder, and autism, raising heretofore unanswered questions not only about the genetics of schizophrenia but also its relationship to other neuropsychiatric disorders. In one study, hiPSCs were generated from clinically sub-typed, genetically-related individuals who harbored a 4 bp deletion in DISC1 [15]. It should be cautioned, however, that the causal relationship of DISC1 (and other genetic associations with psychosis) to sporadic schizophrenia is much more tenuous than is the relationship of familial PD (and the genes implicated in those conditions) to sporadic PD. However, the hope will be that hiPSCs may be used to investigate the phenotype of neural derivatives from clusters of schizophrenia cases harboring candidate genes as identified in genome-wide association studies (GWAS), or through their frequent association with copy number variations (CNV), and via whole genome sequencing efforts [16].
In a series of comprehensive and model free approaches to assess sporadic schizophrenia, Brennand et al. [14] utilized in-depth phenotypic analysis and global mRNA expression in neurons derived from hiPSCs from three patients diagnosed with schizophrenia, two of whom had first degree relatives with related conditions. Subtle yet potentially important differences were found between control and schizophrenia-derived neuronal cultures in the number of neurites, post-synaptic density (PSD95) protein, and neuronal connectivity without a clear effect on synapse number. Neuronal connectivity was assessed using a relatively new and untried but clever method based on the transport of rabies virus proteins between synapses in mixed cultures; rabies transport is limited to synapses, yet not dependent on synaptic or neural activity. Although many control experiments were performed by the authors to document the veracity of this approach, the technique is still in need of rigorous validation by comparison with more standard high-resolution techniques such as electron microscopy to assure that it is actually measuring synaptic connections. In support of the authors‘ conclusions, however, are proteomic and metabolomic studies of post-mortem brains from schizophrenics that have also implicated synaptic and neuritic functions. Unfortunately, spine shape and number were not assessed in this study; these parameters have been reported to be abnormal both in postmortem schizophrenic brains and in models of DISC1 neurons [17]. A strength of the Brennand et al. study was its thorough analysis — albeit negative -- of electrophysiological parameters and calcium transients in well defined hiPSC-derived neurons from the schizophrenia patients.
In an ambitious attempt to interpret the transcriptome of schizophrenia-derived hiPSC neurons in light of prior genetic and expression data in schizophrenia, Brennand et al. found an apparently high incidence in the their neurons of alterations in the expression of genes previously implicated in genetic and postmortem publications. Neuregulin and ANK3 were misexpressed overall while others genes were only misexpressed in a subset of cases. There was also altered expression of genes that had not been previously implicated in schizophrenia, such as Notch, ROBO/slit, and ephrin-A. Such observations are necessarily preliminary because of the small sample of patients used as a source and the variability of results found. Nonetheless, these analyses represent a useful dataset that may help facilitate comparison with other neuropsychiatric patient-derived hiPSC lines as they are developed.
While applying the power of bioinformatics to hiPSCs has the potential to help identify pathways that are associated with schizophrenia, some caveats are worth noting: It is still unclear how the source of the starting somatic cells and their culture conditions, the techniques of hiPSC generation, propagation, and handling, and the passage number of the hIPSCs all impinge on gene expression [20–22]. Variability between lines from the same patient may be large, suggesting that multiple comparisons must be evaluated statistically and replicated with independent samples (see Figure 1a, b). Major sources of genetic and epigenetic variability in hiPSC lines have been identified [17–19[18, 19]]. For example, Laurent et al. found a high frequency of sub-chromosomal CNVs in the 186 human pluripotent stem cell lines they examined, both hESCs and hiPSCs; however, for the latter, the reprogramming process was associated with higher incidence of deletions of tumor-suppressor genes while time in culture was associated with duplications of oncogenic genes. They also observed the emergence of duplications during differentiation. These findings made salient the dynamic nature of genomic abnormalities in pluripotent stem cells and the need for frequent genomic monitoring to assure phenotypic stability if one is going model a disease accurately.
