Abstract
Single-nucleotide substitutions and small in-frame insertions or deletions identified in human breast cancer susceptibility genes BRCA1 and BRCA2 are frequently classified as variants of unknown clinical significance (VUS) due to the availability of very limited information about their functional consequences. Such variants can most reliably be classified as pathogenic or non-pathogenic based on the data of their co-segregation with breast cancer in affected families and/or their co-occurrence with a pathogenic mutation. Biological assays that examine the effect of variants on protein function can provide important information that can be used in conjunction with available familial data to determine the pathogenicity of VUS. In this report, we have used a previously described mouse embryonic stem (mES) cell-based functional assay to characterize eight BRCA2 VUS that affect highly conserved amino acid residues and map to the N-terminal PALB2-binding or the C-terminal DNA-binding domains. For several of these variants, very limited co-segregation information is available, making it difficult to determine their pathogenicity. Based on their ability to rescue the lethality of Brca2-deficient mES cells and their effect on sensitivity to DNA-damaging agents, homologous recombination and genomic integrity, we have classified these variants as pathogenic or non-pathogenic. In addition, we have used homology-based modeling as a predictive tool to assess the effect of some of these variants on the structural integrity of the C-terminal DNA-binding domain and also generated a knock-in mouse model to analyze the physiological significance of a residue reported to be essential for the interaction of BRCA2 with meiosis-specific recombinase, DMC1.
INTRODUCTION
Among the various possible risk factors, inheritance of a mutant BRCA1 or BRCA2 is the single most definitive indicator of increased risk of developing breast cancer (1,2). Women who inherit a mutation in one of these genes have up to 80% risk of developing breast cancer by age 70 (3,4). Germline mutations in these genes account for 20–60% of breast cancer cases in families where multiple individuals are affected (5).
To identify individuals who are at risk of developing breast cancer, sequencing-based genetic tests are now being offered to individuals with a family history of breast and ovarian cancers to identify BRCA mutation carriers (6). Individuals who inherit a mutation known to be pathogenic can benefit by taking aggressive preventive measures such as preventive chemotherapy or prophylactic surgery (7). In addition, there are a significant percentage of women who inherit variants of unknown clinical significance (VUS). A study based on the sequence analyses of 10 000 individuals reported that VUS were present in 13% of the cases (8). Currently, VUS account for 3% of all of the mutations identified in BRCA1/2 due to increased number of individuals being subjected to genetic testing and efforts to determine the pathogenicity of variants (9).
Segregation analysis of BRCA1 and BRCA2 mutations in cancer-afflicted families provides the most reliable information about the nature of these mutations and helps to distinguish between pathogenic (deleterious) and non-pathogenic (neutral or benign) alterations. An assessment of 1433 BRCA1 and BRCA2 VUS was reported based on their co-occurrence in trans with a known pathogenic mutation, detailed analysis of personal and family history of cancer in probands and co-segregation of the variant with disease in pedigrees (10). This study described the odds in favor of neutrality or causality of these variants based on likelihood ratios (LRs), which is an invaluable tool for assessing BRCA variants. More recent efforts to classify VUS have led to the development of a method called the posterior probability model (11). This method combines the LRs with prior probabilities of causalities of a variant determined by sequence conservation and physiochemical properties of the amino acid residue, which is calculated by using the Align-GVGD conservation model (12). Also, a more standardized method of classification of variants has been outlined by the International Agency for Research on Cancer (IARC), which recommends using a five-tier system (classes 1–5, 1 being ‘not pathogenic’ and 5 being ‘definitely pathogenic’) to classify the variants based on their degree of likelihood of pathogenicity (13). A classification of variants for which information is available is listed in the Leiden Open Variation Database (http://brca.iarc.fr/LOVD/home.php) at Leiden University, the Netherlands.
Other methods to assess the clinical significance of variants based on cancer family history, pathology or immunohistochemical analysis of tumors have been developed (14–16). More recently, a simple approach of calculating the LR of a variant being deleterious based on co-segregation analysis using the precise age of onset information has been developed (17). This approach has the advantage in that it takes into account information on gender, genotype, present age and/or age of onset for cancer, though is still limited by the ubiquitous problem of how to account for BRCA phenocopies due to ascertainment bias (18).
In addition to these approaches, numerous biological assays have been developed that assess the consequences of BRCA1 and BRCA2 variants on the protein function (19,20). Assays that examine the effect of BRCA1 variants on the transcriptional activity of the BRCT domain and the effect of BRCA2 variant on homologous recombination (HR) have been shown to be highly sensitive and effective in predicting the pathogenicity of variants (21,22). To complement these methods, we have developed a mouse embryonic stem (mES) cell-based approach to determine the pathogenicity of BRCA1 and BRCA2 variants (23–25). In this approach, we examine the ability of BRCA1 or BRCA2 variants to rescue the lethality of Brca1 or Brca2-null ES cells, respectively (Fig. 1A). Variants that fail to rescue the ES cell lethality are considered pathogenic. Variants that fully or partially rescue the ES cell viability are then tested for sensitivity to DNA-damaging agents, cell proliferation, HR and effect on overall genomic stability. Variants that result in reduced cell viability or have impaired DNA repair function are likely to be pathogenic. In contrast, variants that are indistinguishable from wild-type (WT) BRCA1 or BRCA2 are classified as non-pathogenic or neutral variants.
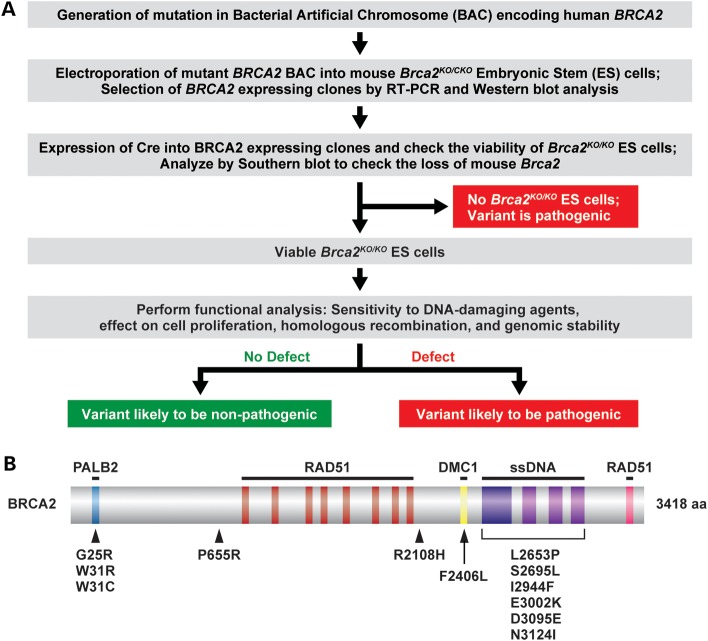
Figure 1.
Scheme to examine human BRCA2 variants using the mES cell-based assay and the location of variants on the protein. (A) Schematic representation of the mES cell-based assay for functional analysis of BRCA2. bacterial artificial chromosome (BAC) DNA containing desired variants was first introduced into mES cells with one knockout allele (KO) and one conditional allele (CKO) of Brca2. After Cre-mediated deletion of a conditional copy of Brca2 depending on the impact of the variants, cells may or may not be viable. The viable ES cells can be functionally similar to WT or defective in some function of BRCA2, depending on the effect of variants. (B) Schematic diagram of the BRCA2 protein showing the different domains and position of the variants analyzed in this study. The BRCA2-interacting partners are shown above the corresponding region of BRCA2 required for their interaction. Below the location of the variants used in this study are shown.
We have used our ES cell-based method to determine the pathogenicity of eight variants (G25R, W31R, W31C, F2406L, S2695L, I2944F, E3002K and N3124I; see Fig. 1B and Table 1) that map to the N-terminal PALB2-binding domain and the C-terminal DNA-binding domain (26). Interaction with PALB2 is essential for BRCA2 function and important for its nuclear localization (26). The C-terminal domain, which spans amino acids 2400–3190 of BRCA2, consists of a helix-turn-helix motif and three oligonucleotide-binding (OB) folds (27). This domain has DNA-binding potential and is the most conserved domain of BRCA2 across metazoans, plants and fungal orthologs (28). These variants are of unknown clinical significance and very limited co-segregation and co-occurrence information is available. We have also examined four other variants (P655R, R2108H, L2653P and D3095E) that are of known pathogenicity and used them as controls for our assay. Of these four variants, two are pathogenic and map to the C-terminal DNA-binding domain and the other two are non-pathogenic/neutral variants that map outside these domains. In this study, we have also used homology-based modeling to predict the effect of variants that map to the C-terminal domain on the structural integrity of BRCA2 and examined the physiological significance of a residue (Phe2406) predicted to be essential for the interaction of BRCA2 with meiosis-specific RecA homolog, DMC1, using a knock-in mouse model.
Table 1.
