Abstract
Recurrent microdeletions of 8p23.1 that include GATA4 and SOX7 confer a high risk of both congenital diaphragmatic hernia (CDH) and cardiac defects. Although GATA4-deficient mice have both CDH and cardiac defects, no humans with cardiac defects attributed to GATA4 mutations have been reported to have CDH. We were also unable to identify deleterious GATA4 sequence changes in a CDH cohort. This suggested that haploinsufficiency of another 8p23.1 gene may contribute, along with GATA4, to the development of CDH. To determine if haploinsufficiency of SOX7—another transcription factor encoding gene—contributes to the development of CDH, we generated mice with a deletion of the second exon of Sox7. A portion of these Sox7Δex2/+ mice developed retrosternal diaphragmatic hernias located in the anterior muscular portion of the diaphragm. Anterior CDH is also seen in Gata4+/− mice and has been described in association with 8p23.1 deletions in humans. Immunohistochemistry revealed that SOX7 is expressed in the vascular endothelial cells of the developing diaphragm and may be weakly expressed in some diaphragmatic muscle cells. Sox7Δex2/Δex2 embryos die prior to diaphragm development with dilated pericardial sacs and failure of yolk sac remodeling suggestive of cardiovascular failure. Similar to our experience screening GATA4, no clearly deleterious SOX7 sequence changes were identified in our CDH cohort. We conclude that haploinsufficiency of Sox7 or Gata4 is sufficient to produce anterior CDH in mice and that haploinsufficiency of SOX7 and GATA4 may each contribute to the development of CDH in individuals with 8p23.1 deletions.
INTRODUCTION
Congenital diaphragmatic hernia (CDH) is characterized by a protrusion of abdominal viscera into the thorax through an abnormal opening or defect in the diaphragm which is present at birth. CDH has an incidence of ∼1 in 4000 births—based on a meta-analysis of 15 studies published between 1976 and 1996 in which the birth incidence of CDH ranged from ∼1/1750 to ∼1/5900 (1). However, this estimate may not accurately reflect the current birth incidence due to changes in prenatal care and the current rates of pregnancy termination. CDH is associated with both a high mortality rate and significant long-term morbidity among survivors. Although CDH can be an isolated defect, additional anomalies are present in 40–60% of cases with cardiac defects being the most common (2–4).
Over 20 copy-number-dependent chromosomal regions that are recurrently associated with the development of CDH have been identified (5–7). Some of these chromosomal regions contain genes that have been clearly implicated in the development of CDH based on both mouse models of CDH and the identification of deleterious changes in individuals with CDH. These genes include the zinc finger protein, multitype 2 (ZFPM2, formerly known as FOG2) gene on 8q22 and the Wilms tumor 1 (WT1) gene on 11p13 (8–13). Other regions contain genes that have been implicated in the development of CDH based on mouse models alone. The nuclear receptor subfamily 2, Group F, Member 2 (NR2F2, formerly known as COUP-TFII) gene, for example, is located in the minimally deleted region for CDH on chromosome 15q26.2. Conditional ablation of Nr2f2 in the foregut mesenchyme, including the post-hepatic mesenchymal plate, is associated with a high incidence of posterolateral CDH in mice (14). However, to date, no clearly deleterious changes in NR2F2 have been identified in individuals with CDH, despite efforts to identify such changes (15).
A similar situation is seen with regards to the GATA binding protein 4 (GATA4) gene. GATA4 encodes a transcription factor and is located in a region of chromosome 8p23.1 that is flanked by segmental duplications 8p-OR-REPD and 8p-OR-REPP. This region is recurrently deleted via non-allelic homologous recombination. CDH and cardiac malformations are common complications of recurrent 8p23.1 microdeletions with one published estimate of penetrance being 22% for CDH and 94% for cardiac malformations, although the actual frequencies will be clarified as additional cases are reported (16). Mouse models suggest that haploinsufficiency of GATA4 is likely to contribute to the development of both of these birth defects. On a pure C57BL6 background, Jay et al. reported that ∼14% (3/21) of mice that were heterozygous for a Gata4 allele in which the second exon has been removed (Gata4+/Δex2) developed overt diaphragmatic herniation and ∼36% (5/14) had cardiac defects that included secundum atrial septal defects, ventricular septal defects and atrioventricular canal defects (17).