Can cultured hiPSCs from patients with neuropsychiatric disorders be used for drug discovery? In the Brennand et al. study [14], 5 antipsychotic medications were tested for their effects on schizophrenia-derived hiPSC neurons and intriguingly, only loxapine reversed deficits in rabies-virus connectivity and partially or fully renormalized gene expression patterns including that of Neuregulin 1. These data raise both hope and questions and clearly need to be confirmed with dose-response curves of the 5 drugs and others with overlapping mechanisms. However, it is likely that having renewable sources of human neurons in culture that bear at least the genetic fingerprint, at some level, of the patient-of-interest is a first step not only to examining therapeutic hypotheses but to personalized medicinei.e., the tailoring a particular drug to a particular patient without the typical trial-and-error that attends the use of most drugs for neurological and neuropsychiatric disorders. If an informative surrogate read-out can be accurately measured in vitrothe use of hiPSCs may even allow clinicians and pharmaceutical companies to test candidate drugs even when the etiology of the disorder in question is unknown.
Promises, caveats, and challenges for the future
The field is just beginning to explore the utility and limitations of using hiPSCs to model aspects of multifactorial disorders in vitro. The results so far are sufficiently encouraging to suggest that we should continue to push this technology to its limits, knowing full well that many technological and biological hurdles – some known, some still to be unveiled – remain to be circumvented if this strategy is to offer clinicians novel mechanistic insights, diagnostics, prognostics, drug targets, and the drugs themselves.
Even the few papers to date in this field, illuminate some of the challenges to overcome: potential variability at each level of cellular differentiation, the inability to draw broader conclusions because of the low number of patients represented for each disease, the lack of extensive amount of clinical and family history on each patient and an absence of analysis of family clusters. Even though evidence suggests that complex disease-related genetic and epigenetic anomalies may be maintained during reprogramming [20, 21], it is yet unclear how genetic instability [22, 23], residual epigenetic memory [24, 25], and variable differentiation efficiency [22, 26, 27] will influence the quality of the data. Fortunately, it does not appear that the process of reprogramming and cell passaging precludes being able to test the efficacy of certain therapeutic interventions on the cells, for example, swapping in a normal version of a defective gene via homologous recombination [28]. Early work suggests that at least karyotypic abnormalities may not overtly affect neuralization of hiPSCs [22]. Although genetic and epigenetic variability may be limiting for transplantation studies, and even the anticipated immunotolerance of grafts composed of cells from the recipient‘s own iPSCs may prove more daunting than hoped [29], hiPSCs should still be useful for disease modeling [30] provided multiple patient-derived and control hiPSCs lines are employed for validation and replication of results.
Characterization, standardization, and an array of “ ‘omic ” tools will be required to enable hiPSCs to illuminate pathogenic mechanisms that are shared by the broad range of disease categories defined in the DSM. A further challenge, however, is not falling into the trap of identifying spurious features because of the massive number of independent measures and variables being assessed. An analogy might be drawn to the field of genomics in the 90‘s and early 2000‘s when genetic associations would be published by individual labs but not reproduced by other labs. Statistical methods to assure adequate power with correction for multiple comparisons and rigorous testing in confirmation sets, a common standard in clinical trials for regulatory approval and now the standard for GWAS studies, should be applied to tests of hiPSCs to avoid spurious and irreproducible results.
An important self-regulatory step would be for the field to avoid drawing conclusions until findings have been validated in large sets of hiPSC lines derived from large samples of patients who, themselves, have been thoroughly and accurately phenotyped and categorized into endophenotypes. For such n values to be feasible, increased attention to the complete detailed clinical and family history, course, evaluation, and past treatments of each patient will be critical before any results can be extrapolated meaningfully.