List of VUS evaluated in this study
| Variant | Protein change | Exon | DNA sequence |
Evolutionary conservationc | No. of BIC entries | |
|---|---|---|---|---|---|---|
| Varianta | BICb | |||||
| G25R | p.Gly25Arg | 3 | c.73G>A | 301G>A | Fully conserved | 1 |
| W31R | p.Try31Arg | 3 | c.91T>C | 319T>C | Fully conserved | 1 |
| W31C | p.Try31Cys | 3 | c.93G>T | 321G>T | Fully conserved | 1 |
| F2406L | p.Phe2406Leu | 14 | c.7218T>G | 7446T>G | Highly conserved, except in Sp | 1 |
| S2695L | p.Ser2695Leu | 18 | c.8084C>T | 8312C>T | Highly conserved, except in Fr, Sp | 2 |
| I2944F | p.Ile2944Phe | 22 | c.8830A>T | 9058A>T | Highly conserved, except in Md, Tn, Fr, Sp | 115 |
| E3002K | p.Glu3002Lys | 23 | c.9004G>A | 9232G>A | Fully conserved | 9 |
| N3124I | p.Asp3124Ile | 25 | c.9371A>T | 9599A>T | Fully conserved | 14 |
Md, Monodelphis domesticus; grey, short-tailed opossum; Tn, Tetraodon nigroviridis, Pufferfish (green spotted); Fr, Fugu rubripes, Pufferfish (fugu); Sp, Strongylocentrotus purpuratus, sea urchin.
aNucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence (NM_000059.3). The initiation codon is codon 1.
bFor BIC nomenclature, +228 corresponds to the ATG translation initiation codon in the reference sequence.
cFully conserved indicates conservation from sea urchin through humans.
RESULTS
BRCA2 variants selected for functional evaluation
For functional analysis, we selected eight BRCA2 VUS (Table 1) that are located in the two known functional domains of BRCA2: the N-terminal PALB2 (partner and localizer of BRCA2)-binding domain and the C-terminal DNA-binding domain. Amino acids 18–40 of BRCA2 are known to bind to PALB2. G25R, W31R and W31C map to the PALB2-binding domain (Fig. 1B). In vitro studies have shown that while W31R and W31C completely disrupt the interaction with PALB2, G25R shows reduced binding (26). The remaining five variants (F2406L, S2695L, I2944F, E3002K and N3124I) are located in the C-terminal DNA-binding domain (Fig. 1B and Table 1). F2406L variant is located within the DMC1-interacting motif of BRCA2 (29). All the eight variants are listed in the Breast Cancer Information Core (BIC) database and the amino acid residues are highly conserved from sea urchins through humans (Table 1). Based on the single-nucleotide polymorphism (SNP) database (http://www.ncbi.nlm.nih.gov/projects/SNP/), only I2944F (rs4987047) has been observed in the sample population. rs4987047 was found in 48 cases (including two homozygous ones) among the 1047 individuals genotyped. In addition to these eight VUS, we also selected four variants (P655R, R2108H, L2653P and D3095E) of known pathogenicity as controls for our functional analysis (Fig. 1B, Table 2). L2653P and D3095E are pathogenic variants that map within the C-terminal domain of BRCA2. P655R and R2108H are non-pathogenic and are not located in any region of known functional importance. The non-pathogenic nature of P655R was predicted based on its prevalence in normal individuals (30). Similarly, an assessment using co-occurrence in trans with known pathogenic mutations, detailed history of personal and family cancer in probands and co-segregation with disease in pedigrees also predicted that R2108H is a non-pathogenic variant (10). Based on their posterior probability of causality, P655R and R2108H are class 1 (non-pathogenic) variants, L2653P is a class 5 (definitely pathogenic) variant and D3095E is a class 4 (likely pathogenic) variant (31).
Table 2.
List of variants with an assigned IARC class evaluated in this study
| Variant | Protein change | Exon | DNA sequence |
No. of BIC entries | Odds in favor of causalityc | Posterior probability of being deleteriousd | Pathogenicity | IARC classd | |
|---|---|---|---|---|---|---|---|---|---|
| Varianta | BICb | ||||||||
| P655R | p.Pro655Arg | 11 | c.1964C>G | 2192C>G | 141 | 3.36 × 10−3 | 6.86 × 10−5 | Not pathogenic | 1 |
| R2108H | p.Arg2108His | 11 | c.6323G>A | 6551G>A | 126 | 1.82 × 10−11 | 3.72 × 10−13 | Not pathogenic | 1 |
| L2653P | p.Leu2653Pro | 17 | c.7958T>C | 8186T>C | 4 | 24.06 | 0.99 | Definitely pathogenic | 5 |
| D3095E | p.Asp3095Glu | 25 | c.9285C>G | 9513C>G | 12 | 22.59 | 9.98 | Likely pathogenic | 4 |
aNucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence (NM_000059.3). The initiation codon is codon 1.
bFor BIC nomenclature, +228 corresponds to the ATG translation initiation codon in the reference sequence.
cFrom Easton et al. (10).
dFrom Lindor et al. (31).
Co-segregation LRs for BRCA2 variants
The familial data consisted primarily of genotyped family members that were first-degree relatives of the proband. Co-segregation data had been collected for more common variants, while very limited or no co-segregation data had been obtained for rare variants. The co-segregation information was used to calculate the LR per family, which was then combined to obtain the LR for each variant (Table 3), as described by Mohammadi et al. (17). They suggest applying the stringency of Goldgar et al. (30) in interpreting the significance of their co-segregation approach, in that an LR > 1000 is necessary to be certain of causality, and an LR < 0.01 is sufficient for neutrality. Therefore, based on this co-segregation method, BRCA2 P655R, R2108H and I2944F are conclusively benign or non-pathogenic variants, whereas BRCA2 L2653P, E3002K, D3095E and N3124I are variants with segregation data suggestive, but not conclusive, of causality or pathogenicity. The Mohammadi et al. (17) LR calculation may not account for phenocopy rate increase due to ascertainment bias, which would have the effect of lowering LRs (18). The LR for BRCA2 W31R, W31C and S2695L are equivocal due to the limited number of families tested for these variants, and no families had been tested for BRCA2 G25R and F2406L, making it difficult to classify them.
Table 3.
Co-segregation LR for BRCA2 variants
| Variant | #families | n+ | n+ | n− | n− | LR variant |
|---|---|---|---|---|---|---|
| G25R | 0 | |||||
| W31R | 1 | 2+ | 0+ | 0− | 0− | 1.9903 |
| W31C | 1 | 2+ | 0+ | 1− | 0− | 0.9777 |
| P655R | 28 | 25+ | 14+ | 8− | 12− | 0.000028 |
| R2108H | 28 | 29+ | 14+ | 4− | 11− | 0.0000000016 |
| F2406L | 0 | |||||
| L2653P | 5 | 8+ | 3+ | 2− | 5− | 9.7549 |
| S2695L | 2 | 3+ | 0+ | 2− | 0− | 0.3456 |
| I2944F | 12 | 16+ | 4+ | 6− | 1− | 0.0000097 |
| E3002K | 14 | 20+ | 5+ | 3− | 7− | 8.2339 |
| D3095E | 24 | 23+ | 32+ | 1− | 17− | 15.3771 |
| N3124I | 20 | 25+ | 7+ | 3− | 13− | 36.9505 |
LR for the variant is the multiplicative product of the LR per individual family when data from more than one family was available, according to the methodology of Mohammadi et al. (17).
#families, Number of families with two or more family members genotyped, including the proband; n+, number of genotyped affected individuals carrying the variant (probands included); n+, number of genotyped unaffected individuals carrying the variant (probands included); n−, number of genotyped affected individuals without the variant; n−, number of genotyped unaffected individuals without the variant.
Predictions based on Align-GVGD score
We used Align-GVGD to assess the effect of the eight variants on BRCA2 function based on sequence conservation from humans through sea urchin (Table 4). Based on the Align-GVGD grade of C55 or C65, G25R, W31R, W31C, E3002K and N3124I variants are predicted to be pathogenic, which is consistent with their co-segregation data. The remaining variants, F2406L, S2695L and I2944F, are grade C0 and thus predicted to be benign or non-pathogenic.
Table 4.