Haploinsufficiency of GATA4 has also been clearly shown to be sufficient to cause cardiac defects in humans. Some of the most compelling evidence for the role of GATA4 in abnormal cardiac development comes from studies in which heterozygous GATA4 mutations were found to segregate with atrial septal defects in seven families with varying levels of penetrance (18–21). Further evidence comes from Nemer et al. who documented de novo Glu216Asp mutations in two individuals with tetralogy of Fallot, and Reamon-Buettner et al. who found a frameshift mutation (Pro226fs) and a nonsense mutation (Arg266Ter) in two patients with atrioventricular septal defects (22, 23). However, none of these individuals—nor over 30 additional individuals with sporadic cardiac defects reported to have non-synonymous changes in GATA4—have been reported to have CDH (16). In this report, we looked for evidence that mutations in GATA4 are sufficient to cause CDH, but no deleterious GATA4 sequence changes were identified in DNA samples from a cohort of 77 individuals with CDH.
These observations and findings led us to hypothesize that haploinsufficiency of another gene in this interval may contribute, along with GATA4, to the development of CDH in individuals with 8p23.1 deletions. Of the 21 other genes located in this region, the sex-determining region Y (SRY)-box 7 (SOX7) gene appeared to be the most likely gene to play a role in diaphragm development (16). SOX7 is a member of the SOX (SRY-related HMG-box) family of transcription factors whose members play key roles in the regulation of embryonic development and in the determination of cell fate (24,25). SOX7 has also been shown to modulate the expression of GATA4 in human embryonic stem cells and in mouse F9 embryonal carcinoma cells and is expressed in both embryonic and adult tissues (26–38).
To determine whether deficiency of SOX7 could contribute to the development of CDH, we generated mice with deletions of the second exon of Sox7, which encodes half of SOX7′s HMG-box DNA binding domain and the entire SOX7 activation domain (28). Up to 14% of Sox7+/Δex2 mice on a mixed C57BL6/SV129 background developed retrosternal CDH that was similar to the CDH seen in GATA4-deficient mice and the anterior CDH described in one patient with an 8p23.1 deletion (16). In contrast to Sox7+/Δex2 mice that are viable and fertile, Sox7Δex2/Δex2 embryos died prior to diaphragm development with signs suggestive of cardiovascular failure.
By immunohistochemistry, we examined the expression of SOX7 in the diaphragm of wild-type mice. We found that SOX7 is expressed in the vascular endothelial cells of the developing diaphragm and may be weakly expressed in some diaphragmatic muscle cells. This pattern of expression is different from that seen for GATA4, making it unlikely that SOX7 directly regulates GATA4 expression during diaphragm development.
Similar to our experience with GATA4, no clearly deleterious SOX7 sequence changes were identified in DNA samples from 77 individuals with CDH. Taken together, these results suggest that haploinsufficiency of Sox7 or Gata4 is sufficient to produce CDH in mice and that haploinsufficiency of SOX7 and GATA4 may each contribute to the development of CDH in individuals with 8p23.1 deletions.
RESULTS
Screening for sequence changes in GATA4 in patients with CDH
In an effort to identify GATA4 sequence changes that contribute to the development of CDH, we screened the coding sequence and intervening intron–exon boundaries of GATA4 in a cohort of 77 patients with CDH. The results of this screening are summarized in Table 1. Twelve individuals were found to have non-synonymous changes—1 with an A346V change inherited from an unaffected father and 11 individuals with an S377G change which, in some cases, were confirmed to have been inherited from an unaffected parent. Both changes have been reported previously by the 1000 Genomes project (http://browser.1000genomes.org/index.html) and in control individuals reported in dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/). These changes were predicted to be ‘Tolerated’ by SIFT (http://sift.jcvi.org/) and ‘Benign’ by PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/). This suggests that they are unlikely to have contributed to the development of CDH in the patients in whom they were identified.