The degree to which experimentally-derived neurons reflect those of the developing and adult brain is unknown and of particular importance to neuropsychiatry in which many of the disorders are developmental, delayed-onset, or neurodegenerative. Can neurons that were essentially newly-born and cultured for a few days to weeks accurately model a disease that develops in vivo at maturity or even with aging (e.g., Alzheimer‘s disease, senile dementia of the non-Alzheimer‘s variety, PD, etc.). Additionally, it is unclear how to faithfully model a relapsing-remitting or cyclic disorder. Indeed many neuropsychiatric illnesses exhibit periods of decompensation and stability. How the state of the patient at the time of the skin biopsy could affect hiPSC epigenetic profiles has not been studied, nor has the influence of medication regimen on fibroblast cells. For instance, valproic acid, a well-documented treatment for seizure and bipolar disorder is known to alter efficiency of reprogramming and epigenetic profiles.
As disease-based cellular models are further developed and as potential pathological phenotypes are uncovered (i.e., findings that can reliably and informatively distinguish a diseased neuron from a well-functioning neuron in a manner relevant to the symptomatology of the disease under study), a growing focus will be placed on “reverse genetics” to understand the underlying molecular mechanisms. These studies will, in turn -- perhaps even in parallel -- lead to combining cellular assays with compound screening as a kick-off for therapeutic development, recently termed “pharmaco-iPSCellomics” [30] (see Figure 1c). At least 4 different approaches to “screenable” phenotypes have recently been suggested. First, morphological phenotypes might be screened via high-content imaging modalities, such as dendritic spine density and cell body size which were noted to deviate from normal in RS hiPSC-derived neurons (findings that mirrored those seen in MeCP2 knockout mice and seemed to respond, as did the mice, to exposure to IGF-1) [10]. A second approach might be to screen for modifiers of biochemical activity aberrantly regulated in cells from a particular disease as was suggested by increased caspase 3/7 activity in Huntington‘s Disease hiPSC-derived neurons [20]. A third approach might be normalization of gene expression, although this path may be fraught with challenge unless a relevant gene can be identified, particularly for complex multigenic diseases where changing even a predictive and informative surrogate gene may not be a clinically-impactful drug target. Fourth and common to all of these approaches may be the need to apply stress to cells in order to expose a screenable phenotype. For example, oxidative stress was necessary to unveil a phenotype in the case of hiPSCs derived from patients with LRRK2-mutated familial PD [12]. Indeed, one might say that stressors of many types – reduced to chemical signals if possible -- may be required to emulate the environmental conditions that may be crucial for the ultimate expression of disease symptomatology in a patient.
Furthermore, all of the above approaches demand that one has a population of hiPSC-derivatives – often neurons -- that are amenable to the instrumentation demanded for high-throughput, high-content, small molecule screening for drug discovery and for advanced comprehensive profiling. None of the extant differentiation strategies to date are capable of providing vast numbers of homogenous neurons rapidly, uniformly, reliably, reproducibly, and inexpensively in monolayer. Immunophenotyping of cell surface neural markers and the rigorous application of fluorescent activated cell sorting (FACS) quantitation may help [31]. Better differentiation protocols may also be required.
Of course, the very discussion of what type of cell to subject to drug discovery algorithms raises an even more fundamental and philosophical question for the entire field of “disease-in-a-dish modeling”: can a disease process be faithfully modeled in vitro -- in isolation from the rest of the body, or does disease expression require the complete in vivo environment, with an intact immune and vascular system as well as multiple, perfectly arranged CNS lineages and cell types?
Highlights.