Summary of functional analysis of BRCA2 variants
| Variant | Align-GVGD |
Protein structure predictiona | Full-length protein detected | Viable Brca2ko/ko ES cells | Sensitivity to DNA-damaging agents | Effect on HR | Effect on splicing | Pathogenicity | ||
|---|---|---|---|---|---|---|---|---|---|---|
| GV | GD | Grade | ||||||||
| G25R | 0.0 | 125.1 | C65 | Unknown | Yes | Yes | Sensitive | 50% reduction | None | Likely pathogenic |
| W31R | 0.0 | 101.3 | C65 | Disruptiveb | Yes | No | N/A | N/A | Skipping of exon 3 | Pathogenic |
| W31C | 0.0 | 214.4 | C65 | Disruptiveb | Yes | No | N/A | N/A | None | Pathogenic |
| P655R | 353.9 | 0.0 | C0 | Unknown | Yes | Yes | No | No | None | Not pathogenic |
| R2108H | 353.9 | 0.0 | C0 | Unknown | Yes | Yes | No | No | None | Not pathogenic |
| F2406L | 112.6 | 4.9 | C0 | Unknown | Yes | Yes | No | No | None | Not pathogenic |
| L2653P | 0.0 | 97.8 | C65 | Disruptive | Yes | Yes, but reduced | Hypersensitive | 90% reduction | None | Likely pathogenic |
| S2695L | 174.6 | 4.9 | C0 | No crystal density | Yes | Yes | No | No | None | Not pathogenic |
| I2944F | 4.9 | 21.3 | C0 | No effect | Yes | Yes | No | No | None | Not pathogenic |
| E3002K | 0.0 | 56.87 | C55 | Disruptive | Yes | No | N/A | N/A | None | Pathogenic |
| D3095E | 0.0 | 44.6 | C65 | Disruptive | Yes | No | N/A | N/A | None | Pathogenic |
| N3124I | 0.0 | 148.9 | C65 | Disruptive | Yes | No | N/A | N/A | None | Pathogenic |
Predictions using homology-based modeling
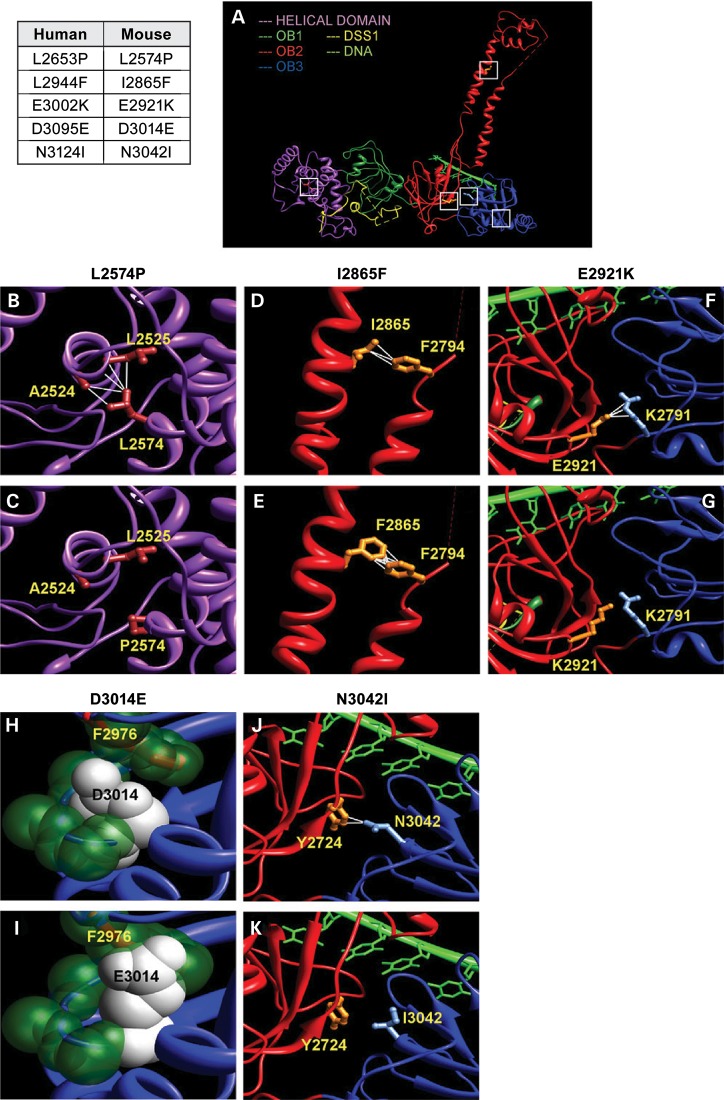
We used the crystal structure of the conserved C-terminal domain of mouse BRCA2 bound to DSS1 and ssDNA (pdb:1MIU, MJE, Fig. 2A) to predict the effect of variants on the structure and function of the human BRCA2 by homology-based modeling (27). This region contains three OB folds and one helix-turn-helix motif. The N-terminal helical domain interacts with an essential co-factor DSS1 to stabilize BRCA2 (27). L2574 (corresponding to human L2653) is placed at the core helix of the N-terminal helical motif and forms long-range hydrophobic contacts with the residues L2525 and A2524 of an adjacent helix (Fig. 2B). Since the proline residue lacks hydrophobicity, mutated P2574 will lose these long-range interactions (Fig. 2C), destabilizing the fold and DSS1 interaction of the domain. Hence, the L2653P variant is predicted to be pathogenic. S2616 (corresponding to S2695 in human BRCA2) is part of a dynamic loop and lacks electron density in the crystal structure. S2616 has no crystal density and hence the original structure has a gap in the region containing this residue. Evidently, the residue does not form any stabilizing contact that could be essential for the protein's fold, and therefore, S2695L is predicted to be a non-pathogenic variant. The OB2 motif has a tower structure formed by two parallel helices, which are maintained by critical contacts such as the hydrophobic contacts between I2865 (corresponding to I2944 in human BRCA2) and F2794 (Fig. 2D). Phenylalanine (F) is also hydrophobic and F2865 maintains the contacts with F2794 (Fig. 2E). Therefore, the I2944F is predicted to have no significant effect. OB2 and OB3 pack tightly against each other to maintain a proper interface that recognizes the ssDNA. Critical interactions between OB2 and OB3 include salt bridges formed between E2921 (E3002 in human BRCA2) and K2791 (Fig. 2F). On the other hand, the E2921K (or E3002K) will reverse the charge of the residue, repulse R2971 and break the salt bridge and other contacts (Fig. 2G). Disruption of contacts between the OB domains can negatively impact ssDNA binding and function, resulting in a deleterious phenotype. D3014 (corresponding to human D3095) residue is present at the core beta-sheet structure of OB3 and placed in between the backbone of a beta-turn and an adjacent phenylalanine (F2976) side chain (Fig. 2H). E3014 has a much longer side chain, which requires 27Å more surface area than D3014. Due to inadequate space in the region, it clashes with other residues in the vicinity, primarily with the adjacent F2976 (Fig. 2G). These clashes could potentially destabilize the core of OB3 domain. N3042 (corresponding to N3124 in the human protein) forms contacts with Y2724 to structure the OB2–OB3 packing. I3042 has a different preferred orientation and loses these contacts. Similar to E3002K mutation, N3124I is predicted to adversely affect ssDNA interaction. In summary, the structural modeling predicts S2695L and I2944F to have no effect on the structural integrity of the C-terminal domain, whereas L2653P, E3002K, D3095E and N3124I are predicted to be disruptive.
Figure 2.
Homology–based modeling of BRCA2 variants based on crystal structure of the mouse protein (A) The crystal structure of mouse C-terminal BRCA2 includes multiple domains: helical (magenta), OB1 (dark green), OB2 (red) and OB3 (blue). While the helical and OB1 domains interact with the co-factor DSS1, OB2 and OB3 domains interact with the ssDNA. The variants studied in this work are indicated by white squares. Human BRCA2 variants and corresponding residues in the mouse protein are indicated in the table on left. (B) L2574 forms hydrophobic contacts with residues A2524 and L2525, and together these residues form a core of the helical domain. (C) P2574 is not hydrophobic and loses these crucial contacts. (D) I2865 is present at the tower structure of OB2 and forms hydrophobic contacts with F2794, which could be crucial for tower structure. (E) Being hydrophobic, F2865 maintains these contacts. (F) At the interface between OB2 and OB3, E2921 forms several contacts and an electrostatic salt bridge with R2971. (G) E2921K mutation reverses the charge of the residue, introducing repulsive interactions and hence disrupting the crucial salt bridge and other contacts. (H) D3014 is placed on a beta-strand at the core of OB3. The side chain of D3014 is stacked up between the backbone of the beta-strand and the F2976 of an adjacent helix. (I) E3014 has a larger chain length and surface area, which clashes with the F2976. (J) N3042 is present at the beta-sheet interface between OB2 and OB3 and has two contacts across the interface with Y2724. (K) I3042 loses these contacts, which could disturb the proper packing arrangement of these domains. Except (H) and (I), the side chains of the helical domain are colored brown, OB2 are colored orange and OB3 are colored light blue. In (H) and (I), key atoms are shown as spheres with Van der Waals radii. Atoms of D3014 and E3014 are colored white, whereas the other key atoms are colored dark green.