Table 1.
GATA4 sequence changes observed in 77 patients with CDH
| Change | Number of patients (inheritance) | 1000 Genomes/dbSNP/DNA source–number of patient samples | Predicted effect (SIFT) | Predicted effect (PolyPhen2) |
|---|---|---|---|---|
| Non-synonymous changes | ||||
| p.Ala346Val c.1037C>T | 1 (Paternal) | Yes/yes (Het 0.010)/L-1 | Tolerated | Benign |
| p.Ser377Gly c.1129A>G | 11 | Yes/yes (Het 0.113, MAF 0.0517)/WB-8, L-3 | Tolerated | Benign |
| Synonymous changes | ||||
| p.Gly112=c.336G>A | 1 | No/no/L-1 | N/A | N/A |
| p.Phe154=c.462C>T | 1 | Yes/yes/WB-1 | N/A | N/A |
| p.Tyr244=c.732C>T | 1 | No/yes (Het 0.003)/L-1 | N/A | N/A |
| p.Pro341=c.1023T>C | 1 | Yes/yes (Het 0.024)/L-1 | N/A | N/A |
| p.Asn352=c.1056C>T | 2 | Yes/yes (Het 0.221 MAF 0.0631)/WB-2 | N/A | N/A |
| p.Ser371=c.1113A>G | 2 | Yes/yes (Het 0.500 MAF 0.0259)/WB-2 | N/A | N/A |
Data accessed on October 2011.
Het, heterozygosity reported by dbSNP; MAF, minor allele frequency reported by dbSNP; WB, whole blood; F, fibroblasts cultures; L, EBV transformed lymphocytes; N/A, not applicable.
Haploinsufficiency of Sox7 causes retrosternal CDH
To determine if deficiency of SOX7 could cause CDH in mice, we generated mice with deletions of the second exon of Sox7 which encodes for half of SOX7′s HMG-box DNA binding domain and the entire SOX7 activation domain, which is located in the C-terminal region of the protein (Fig. 1) (28). Sox7+/Δex2 mice were viable and their average weight at 2, 4 and 6 weeks after birth was indistinguishable from wild-type mice (data not shown). However, on a mixed C57BL6/SV129 background, ∼14% (10/71) of Sox7+/Δex2 mice aged P28 to adult had CDH located directly behind the sternum in the ventral midline.
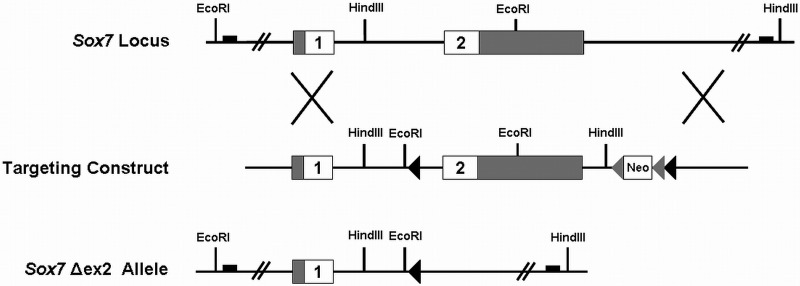
Figure 1.
Targeting strategy for generating the Sox7Δex2 allele. The coding (white) and non-coding (grey) portions of the Sox7 gene are shown along with the location of relevant restriction sites in the genomic DNA. The targeting construct contained a single loxP site (black triangle) upstream of exon 2 and a neomycin resistance cassette (Neo) flanked by FRT sites (grey triangles) and a 3′ loxP site (black triangle) downstream of exon 2. Proper targeting was confirmed by Southern blot using novel EcoRI and HindIII restriction sites contained in the targeting vector. The approximate locations of the Southern blot probes are represented by black boxes. The second exon of Sox7 and the neomycin resistance cassette were deleted by collapsing the two loxP sites in vivo. This was accomplished by crossing mice carrying a properly targeted Sox7 gene to mice expressing Cre in the female germline.