-
>
Differentiated iPS lines from neuropsychiatric disease show neuronal abnormalities
-
>
Neuritic, synaptic and gene expression defects arise in culture and after cellular stress
-
>
Drug treatments can largely reverse neuronal abnormalities in iPS cultures
-
>
Variability and cellular instability combined with ‘omics technologies raise challenges
-
>
A roadmap for validating models and finding personalized therapies is proposed
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nestler EJ, Hyman SE. Animal models of neuropsychiatric disorders. Nature neuroscience. 2010;13(10):1161–1169. doi: 10.1038/nn.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 1b.Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008;134(5):877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463(7284):1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim J, Efe JA, Zhu S, Talantova M, Yuan X, Wang S, Lipton SA, Zhang K, Ding S. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci U S A. 2011;108(19):7838–7843. doi: 10.1073/pnas.1103113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim KS, Lee HJ, Jeong HS, Li J, Teng YD, Sidman RL, Snyder EY, Kim SU. Self-renewal induced efficiently, safely, and effective therapeutically with one regulatable gene in a human somatic progenitor cell. Proc Natl Acad Sci U S A. 2011;108(12):4876–4881. doi: 10.1073/pnas.1019743108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mattis VB, Svendsen CN. Induced pluripotent stem cells: A new revolution for clinical neurology? Lancet neurology. 2011;10(4):383–394. doi: 10.1016/S1474-4422(11)70022-9. [DOI] [PubMed] [Google Scholar]
- 6. Hargus G, Cooper O, Deleidi M, Levy A, Lee K, Marlow E, Yow A, Soldner F, Hockemeyer D, Hallett PJ, et al. Differentiated parkinson patient-derived induced pluripotent stem cells grow in the adult rodent brain and reduce motor asymmetry in parkinsonian rats. Proc Natl Acad Sci U S A. 2010;107(36):15921–15926. doi: 10.1073/pnas.1010209107. * This is the first study to demonstrate integration of disease-derived hiPSC neurons in vivo after transplantation to the murine brain
- 7. Ebert AD, Yu J, Rose FF, Jr, Mattis VB, Lorson CL, Thomson JA, Svendsen CN. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457(7227):277–280. doi: 10.1038/nature07677. * The study examines the effects of drug action on protein expression and motorneuron numbers using hiPSCs from SMA patients
- 8. Lee G, Papapetrou EP, Kim H, Chambers SM, Tomishima MJ, Fasano CA, Ganat YM, Menon J, Shimizu F, Viale A, et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461(7262):402–406. doi: 10.1038/nature08320. ** A thorough investigation of neural crest differentiation with hypothesis-based small molecule testing using hiPSCs generated from IKBKAP linked dysautonomia is presented. The study also introduces the importance of examining differentiation to other lineages
- 9.Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH. L1 retrotransposition in neurons is modulated by mecp2. Nature. 2010;468(7322):443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. A model for neural development and treatment of rett syndrome using human induced pluripotent stem cells. Cell. 2010;143(4):527–539. doi: 10.1016/j.cell.2010.10.016. ** The study utilizes post-mortem data and mouse models as a guide to examining relevant morphological phenotypes of hiPSC derived neurons from mutant MecP2 Rett Syndrome patients and tests the effect of putative therapeutics on these phenotypes
- 11.Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136(5):964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen HN, Byers B, Cord B, Shcheglovitov A, Byrne J, Gujar P, Kee K, Schule B, Dolmetsch RE, Langston W, et al. Lrrk2 mutant ipsc-derived da neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell. 2011;8(3):267–280. doi: 10.1016/j.stem.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seibler P, Graziotto J, Jeong H, Simunovic F, Klein C, Krainc D. Mitochondrial parkin recruitment is impaired in neurons derived from mutant pink1 induced pluripotent stem cells. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31(16):5970–5976. doi: 10.1523/JNEUROSCI.4441-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, Li Y, Mu Y, Chen G, Yu D, McCarthy S, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473(7346):221–5. doi: 10.1038/nature09915. ** A comprehensive characterization of hiPSC derived neurons from Schizophrenia patients is presented including electrophysiology, mRNA expression profiling, and phenotypic analyses with cell culture testing of clinical anti-psychotics in context of data mining of relevant GWAS databases in psychiatric illness