Functional evaluation of variants based on rescue of Brca2KO/KO ES cells
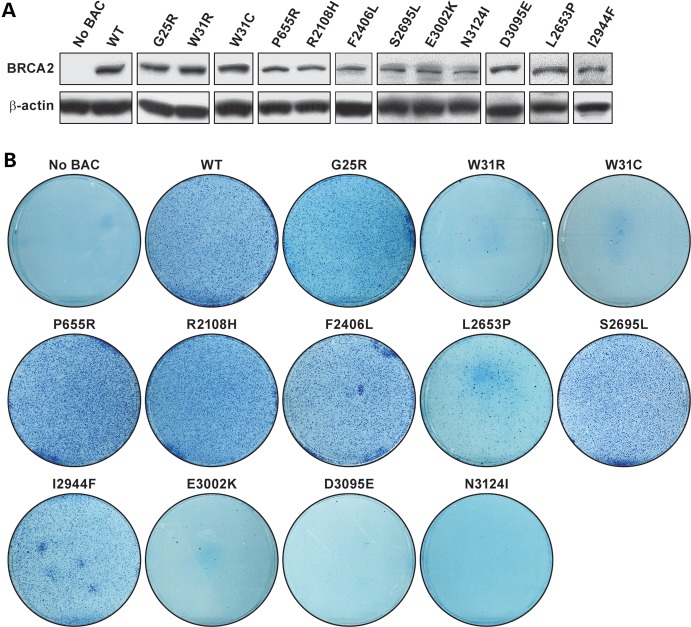
We next used our mES cell-based assay to experimentally test the validity of these predictions and to examine the consequences of these variants on BRCA2 function. We generated each variant in a bacterial artificial chromosome (BAC) clone with a 127 kb insert containing a c-myc-tagged full-length human BRCA2. Individual BACs containing these variants were electroporated into PL2F7 ES cells containing a functionally null and a conditional allele of Brca2 (Brca2CKO/KO) (24). To identify the clones expressing the variants, BAC-positive ES cells were screened by reverse transcription–polymerase chain reaction (RT–PCR, data not shown), followed by western blot analysis (Fig. 3A). At least two independent clones expressing each variant were used for further analysis. One of the assays to assess BRCA2 function is its ability to rescue the lethality of Brca2KO/KO ES cells upon Cre-mediated deletion of the conditional allele. Deletion of this allele also generates a functional human HPRT1 mini-gene that allows selection of the recombinant clones in the presence of hypoxanthine–aminopterin–thymidine (HAT). To assess this, we transiently expressed Cre and selected the recombinant clones in HAT media. HATr colonies were genotyped by Southern analysis to confirm the loss of the conditional allele of Brca2 (Supplementary Material, Fig. S1). ES cells expressing G25R, P655R, R2108H, F2406L, S2695L and I2944F resulted in HATr colonies, and the number of colonies was similar to those expressing WT BRCA2 (Fig. 3B). In contrast, W31R, W31C, D3095E, E3002K and N3124I did not produce any HATr colonies after Cre expression, suggesting these variants are pathogenic (Fig. 3B). L2653P resulted in viable Brca2KO/KO ES cells, but the number of HATr colonies was reduced by 60–70% (Fig. 3B), suggesting that there was a defect in BRCA2 function. This was further supported by the observation that these rescued (Brca2KO/KO;TgL2653P) ES cells were defective in growth when compared with WT cells and had reduced plating efficiency (data not shown). In contrast, the other rescued ES cells were comparable to WT cells in growth and plating efficiencies (data not shown).
Figure 3.
Mouse ES cell-based assay for functional assay of BRCA2 variants: (A) western blot showing the expression of human BRCA2 variants in mES cells before removal of the conditional copy of Brca2. Antibody against c-myc epitope was used to detect the c-myc-tagged BRCA2. β-Actin was used as control. (B) Methylene blue staining of HATr colonies of ES cells expressing no BAC, WT or different variants of BRCA2.
Effect of variants on DNA repair function of BRCA2
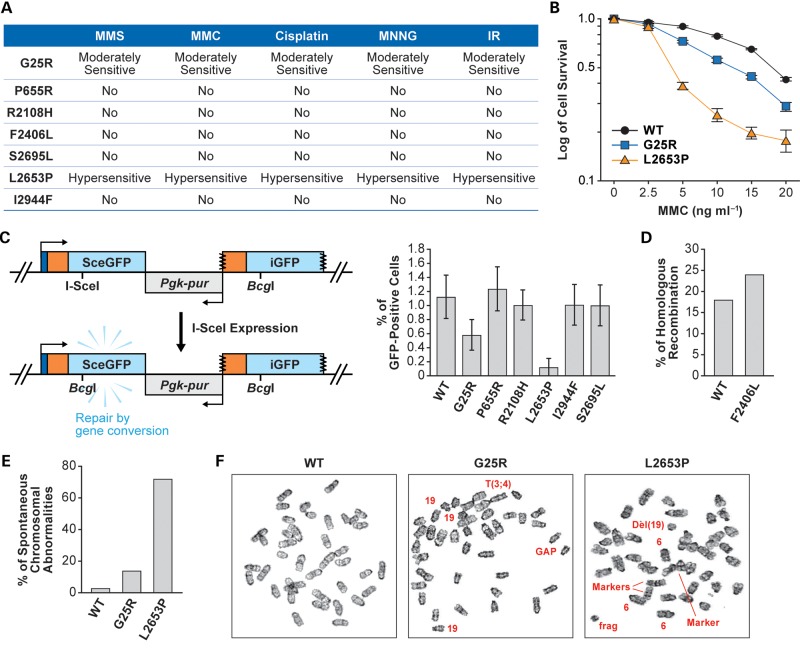
Variants that rescued the lethality of Brca2KO/KO ES cells were next tested to see if they were fully functional or had any defects in the DNA repair function of BRCA2. BRCA2 plays a key role in DNA double-strand break (DSB) repair by HR and BRCA2-mutant cells showed hypersensitivity to different DNA-damaging agents (32–36). We examined the sensitivity of the rescued ES cells to cisplatin, mitomycin C (MMC), methyl methanesulfonate, methyl-N′-nitro-N-nitrosoguanidine and ionizing radiation (IR). Brca2KO/KO ES cells expressing P655R, R2108H, F2406L, S2695L and I2944F variants were indistinguishable from the WT BRCA2-expressing cells in their sensitivity (Fig. 4A). In contrast, ES cells expressing G25R showed a moderate but significant sensitivity (P = 0.0037), and those expressing L2653P exhibited hypersensitivity to all the genotoxins tested (Fig. 4A and B and data not shown).
Figure 4.
Functional evaluation of BRCA2 variants: (A) sensitivity of the Brca2KO/KO ES cells expressing human BRCA2 variants to different DNA-damaging agents. No, no significant difference in the sensitivity compared with the Brca2KO/KO ES cells expressing WT human BRCA2. (B) Survival of ES cells expressing WT, BRCA2G25R or BRCA2L2653P exposed to MMC. P-values for BRCA2G25R is 0.0198 and for BRCA2L2653P is 0.0098 at 10 ng/ml MMC. (C) HR assay using direct repeats of mutated green fluorescent protein (DR-GFP) (37). Upon expression of I-SceI in the cells, a DSB is induced in the SceGFP construct. HR using the promoterless downstream iGFP as template generates an intact GFP protein and can be monitored by cellular green fluorescence. Right panel shows the percentages of GFP-positive cells after expressing I-SceI. (D) HR efficiency as measured by gene targeting at the Rosa26 locus (24). (E) Histogram showing the total number of spontaneous chromosomal abnormalities in cells expressing WT, G25R or L2653P BRCA2. Chromosomal abnormalities observed in WT: breaks/gaps 1%, fragments 2%; in G25R: breaks/gaps 5%, dicentric chromosomes 2%, fragments 4% and marker chromosomes 3%; in L2653P: breaks/gaps 21%, dicentric chromosomes 11%, fragments 17%, radial 2% and marker chromosomes 21%. A marker chromosome is an abnormal chromosome that is distinctive in appearance but not fully identified. Euploidy is not included in these abnormalities. (F) Panels showing representative metaphase spread of cells expressing WT, G25R and L2653P BRCA2. Arrows mark the chromosomal abnormalities; 19 refers to three copies of chromosome 19 in G25R and 6 refers to three copies of chromosome 6 in L2653P. To rule out the possibility of secondary mutations, each experiment was conducted using at least two independent clones.
Next, to determine whether these variants directly affected the DSB repair, we examined the repair of a single DSB using direct repeats of mutated green fluorescent protein (DR-GFP) assay (Fig. 4C, left panel) (37). The ES cells analyzed had DR-GFP integrated into their genome at the Pim1 locus. Expression of I-SceI in cells containing the DR-GFP cassette results in a single DSB. The repair of this DSB by HR results in functional expression of green fluorescent protein (GFP) that can be measured by flow cytometry (37). Because the ES cells expressing F2406L lacked the DR-GFP cassette, effect of this variant on HR was examined by measuring the gene-targeting efficiency as described previously (24). In this study, no other variant was examined by gene-targeting method. Analysis of the ES cells that expressed BRCA2 variants by DR-GFP assay showed that P655R, R2108H, S2695L and I2944F variants had similar levels of HR repair compared with the cells expressing WT BRCA2 (Fig. 4C, right panel). In contrast, HR repair was reduced by 50% in the cells expressing the G25R variant and by 90% in L2653P-expressing cells, implying a defect in HR (Fig. 4C, right panel). Cells expressing F2406L variant exhibited a gene-targeting frequency similar to the cells expressing WT BRCA2, suggesting that this variant is not defective in HR (Fig. 4D).