In the most severe of the anterior retrosternal hernias seen in Sox7+/Δex2 mice, the gallbladder and a pedunculated portion of the liver were found in the thoracic cavity (Fig. 2A–C). These herniated viscera were encased in a thin, membranous sac. The pedunculated nature of the liver mass often made it difficult to reduce the contents of the hernia sac into the abdominal cavity. Both gross and histological analyses revealed the presence of a muscular ring around the diaphragmatic defect (Fig. 2C, F and G).
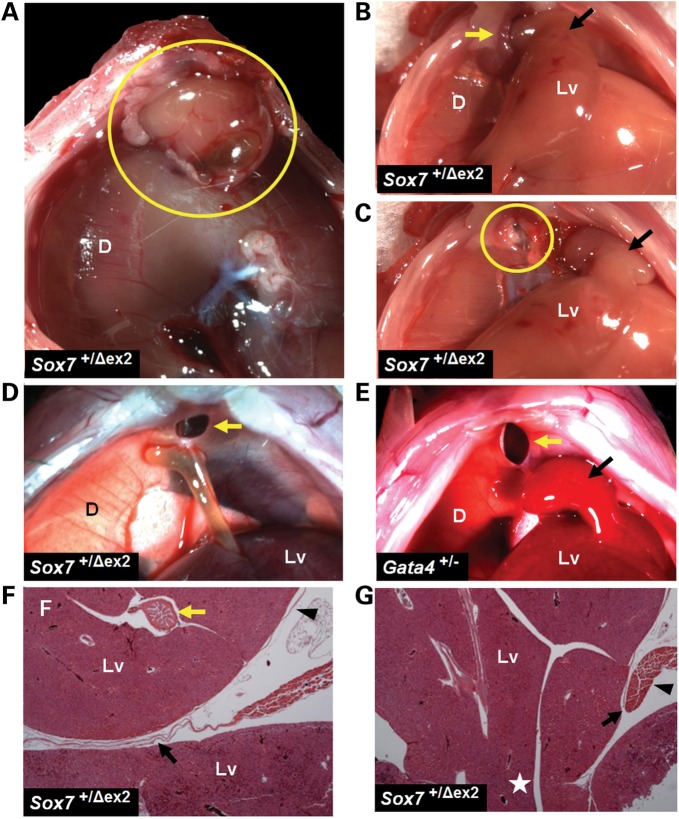
Figure 2.
A portion of Sox7+/Δex2 mice develop retrosternal CDH that is similar to those seen in Gata4+/− mice. (A) A retrosternal CDH in a Sox7+/Δex2 mouse as viewed from the thorax (yellow circle). The herniated gallbladder and liver tissue are covered by a thin membranous sac. The liver (Lv) can be seen through the intact portion of the transparent diaphragm. (B and C) A retrosternal CDH as viewed from the abdomen. A pedunculated mass of liver tissue (Lv, black arrow) was reduced through a muscular ring at the border of the diaphragmatic defect (yellow arrow and circle). (D) A retrosternal CDH (yellow arrow) viewed from the abdomen. The gallbladder is abnormally fused to the underside of the diaphragm. (E) A retrosternal CDH (yellow arrow) seen from the abdomen in a Gata4+/− mouse. A pedunculated mass of liver tissue (Lv, black arrow) was reduced into the abdomen. (F) H&E-stained section through the diaphragm and the retrosternal hernial sac of a Sox7+/Δex2 mouse reveals a mass of herniated liver tissue (Lv) and the gallbladder (yellow arrow) surrounded by a thin membranous sac (black arrowhead). This section was taken in a region located away from the pedicle so a portion of the membrane (black arrow) appears between the herniated liver (above) and the unherniated liver (below). (G) H&E-stained section through the same hernial sac shown in panel (F). The pedicle of the herniated liver mass (white star) joins the unherniated liver in the abdomen. There is a sharp demarcation between the diaphragmatic musculature and the membranous sac (black arrow) and evidence of muscular thickening at the edge of the diaphragmatic defect (black arrowhead). D = diaphragm; Lv = liver.