- 15.Chiang CH, Su Y, Wen Z, Yoritomo N, Ross CA, Margolis RL, Song H, Ming GL. Integration-free induced pluripotent stem cells derived from schizophrenia patients with a disc1 mutation. Molecular psychiatry. 2011;16(4):358–360. doi: 10.1038/mp.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang J, Perlis RH, Lee PH, Rush AJ, Fava M, Sachs GS, Lieberman J, Hamilton SP, Sullivan P, Sklar P, et al. Cross-disorder genomewide analysis of schizophrenia, bipolar disorder, and depression. The American journal of psychiatry. 2010;167(10):1254–1263. doi: 10.1176/appi.ajp.2010.09091335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishizuka K, Kamiya A, Oh EC, Kanki H, Seshadri S, Robinson JF, Murdoch H, Dunlop AJ, Kubo KI, Furukori K, et al. Disc1-dependent switch from progenitor proliferation to migration in the developing cortex. Nature. 2011;473(7345):92–6. doi: 10.1038/nature09859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laurent LC, Ulitsky I, Slavin I, Tran H, Schork A, Morey R, Lynch C, Harness JV, Lee S, Barrero MJ, Ku S, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human escs and ipscs during reprogramming and time in culture. Cell Stem Cell. 2011;8(1):106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pera MF. Stem cells: The dark side of induced pluripotency. Nature. 2011;471(7336):46–47. doi: 10.1038/471046a. [DOI] [PubMed] [Google Scholar]
- 20.Zhang N, An MC, Montoro D, Ellerby LM. Characterization of human Huntington's disease cell model from induced pluripotent stem cells. PLoS currents. 2010;2:RRN1193. doi: 10.1371/currents.RRN1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chamberlain SJ, Chen PF, Ng KY, Bourgois-Rocha F, Lemtiri-Chlieh F, Levine ES, Lalande M. Induced pluripotent stem cell models of the genomic imprinting disorders angelman and prader-willi syndromes. Proc Natl Acad Sci U S A. 2010;107(41):17668–17673. doi: 10.1073/pnas.1004487107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boulting GL, Kiskinis E, Croft GF, Amoroso MW, Oakley DH, Wainger BJ, Williams DJ, Kahler DJ, Yamaki M, Davidow L, et al. A functionally characterized test set of human induced pluripotent stem cells. Nature biotechnology. 2011;29(3):279–286. doi: 10.1038/nbt.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayshar Y, Ben-David U, Lavon N, Biancotti JC, Yakir B, Clark AT, Plath K, Lowry WE, Benvenisty N. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7(4):521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 24.Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nature biotechnology. 2010;28(8):848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467(7313):285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu BY, Weick JP, Yu J, Ma LX, Zhang XQ, Thomson JA, Zhang SC. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci U S A. 2010;107(9):4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim JE, O'Sullivan ML, Sanchez CA, Hwang M, Israel MA, Brennand K, Deerinck TJ, Goldstein LS, Gage FH, Ellisman MH, Ghosh A. Investigating synapse formation and function using human pluripotent stem cell-derived neurons. Proc Natl Acad Sci U S A. 2011;108(7):3005–3010. doi: 10.1073/pnas.1007753108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howden SE, Gore A, Li Z, Fung HL, Nisler BS, Nie J, Chen G, McIntosh BE, Gulbranson DR, Diol NR, et al. Genetic correction and analysis of induced pluripotent stem cells from a patient with gyrate atrophy. Proc Natl Acad Sci U S A. 2011;108(16):6537–6542. doi: 10.1073/pnas.1103388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao T, Zhang ZN, Rong Z, Xu Y. Immunogenicity of induced pluripotent stem cells. Nature. 2011 doi: 10.1038/nature10135. [DOI] [PubMed] [Google Scholar]
- 30.Inoue H, Yamanaka S. The use of induced pluripotent stem cells in drug development. Clinical pharmacology and therapeutics. 2011;89(5):655–661. doi: 10.1038/clpt.2011.38. [DOI] [PubMed] [Google Scholar]
- 31.Yuan SH, Martin J, Elia J, Flippin J, Paramban RI, Hefferan MP, Vidal JG, Mu Y, Killian RL, Israel MA, et al. Cell-surface marker signatures for the isolation of neural stem cells, glia and neurons derived from human pluripotent stem cells. PloS one. 2011;6(3):e17540. doi: 10.1371/journal.pone.0017540. [DOI] [PMC free article] [PubMed] [Google Scholar]