To examine the effect of G25R and L2653P on overall genomic integrity, we examined the chromosome spreads of the ES cells expressing these variants. We found G25R-expressing cells to exhibit a moderate increase in spontaneous chromosomal aberrations (Fig. 4E and F), which is consistent with the moderate defect in DNA repair observed above (Fig. 4A and B). ES cells expressing L2653P showed a marked increase in genomic instability based on the number of chromosomal aberrations observed in these cells (Fig. 4E and F). Taken together, these functional studies suggest that P655R, R2108H, F2406L, S2695L and I2944F variants are functionally similar to WT BRCA2, whereas G25R and L2653P are defective in DNA repair.
Effect of the BRCA2 variants on exon inclusion/exclusion
Although full-length BRCA2 protein was detected for all the variants analyzed in this study, small deletions due to exon skipping or generation of cryptic splice sites may not be detectable by western blot analysis. Therefore, we examined these variants for any effect on exonic splicing regulator (ESR) sequences using ESEfinder (http://rulai.cshl.edu/cgibin/tools/ESE3/esefinder.cgi?process=home) and the FAS-ESS web server (http://genes.mit.edu/fas-ess/) (38–40). Three of the variants, G25R (301G>A), W31R (319T>C) and W31C (321G>T) are located in exon 3. We did not detect a change in any ESR sequences due to the 301G>A (G25R) mutation. In contrast, 319T>C (W31R) is predicted to result in a putative SRp40-binding site (TTAATCG) and 321G>T (W31C) may generate a putative exonic silencer sequence (ESS) site (TTGTTT) (Supplementary Material, Table S2). RT–PCR analysis of the ES cells expressing these variants using the primers from exons 2 and 4 showed the presence of a 368 bp product containing exon 3. An additional 119 bp fragment (59.9% of total transcripts, Supplementary Material, Fig. S2A) was observed in ES cells expressing 321G>T (W31C). This fragment was also present at greatly reduced levels in cells expressing WT BRCA2 (8.34% of total transcripts) and the 301G>A (G25R) variant (5.5% of total transcripts) but was absent in cells expressing 319T>C (W31R) (Supplementary Material, Fig. S2A). Sequence analysis revealed that this 119 bp product lacked exon 3 (Supplementary Material, Fig. S2B). These results suggest that c.321G>T (W31C) causes increased skipping of exon 3, which may be due to the generation of a putative ESS. This observation is consistent with the recent finding by Sanz et al. (41) who used a splicing mini-gene construct to show that 321G>T (W31C) variant caused an increase in exon 3 exclusion. Skipping of exon 3 is predicted to generate an in-frame deletion of 83 amino acids, essential for BRCA2 interaction with PALB2 (26). Furthermore, loss of exon 3 is associated with an increased risk of breast/ovarian cancer (42).
Among the seven (F2406L, L2653P, S2695L, I2944F, E3002K, D3095E and N3124I) other variants, 9058A>T (I2944F) is predicted to generate a putative SF2/ASF-binding site (CAGAAGG) and 9599A>T (N3124I) is predicted to result in one putative SRp55-binding site (AGCATC) (Supplementary Material, Table S2). On the other hand, 9513C>G (D3095E) is predicted to result in a loss of one putative SF2/ASF(IgM-BRCA1)-binding site (CGAATGT) and 9232G>A (E3002K) caused a loss of one putative SF2/ASF-binding site (CAGAAGG) (Supplementary Material, Table S2). Previously, 9513C>G (D3095E) was shown to localize to a predicted exonic splicing enhancer site and was predicted to alter SF2/ASF binding (43). None of these variants affected any putative ESS sequences as predicted by the FAS-ESS web server (Supplementary Material, Table S2). RT–PCR analysis of ES cells expressing these seven variants showed no effect on exon skipping or alternative splicing of the transcripts compared with the WT BRCA2-expressing cells (Supplementary Material, Fig. S2C–E).
The remaining two variants [2192C>G (P655R) and 6551G>A (R2108H)] are located in exon 11 and the potential loss of this large exon due to exon skipping is predicted to result in a much smaller protein. Since we did not observe any detectable change in the size of the proteins encoded by these two mutants (Fig. 3A), we concluded that these variants have no effect on exon 11 exclusion.
In vivo role of Phe2406 of BRCA2
Among the eight residues of BRCA2 analyzed, Phe2406 is located in a conserved PhePP motif (amino acids 2386–2411) of BRCA2 and was inferred to be important because of its interaction with meiosis-specific RecA homolog DMC1, but not with RAD51 based on in vitro studies (29). Substituting Phe2406 in this motif with any other amino acid except the two other aromatic amino acids was shown to disrupt this interaction (29). Since the effect of losing this interaction with DMC1 will be visible only in meiotic cells, we generated a knock-in mouse model where the hydrophobic phenylalanine at position 2351 of mouse BRCA2 (corresponding to human 2406 residue) was replaced by a hydrophilic and charged aspartic acid residue. Although, leucine at this position is predicted to have a detrimental effect on the interaction, substitution to aspartic acid residue is predicted to severely disrupt the interaction. To have a clear phenotype in mice, we chose to examine the effect of F2351D instead of F2351L. Based on functional studies in mES cells, F2406D was functionally indistinguishable from WT BRCA2 as well as F2406L (data not shown).
Using a gene-targeting approach, the desired mutation (TTC → GAC) was generated in exon 14 of mouse Brca2 in ES cells (see Materials and Methods and Supplementary Material, Fig. S4 for details). Heterozygous (Brca2F2351D/+) mice carrying the mutant allele (Brca2F2351D, referred to as M allele for simplicity) were intercrossed and also crossed with mice heterozygous for the null allele [Brca2tmBrd1 or knockout allele (KO) for simplicity]. Homozygous mutant (Brca2F2351D/Brca2F2351D or M/M) and compound heterozygous (KO/M) offspring were obtained at the expected Mendelian ratio (data not shown). The gonad sizes, fecundity and fertility of homozygous (M/M) and compound heterozygous (KO/M) mice were comparable to the WT or heterozygous littermates (data not shown). The histological analysis of 4-week-old mutant male testes and 3-week-old mutant female ovaries were indistinguishable from WT (Supplementary Material, Fig. S4A). To determine whether DMC1 was properly recruited to chromosomes during prophase I, we analyzed surface spreads of spermatocytes and stained them with SYCP3 and DMC1. SYCP3, a component of the lateral elements of the synaptonemal complex, was used to identify cells at different stages of meiotic prophase based on the degree of chromosome condensation and synaptonemal complex formation (44). Our results show that DMC1 focus formation was not affected by this mutation (Supplementary Material, Fig. S4B and C). Taken together, these results indicate that mutating the conserved phenylalanine residues of the PhePP motif to aspartate does not have an effect on meiotic progression and gametogenesis.
DISCUSSION
Interpreting the results of BRCA1 and BRCA2 genetic tests to predict breast cancer risk is of significant clinical importance. Determining the functional significance of variants that clearly disrupt the gene or variants for which sufficient epidemiological data are available can be relatively easy. In spite of the successful classification of numerous variants, hundreds of others remain unclassified due to the availability of limited familial or functional data. In this study, we have used our mES cell-based approach to evaluate the functional significance of eight variants listed in the BIC database that are of unknown clinical significance (Table 4). In addition, we have examined four variants that are of known pathogenicity that served as controls. As a first step towards understanding the consequences of these variants, we used co-segregation data to determine their pathogenicity based on the LR for each variant. The data supported the non-pathogenic nature of P655R, R2108H and I2944F. The LRs supported the likely pathogenic effect of L2653P, E3002K, D3095E and N3124I. These results were also supported by their Align-GVGD grade (Tables 3 and 4). Very limited or no information for other variants was available, making it difficult to determine their pathogenicity.
Our efforts to classify the BRCA2 variants using the ES cell-based functional assay revealed that the two known non-pathogenic variants, P655R and R2108H, had no deleterious effects on BRCA2 function. They behaved like WT BRCA2 in their ability to rescue the lethality of Brca2KO/KO ES cells as well as other DNA repair assays and are considered non-pathogenic. Our evaluation of D3095E revealed its failure to rescue the lethality of Brca2KO/KO ES cells, which is consistent with its classification based on posterior probability value of being likely pathogenic. Interestingly, L2653P (classified as class 5 based on posterior probability) resulted in viable Brca2KO/KO ES cells, albeit their number was significantly reduced compared with WT BRCA2. Furthermore, these rescued cells were hypersensitive to DNA-damaging agents and exhibited a marked increase in genomic instability suggesting a significant defect in BRCA2 DNA repair function suggesting that this variant is likely to be pathogenic. Based on our previous analysis of more than 25 variants using this approach, we have identified at least four variants (Y3308X, E3309X, R2336H and L2510P) that exhibited a similar phenotype and in every case the available familial data suggested that they are pathogenic (24,45). Taken together, the results of our functional evaluation of these four variants are consistent with their known classification (Tables 2 and 4).