Less severe hernias consisted of a retrosternal opening in the diaphragm without evidence of liver incarceration through the diaphragm defect (Fig. 2D). These diaphragmatic defects lacked a membranous covering. Occasionally, the gallbladder was abnormally fused to the underside of the anterior diaphragm (Fig. 2D). In all cases, the defect was located in a region of the anterior diaphragm that would typically be muscularized.
This type of CDH was similar to that described by Jay et al. in Gata4+/Δex2 mice on a congenic C57BL6 background (17). Occasionally, we also observed this type of CDH in a different Gata4+/− mouse strain described by Molkentin et al.—in which a Gata4 null allele was generated by replacing exons encoding GATA4′s two DNA-binding zinc finger domains with a neomycin resistance gene—after it had been backcrossed through six or more generations onto a C57BL6 background (Fig. 2E) (39). In contrast, CDH was not seen in Sox7+/Δex2 mice (0/41) backcrossed through eight generations onto a C57BL6 background.
Expression pattern of SOX7 and GATA4
To determine when and where SOX7 was expressed in the diaphragm, we performed immunohistochemistry for SOX7 in wild-type embryos and pups at E12.5, E14.5, E15.5, E16.5 (embryonic days) and P1. SOX7 was not detected in the pleuroperitoneal fold (PPF) at E12.5 or in the anterior, middle or posterior diaphragm at this time point (Supplementary Material, Figs S1 and S2). Between E14.5 and E16.5, SOX7 was not detected in the anterior, middle or posterior diaphragm except in the nuclei of vascular endothelial cells (Fig. 3A and B; Supplementary Material, Figs S3 and S4). In sections from P1 pups, SOX7 expression was not seen in the non-muscular diaphragm (Fig. 3C; Supplementary Material, Fig. S5). In the muscularized portion of the diaphragm, strong nuclear staining for SOX7 was again seen in the vascular endothelium (Fig. 3C and D). In P1 pups, the expression of SOX7 in diaphragmatic muscle cells was more difficult to evaluate, with some cells appearing to have a subtle blush of SOX7 nuclear staining (Fig. 3C and D; Supplementary Material, Fig. S5).
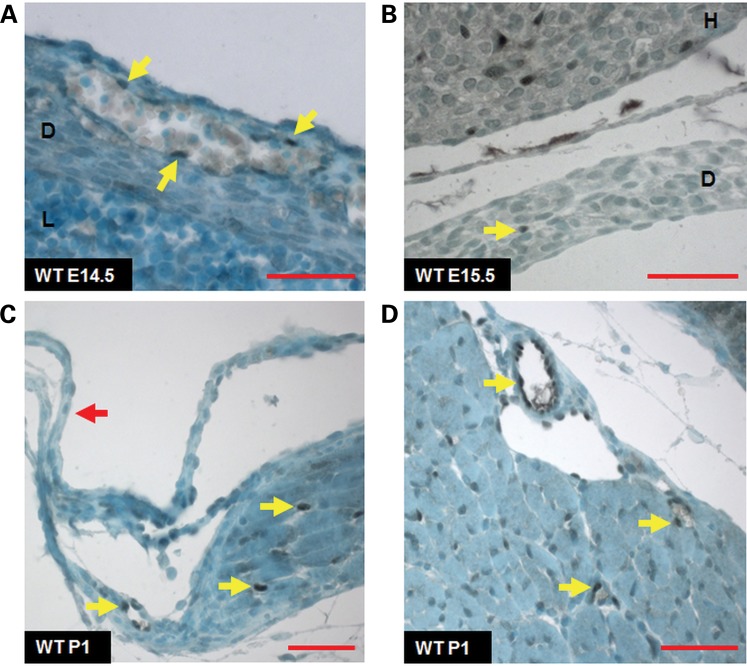
Figure 3.