Our evaluation of the eight VUS revealed that similar to the D3095E variant, W31R, W31C, E3002 and N3124I failed to result in viable Brca2KO/KO ES cells suggesting that these variants clearly disrupt BRCA2 function and are pathogenic. W31R and W31C are located in the PALB2-binding domain of BRCA2 (26). One of the core residues essential for this interaction is a tryptophan residue at position 31 of BRCA2 that forms a polar bridge with Ser1065 of PALB2 (46). Change of this residue to arginine or cysteine has been shown to abolish the interaction with PALB2. Moreover, the nucleotide change for W31C also affects the ESR sequence, which is likely to cause skipping of exon 3 (41). Our functional evaluation of E3002K confirms a recent study of 58 French Canadian families with at least three cases of breast and/or ovarian cancer and 960 cases not selected for family history of cancer, which found this variant to be pathogenic (47).
The remaining four variants (G25R, F2406L, S2695L and I2944F) resulted in viable Brca2KO/KO ES cells and the number of rescued colonies was comparable to WT, P655R and R2108H. Further studies revealed that F2406L, S2695L and I2944F were fully proficient in DNA repair based on their sensitivity to various DNA-damaging agents as well efficiency of HR suggesting that these variants have no defect in BRCA2 function and are non-pathogenic. We obtained information from only two families carrying S2695L variant and none for F2406L, which makes it difficult to determine their pathogenicity based on co-segregation data. The phenylalanine residue at position 2406 is located in the DMC1-binding motif of BRCA2 (2404KVFVPPFK2411), which is essential for its interaction with DMC1 based on in vitro studies using a mutant peptide array (29). The fact that this residue interacts with a meiosis-specific protein may explain why we did not observe any defect in mitotic cells when this residue was changed to leucine. Surprisingly, in a knock-in mouse model, in which we substituted the hydrophobic phenylalanine residue with hydrophilic and charged aspartate residue (F2351D, equivalent to F2406D in humans), we observed no apparent defect in meiotic cells as well. The non-pathogenic nature of I2944F is supported by the LRs calculated based on co-segregation data obtained from 12 families (Table 3). I2944F was found in unaffected individuals based on SNP database as well as in an epidemiological study using 71 breast cancer families and 95 control individuals (48,49). These observations strongly suggest the non-pathogenic nature of I2944F variant, which is confirmed by the results of our functional analysis.
Unlike F2406L, S2695L and I2944F, G25R exhibited moderate sensitivity to all DNA-damaging agents suggesting that BRCA2 function is compromised. Compared with W31C and W31R, the relatively mild effect of G25R may result from its moderate effect on PALB2 binding (26). This intermediate phenotype makes it difficult to determine the precise pathogenicity of G25R. Previously, while assessing the functional consequences of R3052Q, we observed a phenotype similar to G25R. We observed no effect on viability of ES cells; however, a very mild sensitivity to a few DNA-damaging agents (24). Although we did not observe a significant increase in genomic instability, we did observe some chromosomal translocations in the ES cells (Kuznetsov and Sharan, unpublished data). These observations led us to conclude that this variant might be a low-risk variant. Based on posterior priority score, R3052Q is considered likely to be non-pathogenic (class 2 variant) (31). In the case of G25R, although we observed no effect on the viability of ES cells, the cells exhibited consistent moderate sensitivity to all DNA-damaging agents, reduction in HR and an increase in genomic instability suggesting that this variant is defective in DSB repair and is likely to be pathogenic. However, the risk of developing the disease may be lower than other variants that show a marked reduction in cell survival and exhibit hypersensitivity to genotoxins.
In addition to the Align-GVGD score that takes into account the evolutionary conservation of amino acids, we have used homology-based modeling to predict the effect of variants (L263P, S2695L, I2944F, E3002K, D3095E and N3124I) on the structure of the C-terminal domain of BRCA2 (Fig. 2). The results of our functional studies confirm the predictions and support the use of homology-based modeling as a predictive tool to determine the functional consequences of variants mapping to this region. A recent prediction of human C-terminal BRCA2 mutants based on a computational prediction that uses a probabilistic ratio to predict the functional impairment of the protein by any variant matches very well with our results in all cases except for S2695L, which was considered inconclusive in this protein LR model (49). Our functional studies in mES cells show that S2695L is functionally indistinguishable from WT BRCA2.
In conclusion, the comprehensive functional characterization of 12 variants described here using our mES cell-based approach has allowed us to experimentally determine their effect on BRCA2 function and to classify them based on their pathogenicity. We have extended the functional characterization to include structural predictions using homology-based modeling. As we expand the number of variants that are evaluated, we will have a better understanding of the full range of phenotypic heterogeneity that can be observed between different variants in the ES cells, which can be utilized to calculate LR. While the mES cell-based assay has not yet been fully validated for use in variant reclassification by commercial diagnostic laboratories, the results reported here further illustrate the utility of the mES cells for the functional evaluation of human BRCA2 variants of unknown pathogenicity.
MATERIALS AND METHODS
Reagents
BAC CTD-2342K5 with a 127 kb insert containing the full-length human BRCA2 gene was used to generate mutations. All oligonucleotides were obtained from Invitrogen. Antibodies used are c-myc tag (ab18185, Abcam) and actin (Ab-5, NeoMarkers).
Selection of BRCA2 variants
Variants were selected from the list of BRCA2 variants deposited in the BIC database (http://research.nhgri.nih.gov/bic/). VUS that mapped to the PALB2-binding domain and the C-terminal DNA-binding domain were selected for evaluation. GenBank accession number of BRCA2 is NM_000059.3. As per human genome variation society nomenclature, nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence. The initiation codon is codon 1. For BIC nomenclature, +228 corresponds to the ATG translation initiation codon in the reference sequence.
Co-segregation LRs
Probands were BRCA1 and BRCA2 sequenced at Myriad Genetic Laboratories, Inc. Only probands absent for any other known deleterious mutations or uncertain variants were included in co-segregation analysis. With few exceptions, family members were only tested for the presence or absence of the variant identified in the proband. Co-segregation analysis LRs were determined as described by Mohammadi et al. (17).
Align-GVGD
All variants were examined for evolutionary conservation using Align-GVGD (http://www.agvgd.iarc.fr/), which combines protein multiple sequence alignments and the physiochemical properties of the amino acids to determine Grantham variation (GV) and Grantham deviation (GD) scores. The GV and GD scores are used to determine the Align-GVGD grade (C0, C15, C25, C35, C45, C55, C65), with C0 being most likely neutral and C65 being most likely pathogenic.
Generation of BRCA2 variants in a bacterial artificial chromosome clone
BRCA2 was N-terminally tagged with the c-myc epitope in the BAC using the mini-lambda-based ‘recombineering’ (recombination-mediated genetic engineering) system, as described previously (50). Mutations were introduced either by the recombineering-based ‘hit and fix’ method (50) or by the galK selection and counter-selection method (51). Oligonucleotide sequences are available upon request.
Generation of BRCA2 transgenic ES cells
PL2F7 ES cells were maintained as described previously (24). Thirty micrograms of BAC DNA carrying various mutant alleles of BRCA2 were electroporated into 1.1 × 107 PL2F7 ES cells, selected in the presence of G418 (Invitrogen) and characterized as described previously (24). BRCA2 expression was checked using the Titan one-tube RT–PCR kit (Roche) according to the manufacturer's protocol. The primers used for RT–PCR are forward primer 5′-ACATGTCCCGAAAATGAGGA-3′ and a reverse primer 5′-GCCGATCTTCTGCTTCTATCA-3′ specific to exons 11 and 18, respectively. The amplified product is a 1250 bp fragment as determined by agarose gel electrophoresis.
Expression analysis
Proteins were extracted in radioimmunoprecipitation assay buffer (50 mm Tris–HCl, pH 7.4, 1 mm ethylenediaminetetraacetic acid, 150 mm NaCl, 0.1% sodium dodecyl sulphate, 1% Triton X-100, 0.25% sodium deoxycholate, 1 mm sodium fluoride, 1 mm orthovanadate) and separated using NuPAGE 4–12% gradient gel (Invitrogen) electrophoresis for western blot analysis. ECL plus western blotting detection system (Amersham) was used for chemiluminescent detection.
BRCA2 functional assays
A number of functional assays that examine the effect of the variants on BRCA2 protein function were performed. To examine the effect of BRCA2 variants on ES cell viability, the conditional allele of Brca2 in ES cells expressing each variant was deleted by electroporating 20 μg of Pgk-Cre plasmid into BRCA2-expressing clones. Recombinant colonies were selected in the HAT media (Gibco) after seeding 1 × 106 cells in a 100 mm dish. HATr colonies were visualized by staining with methylene blue (2% methylene blue (wt/vol) in 70% ethanol for 15 min followed by washing in 70% ethanol). Sensitivity assays to different drugs and IR were performed as described previously (24). For the HR assay using DR-GFP reporter construct, ES cells with stable DR-GFP integration were electroporated with 80 μg of empty vector (pCAGGS) or the I-SceI expression vector pCABSce (37). GFP-positive cells were counted by flow cytometric analysis 72 h after transfection. HR efficiency was tested by gene targeting at Rosa26 locus as described previously (24). Karyotyping of the ES cells was performed after colcemid (Invitrogen) treatment for 1.5 h, as described previously (24). We randomly selected 100 well-spread metaphases containing at least 40 chromosomes from each genotype and examined them for structural aberrations, blind of the BRCA2 genotype.
Effect of variants on splicing
We examined the effect of variants on aberrant splicing by RT–PCR, and total RNA was extracted using RNA-BEE (Tel-Test, Inc.) according to the manufacturer's protocol. To detect an alternatively spliced form of BRCA2, RT–PCR analysis was performed using Titan one-step RT–PCR kit (Roche) according to the manufacturer's protocol. Sequence of primers used is listed in Supplementary Material, Table S1. Each detectable splice variant was cloned into TOPO cloning vector (Invitrogen) and sequenced.
Generation of Brca2F2351D knock-in mice
The Brca2F2351D allele was generated in embryonic stem cell using a targeting construct that represents 15.2 kb of mouse genomic DNA that contains the region from intron 11 to exon 21 of Brca2 gene. This 15.2 kb fragment was retrieved from BAC421 using recombineering. The targeting vector contains the positively selectable gene neor flanked by two loxP sites inserted into intron 14 of the Brca2 gene. The F2351D mutation, encoded by a TTC → GAC change, is located in exon 14. This replacement generates a new SalI restriction site. The targeting construct contains also a copy of the negatively selectable marker, herpes simplex thymidine kinase gene (TK) (Supplementary Material, Fig. S3A). The correctly targeted ES clones were selected by Southern blot and presence of the mutation was confirmed by sequencing and restriction digestion of the PCR-amplified fragment (484 bp) surrounding the mutated region by SalI (Supplementary Material, Fig. S3B–F). Resulting chimeras from these ES cells successfully transmitted the mutated allele after breeding with WT mice. Heterozygous offspring in the C57BL/6J × 129/Sv mixed genetic background were crossed with β-actin–Cre transgenic mice (52) to remove the neor gene from intron 14.
Histology
Testes and ovaries were fixed in 10% neutral buffered formalin. After dehydration in ethanol, series samples were embedded in paraffin, serially sectioned and stained with hematoxylin and eosin. Slides were examined using bright field microscopy.
Spermatocyte spread preparation and immunofluorescence
Surface spreads of spermatocytes from the testes of 6-week-old mutant and control animals were prepared and stained as described previously (53). The primary antibodies for immunofluorescence were used for SYCP3 at 1:500 dilution and for DMC1 at 1:200 dilution. Secondary antibodies used were goat anti-rabbit Ig-G Alexa Fluor 488 and goat anti-mouse Ig-G Alexa Fluor 594 (Invitrogen). Secondary antibodies were used at a 1:1000 dilution.
Crystal structure modeling
The crystal structure of C-terminal portion of mouse BRCA2 was retrieved from the protein data bank at http://www.rcsb.org/ (accession code 1MIU). Mutations of the critical residues were carried out in Crystallographic Object-Oriented Toolkit (Coot), followed by selection of the highest probable rotamer orientation and regularization (54). The resultant molecules are analyzed and displayed with the software UCSF Chimera (55).
Statistical analyses
All data are expressed as a mean ± SD. Differences between two groups were compared using two-tailed unpaired Student's t-test (Microsoft Excel for Mac). P < 0.05 was considered significant.
SUPPLEMENTARY MATERIAL
FUNDING
Research supported by intramural funds from US National Cancer Institute, National Institutes of Health.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Jairaj Acharya, Suhwan Chang, Rajanikant Chittela, Ira Daar and Eswary Thirthagiri for helpful discussions and critical review of the manuscript. We also thank Jiro Wada (SAIC-Frederick, Inc., Scientific Publications, Graphics & Media Department) for illustrations. We thank Dr Maria Jasin for the DR-GFP plasmid and I-SceI expression vector. The research was sponsored by the Center for Cancer Research, National Cancer Institute, US National Institutes of Health.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Fackenthal J.D., Olopade O.I. Breast cancer risk associated with BRCA1 and BRCA2 in diverse populations. Nat. Rev. Cancer. 2007;7:937–948. doi: 10.1038/nrc2054. [DOI] [PubMed] [Google Scholar]
- 2.Ford D., Easton D.F., Stratton M., Narod S., Goldgar D., Devilee P., Bishop D.T., Weber B., Lenoir G., Chang-Claude J., et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am. J. Hum. Genet. 1998;62:676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen S., Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. J. Clin. Oncol. 2007;25:1329–1333. doi: 10.1200/JCO.2006.09.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rahman N., Stratton M.R. The genetics of breast cancer susceptibility. Annu. Rev. Genet. 1998;32:95–121. doi: 10.1146/annurev.genet.32.1.95. [DOI] [PubMed] [Google Scholar]
- 5.Nathanson K.L., Wooster R., Weber B.L. Breast cancer genetics: what we know and what we need. Nat. Med. 2001;7:552–556. doi: 10.1038/87876. [DOI] [PubMed] [Google Scholar]
- 6.Palma M., Ristori E., Ricevuto E., Giannini G., Gulino A. BRCA1 and BRCA2: the genetic testing and the current management options for mutation carriers. Crit. Rev. Oncol. Hematol. 2006;57:1–23. doi: 10.1016/j.critrevonc.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Domchek S.M., Friebel T.M., Singer C.F., Evans D.G., Lynch H.T., Isaacs C., Garber J.E., Neuhausen S.L., Matloff E., Eeles R., et al. Association of risk-reducing surgery in BRCA1 or BRCA2 mutation carriers with cancer risk and mortality. JAMA. 2010;304:967–975. doi: 10.1001/jama.2010.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank T.S., Deffenbaugh A.M., Reid J.E., Hulick M., Ward B.E., Lingenfelter B., Gumpper K.L., Scholl T., Tavtigian S.V., Pruss D.R., et al. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J. Clin. Oncol. 2002;20:1480–1490. doi: 10.1200/JCO.2002.20.6.1480. [DOI] [PubMed] [Google Scholar]
- 9.Eggington J.M. National Society of Genetic Counselors 2011 Annual Education Conference. San Diego, CA: San Diego Marriott; 2011. Perplexing variants of uncertain significance; p. 13. [Google Scholar]
- 10.Easton D.F., Deffenbaugh A.M., Pruss D., Frye C., Wenstrup R.J., Allen-Brady K., Tavtigian S.V., Monteiro A.N., Iversen E.S., Couch F.J., et al. A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am. J. Hum. Genet. 2007;81:873–883. doi: 10.1086/521032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldgar D.E., Easton D.F., Byrnes G.B., Spurdle A.B., Iversen E.S., Greenblatt M.S. Genetic evidence and integration of various data sources for classifying uncertain variants into a single model. Hum. Mutat. 2008;29:1265–1272. doi: 10.1002/humu.20897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tavtigian S.V., Byrnes G.B., Goldgar D.E., Thomas A. Classification of rare missense substitutions, using risk surfaces, with genetic- and molecular-epidemiology applications. Hum. Mutat. 2008;29:1342–1354. doi: 10.1002/humu.20896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plon S.E., Eccles D.M., Easton D., Foulkes W.D., Genuardi M., Greenblatt M.S., Hogervorst F.B., Hoogerbrugge N., Spurdle A.B., Tavtigian S.V. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum. Mutat. 2008;29:1282–1291. doi: 10.1002/humu.20880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomez Garcia E.B., Oosterwijk J.C., Timmermans M., van Asperen C.J., Hogervorst F.B., Hoogerbrugge N., Oldenburg R., Verhoef S., Dommering C.J., Ausems M.G., et al. A method to assess the clinical significance of unclassified variants in the BRCA1 and BRCA2 genes based on cancer family history. Breast Cancer Res. 2009;11:R8. doi: 10.1186/bcr2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spearman A.D., Sweet K., Zhou X.P., McLennan J., Couch F.J., Toland A.E. Clinically applicable models to characterize BRCA1 and BRCA2 variants of uncertain significance. J. Clin. Oncol. 2008;26:5393–5400. doi: 10.1200/JCO.2008.17.8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spurdle A.B., Lakhani S.R., Healey S., Parry S., Da Silva L.M., Brinkworth R., Hopper J.L., Brown M.A., Babikyan D., Chenevix-Trench G., et al. Clinical classification of BRCA1 and BRCA2 DNA sequence variants: the value of cytokeratin profiles and evolutionary analysis—a report from the kConFab Investigators. J. Clin. Oncol. 2008;26:1657–1663. doi: 10.1200/JCO.2007.13.2779. [DOI] [PubMed] [Google Scholar]
- 17.Mohammadi L., Vreeswijk M.P., Oldenburg R., van den Ouweland A., Oosterwijk J.C., van der Hout A.H., Hoogerbrugge N., Ligtenberg M., Ausems M.G., van der Luijt R.B., et al. A simple method for co-segregation analysis to evaluate the pathogenicity of unclassified variants; BRCA1 and BRCA2 as an example. BMC Cancer. 2009;9:211. doi: 10.1186/1471-2407-9-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldgar D., Venne V., Conner T., Buys S. BRCA phenocopies or ascertainment bias? J. Med. Genet. 2007;44:e86. author reply e88. [PMC free article] [PubMed] [Google Scholar]