Immunohistochemical staining for SOX7 in the developing diaphragm. SOX7 expression in the anterior diaphragms of wild-type mice at E14.5 (panel A) and E15.5 (panel B) is limited to the vascular endothelium (yellow arrows). SOX7 immunoreactivity is also seen in the vascular endothelium of the heart (panel B). At P1, SOX7 is not expressed in the non-muscular diaphragm (red arrow, panel C). Strong nuclear staining for SOX7 is seen in the vascular endothelium (yellow arrows) in the muscular portions of the anterior (panel C) and the posterior diaphragm (panel D) at P1. Subtle SOX7 staining is also seen in the nuclei of some diaphragmatic muscle cells in the anterior and posterior diaphragm. D = diaphragm; H = heart; L = liver; red scale bars = 50 µm.
SOX7-positive cells were also seen in the endocardium and in the vascular endothelium of the heart and lungs between E12.5 and P1 (Fig. 3B; Supplementary Material, Fig. S6). The specificity of the SOX7 antibody was confirmed in sections from a rare E15.5 Sox7Δex2/Δex2 embryo which showed no SOX7 staining in these tissues (Supplementary Material, Fig. S7).
We also determined the expression pattern of GATA4 during diaphragm development. Nuclear GATA4 immunoreactivity was detected in a subset of cells in the PPF at E12.5 and in a subset of cells in the anterior, middle and posterior regions of the diaphragm from E12.5 to E16.5 (Supplementary Material, Figs S1–S4). In sections from P1 pups, GATA4 nuclear staining was not detected in the muscular and non-muscular regions of the diaphragm with the exception of the outermost cell layers—corresponding to the diaphragmatic pleura and peritoneum—and in rare vascular endothelial cells (Supplementary Material, Fig. S5). These data are consistent with previously published reports in which GATA4 expression was shown to be restricted to the non-muscular, mesenchymal component of the PPF and the MyoD-negative mesenchymal cells of the diaphragm at E16.5, with little GATA4 expression being seen in the muscular diaphragm at P1 (17,40).
Comparison of the diaphragmatic vasculatures of Sox7+/Δex2 and wild-type embryos and mice
To determine if Sox7 haploinsufficiency had a deleterious effect on the formation of blood vessels in the developing anterior diaphragm, we looked for evidence of vascularization in sagittal histological sections of Sox7+/Δex2 (n = 4) and wild-type (n = 4) embryos at E14.5. Blood vessels were observed in midline sections of the anterior diaphragm of all embryos at this time point (Supplementary Material, Fig. S8).
We then compared the diaphragmatic vascular patterns of Sox7+/Δex2 mice on a mixed C57BL6/SV129 background (n = 6) with the vascular patterns of wide-type mice on a mixed C57BL6/SV129 background (n = 6), a pure SV129 background (n = 2) and a pure C57BL6 background (n = 2). Particular attention was given to the vessels leading to the anterior diaphragm. Although variations in the branching patterns, connections and the diameters of vessels were seen in Sox7+/Δex2 mice, in each case, similar variations were seen in at least one of the wild-type controls (Supplementary Material, Fig. S9).
Homozygous Sox7Δex2/Δex2 embryos die in utero with signs of cardiovascular failure
In crosses between Sox7+/Δex2 mice, heterozygous progeny were recovered in expected Mendelian ratios and in appropriate male/female proportions at weaning (P28), but no Sox7Δex2/Δex2 homozygous mice were identified. To determine the time point at which Sox7Δex2/Δex2 homozygous embryos/mice die, embryos from Sox7+/Δex2 intercrosses were harvested at E8.5, E9.5, E10.5 and E14.5. Sox7Δex2/Δex2 embryos were recovered in Mendelian ratios through E10.5 but no Sox7Δex2/Δex2 embryos were seen at E14.5 (Table 2).
Table 2.
Sox7Δex2/Δex2 embryos die between E10.5 and E14.5
| Embryonic day | WT | Sox7+/Δex2 | Sox7Δex2/Δex2 | Total |
|---|---|---|---|---|
| E8.5 | 31 (25%) | 60 (49%) | 32 (26%) | 123 |
| E9.5 | 24 (17%) | 77 (56%) | 37 (27%) | 138 |
| E10.5 | 35 (27%) | 66 (50%) | 30 (23%) | 131 |
| E14.5 | 10 (45%) | 12 (55%) | 0 (0%) | 22 |
At E8.5 all embryos were alive and wild-type, Sox7+/Δex2, and Sox7Δex2/Δex2 embryos were indistinguishable. In contrast, by E10.5, 79% (22/28) of Sox7Δex2/Δex2 embryos showed evidence of delayed development, including one embryo that had died, 61% (17/28) had pericardial edema and 79% (22/28) had failure of yolk sac remodeling. These findings are suggestive of cardiovascular failure (Fig. 4).
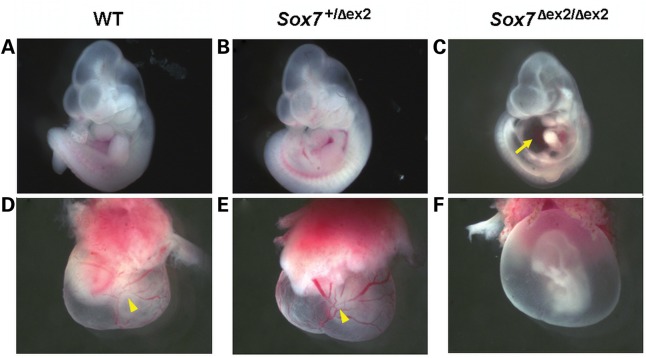
Figure 4.
The majority of Sox7Δex2/Δex2 embryos at E10.5 show signs of delayed development and cardiovascular failure. (A–C) Wild-type and Sox7+/Δex2 embryos appeared normal at E10.5. In contrast, the majority of Sox7Δex2/Δex2 embryos showed signs of developmental delay and pericardial edema (yellow arrow). (D–F) The yolk sacs of wild-type and Sox7+/Δex2 mice show evidence of normal vascular remodeling (yellow arrow heads). In contrast, the majority of Sox7Δex2/Δex2 embryos at E10.5 show failure of yolk sac remodeling.
Although the great majority of Sox7Δex2/Δex2 embryos die before E14.5, genotyping of embryos from Sox7+/Δex2 intercrosses used for subsequent studies has revealed the existence of rare Sox7Δex2/Δex2 embryos that survive beyond E14.5.
Screening for sequence changes in SOX7 in patients with CDH
In an effort to identify SOX7 sequence changes that contribute to the development of CDH, we screened the coding sequence and intervening intron–exon boundaries of SOX7 in 77 patients with CDH. The results of this screen are summarized in Table 3. Five individuals with non-synonymous changes were identified; two with a T169S change inherited from an unaffected father, one with an R233H change inherited from an unaffected father and two with a G267S change that was not found in their mothers but for whom the paternal DNA was not available for analysis. None of these individuals had non-synonymous changes in GATA4. Only the T169S change has been reported previously in data from the 1000 Genomes project and in dbSNP. However, all of these changes were predicted to be ‘Tolerated’ by SIFT and ‘Benign’ by PolyPhen2. This suggests that they are unlikely to have contributed to the development of CDH in the patients in whom they were identified.
Table 3.