- 19.Carvalho M.A., Couch F.J., Monteiro A.N. Functional assays for BRCA1 and BRCA2. Int. J. Biochem. Cell Biol. 2007;39:298–310. doi: 10.1016/j.biocel.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Couch F.J., Rasmussen L.J., Hofstra R., Monteiro A.N., Greenblatt M.S., de Wind N. Assessment of functional effects of unclassified genetic variants. Hum. Mutat. 2008;29:1314–1326. doi: 10.1002/humu.20899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee M.S., Green R., Marsillac S.M., Coquelle N., Williams R.S., Yeung T., Foo D., Hau D.D., Hui B., Monteiro A.N., et al. Comprehensive analysis of missense variations in the BRCT domain of BRCA1 by structural and functional assays. Cancer Res. 2010;70:4880–4890. doi: 10.1158/0008-5472.CAN-09-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farrugia D.J., Agarwal M.K., Pankratz V.S., Deffenbaugh A.M., Pruss D., Frye C., Wadum L., Johnson K., Mentlick J., Tavtigian S.V., et al. Functional assays for classification of BRCA2 variants of uncertain significance. Cancer Res. 2008;68:3523–3531. doi: 10.1158/0008-5472.CAN-07-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L., Biswas K., Habib L.A., Kuznetsov S.G., Hamel N., Kirchhoff T., Wong N., Armel S., Chong G., Narod S.A., et al. Functional redundancy of exon 12 of BRCA2 revealed by a comprehensive analysis of the c.6853A>G (p.I2285V) variant. Hum. Mutat. 2009;30:1543–1550. doi: 10.1002/humu.21101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuznetsov S.G., Liu P., Sharan S.K. Mouse embryonic stem cell-based functional assay to evaluate mutations in BRCA2. Nat. Med. 2008;14:875–881. doi: 10.1038/nm.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang S., Biswas K., Martin B.K., Stauffer S., Sharan S.K. Expression of human BRCA1 variants in mouse ES cells allows functional analysis of BRCA1 mutations. J. Clin. Invest. 2009;119:3160–3171. doi: 10.1172/JCI39836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia B., Sheng Q., Nakanishi K., Ohashi A., Wu J., Christ N., Liu X., Jasin M., Couch F.J., Livingston D.M. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 27.Yang H., Jeffrey P.D., Miller J., Kinnucan E., Sun Y., Thoma N.H., Zheng N., Chen P.L., Lee W.H., Pavletich N.P. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 28.Holloman W.K. Unraveling the mechanism of BRCA2 in homologous recombination. Nat. Struct. Mol. Biol. 2011;18:748–754. doi: 10.1038/nsmb.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thorslund T., Esashi F., West S.C. Interactions between human BRCA2 protein and the meiosis-specific recombinase DMC1. EMBO J. 2007;26:2915–2922. doi: 10.1038/sj.emboj.7601739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldgar D.E., Easton D.F., Deffenbaugh A.M., Monteiro A.N., Tavtigian S.V., Couch F.J. Integrated evaluation of DNA sequence variants of unknown clinical significance: application to BRCA1 and BRCA2. Am. J. Hum. Genet. 2004;75:535–544. doi: 10.1086/424388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindor N.M., Guidugli L., Wang X., Vallee M.P., Monteiro A.N., Tavtigian S., Goldgar D.E., Couch F.J. A review of a multifactorial probability-based model for classification of BRCA1 and BRCA2 variants of uncertain significance (VUS) Hum. Mutat. 2012;33:8–21. doi: 10.1002/humu.21627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramus S.J., Gayther S.A. The contribution of BRCA1 and BRCA2 to ovarian cancer. Mol. Oncol. 2009;3:138–150. doi: 10.1016/j.molonc.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel K.J., Yu V.P., Lee H., Corcoran A., Thistlethwaite F.C., Evans M.J., Colledge W.H., Friedman L.S., Ponder B.A., Venkitaraman A.R. Involvement of Brca2 in DNA repair. Mol. Cell. 1998;1:347–357. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- 34.Morimatsu M., Donoho G., Hasty P. Cells deleted for Brca2 COOH terminus exhibit hypersensitivity to gamma-radiation and premature senescence. Cancer Res. 1998;58:3441–3447. [PubMed] [Google Scholar]
- 35.Marple T., Kim T.M., Hasty P. Embryonic stem cells deficient for Brca2 or Blm exhibit divergent genotoxic profiles that support opposing activities during homologous recombination. Mutat. Res. 2006;602:110–120. doi: 10.1016/j.mrfmmm.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 36.Godthelp B.C., van Buul P.P., Jaspers N.G., Elghalbzouri-Maghrani E., van Duijn-Goedhart A., Arwert F., Joenje H., Zdzienicka M.Z. Cellular characterization of cells from the Fanconi anemia complementation group, FA-D1/BRCA2. Mutat. Res. 2006;601:191–201. doi: 10.1016/j.mrfmmm.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 37.Pierce A.J., Johnson R.D., Thompson L.H., Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z., Rolish M.E., Yeo G., Tung V., Mawson M., Burge C.B. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 39.Smith P.J., Zhang C., Wang J., Chew S.L., Zhang M.Q., Krainer A.R. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum. Mol. Genet. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- 40.Cartegni L., Wang J., Zhu Z., Zhang M.Q., Krainer A.R. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanz D.J., Acedo A., Infante M., Duran M., Perez-Cabornero L., Esteban-Cardenosa E., Lastra E., Pagani F., Miner C., Velasco E.A. A high proportion of DNA variants of BRCA1 and BRCA2 is associated with aberrant splicing in breast/ovarian cancer patients. Clin. Cancer Res. 2010;16:1957–1967. doi: 10.1158/1078-0432.CCR-09-2564. [DOI] [PubMed] [Google Scholar]
- 42.Nordling M., Karlsson P., Wahlstrom J., Engwall Y., Wallgren A., Martinsson T. A large deletion disrupts the exon 3 transcription activation domain of the BRCA2 gene in a breast/ovarian cancer family. Cancer Res. 1998;58:1372–1375. [PubMed] [Google Scholar]
- 43.Pettigrew C.A., Wayte N., Wronski A., Lovelock P.K., Spurdle A.B., Brown M.A. Colocalisation of predicted exonic splicing enhancers in BRCA2 with reported sequence variants. Breast Cancer Res. Treat. 2008;110:227–234. doi: 10.1007/s10549-007-9714-5. [DOI] [PubMed] [Google Scholar]
- 44.Dobson M.J., Pearlman R.E., Karaiskakis A., Spyropoulos B., Moens P.B. Synaptonemal complex proteins: occurrence, epitope mapping and chromosome disjunction. J. Cell Sci. 1994;107:2749–2760. doi: 10.1242/jcs.107.10.2749. [DOI] [PubMed] [Google Scholar]
- 45.Biswas K., Das R., Alter B.P., Kuznetsov S.G., Stauffer S., North S.L., Burkett S., Brody L.C., Meyer S., Byrd R.A., et al. A comprehensive functional characterization of BRCA2 variants associated with Fanconi anemia using mouse ES cell-based assay. Blood. 2011;118:2430–2442. doi: 10.1182/blood-2010-12-324541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oliver A.W., Swift S., Lord C.J., Ashworth A., Pearl L.H. Structural basis for recruitment of BRCA2 by PALB2. EMBO Rep. 2009;10:990–996. doi: 10.1038/embor.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cote S., Arcand S.L., Royer R., Nolet S., Mes-Masson A.M., Ghadirian P., Foulkes W.D., Tischkowitz M., Narod S.A., Provencher D., et al. The BRCA2 c.9004G>A (E2003K) variant is likely pathogenic and recurs in breast and/or ovarian cancer families of French Canadian descent. Breast Cancer Res. Treat. 2012;131:333–340. doi: 10.1007/s10549-011-1796-4. [DOI] [PubMed] [Google Scholar]
- 48.Wagner T.M., Hirtenlehner K., Shen P., Moeslinger R., Muhr D., Fleischmann E., Concin H., Doeller W., Haid A., Lang A.H., et al. Global sequence diversity of BRCA2: analysis of 71 breast cancer families and 95 control individuals of worldwide populations. Hum. Mol. Genet. 1999;8:413–423. doi: 10.1093/hmg/8.3.413. [DOI] [PubMed] [Google Scholar]
- 49.Karchin R., Agarwal M., Sali A., Couch F., Beattie M.S. Classifying variants of undetermined significance in BRCA2 with protein likelihood ratios. Cancer Inform. 2008;6:203–216. doi: 10.4137/cin.s618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Y., Sharan S.K. A simple two-step, ‘hit and fix’ method to generate subtle mutations in BACs using short denatured PCR fragments. Nucleic Acids Res. 2003;31:e80. doi: 10.1093/nar/gng080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warming S., Costantino N., Court D.L., Jenkins N.A., Copeland N.G. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lewandoski M., Meyers E.N., Martin G.R. Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harb. Symp. Quant. Biol. 1997;62:159–168. [PubMed] [Google Scholar]
- 53.Romanienko P.J., Camerini-Otero R.D. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell. 2000;6:975–987. doi: 10.1016/s1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- 54.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 55.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.