SOX7 sequence changes observed in 77 patients with CDH
| Change | Number of patients (inheritance) | 1000 Genomes/dbSNP/DNA source–number of patient samples | Predicted effect (SIFT) | Predicted effect (PolyPhen2) |
|---|---|---|---|---|
| Non-synonymous changes | ||||
| p.Thr169Ser c.506C>G | 2 (Paternal) | Yes/yes (MAF 0.0533)/WB-2 | Tolerated | Benign |
| p.Arg233His c.698G>A | 1 (Paternal) | No/no/WB-1 | Tolerated | Benign |
| p.Gly267Ser c.799G>A | 2 (Not maternal, fathers not available) | Yes/yes/L-2 | Tolerated | Benign |
| Synonymous changes | ||||
| p.Pro116=c.348G>A | 1 | Yes/yes (MAF 0.0609)/WB-1 | N/A | N/A |
| p.Gly239=c.717G>A | 1 | Yes/yes (Het 0.008, MAF 0.0018)/L-1 | N/A | N/A |
| p.Leu303=c.909T>C | 36 | Yes/yes (MAF 0.4579)/WB-27, F-1, L-8 | N/A | N/A |
Data accessed on October 2011.
Het, heterozygosity reported by dbSNP; MAF, minor allele frequency reported by dbSNP; WB, whole blood; F, fibroblasts cultures; L, EBV transformed lymphocytes; N/A, not applicable.
DISCUSSION
Haploinsufficiency of SOX7 and GATA4 is likely to contribute to the development of CDH
The paucity of families with multiple affected individuals makes it difficult to use a linkage-based approach to identify genes responsible for sporadic birth defects like CDH. However, the location of copy-number-sensitive genes that contribute to these defects can often be inferred by identifying regions of the genome that are recurrently deleted or duplicated in affected individuals (5). Using this cytogenetically based approach, a recurrently deleted region bounded by segmental duplications was identified as the location of the genetic factors contributing to CDH in patients with 8p23.1 deletions (16,41,42). This region contains both SOX7 and GATA4.
In this report, we have shown that a portion of Sox7+/Δex2 mice develop retrosternal diaphragmatic hernias that are covered with a membranous sac. These hernias are located in the anterior midline behind the sternum. Similar hernias are also seen in Gata4+/− mice. Although most reported cases of CDH in patients with 8p23.1 deletions are described simply as left-sided, and are likely to be posterolateral, we have previously reported one patient with a de novo 8p23.1 deletion who had a large anterior CDH with herniation of the liver (16). These observations lead us to conclude that haploinsufficiency of SOX7 and haploinsufficiency of GATA4 are likely to contribute to the development of CDH in patients with 8p23.1 deletions. We recognize, however, that our results do not exclude the possibility that haploinsufficiency of another gene or genes may also be contributing to the risk of CDH seen in patients with 8p23.1 deletions.
Over 20 copy-number-dependent chromosomal regions that are recurrently associated with the development of CDH have been described (5–7). The dosage-sensitive genes that contribute to the development of CDH associated with the majority of these regions have yet to be identified. Although copy number changes of a single gene may be found to be responsible for CDH at some of these loci, our results underscore the importance of considering the possibility that two or more genes contribute to this phenotype. This is particularly important when mutation screens conducted in cohorts of CDH patients fail to identify deleterious changes in individual candidate genes.
Deleterious changes in either SOX7 or GATA4 alone are likely to contribute to only a small portion of CDH cases
Although haploinsufficiency of either Sox7 or Gata4 is sufficient to produce CDH in mice, no deleterious changes in either of these genes were identified in the cohort of CDH patients screened in this report. Although sequencing of the coding regions and associated intron/exon boundaries of SOX7 and GATA4 is likely to detect most deleterious changes in these genes, the exact sensitivities of the mutation screens employed in our studies are not known. However, if we assume that these screens are sufficient to identify 80% of deleterious changes in these genes, then a screen of 77 individuals with CDH would have a >95% chance of identifying at least one deleterious change, if such changes contributed to at least 5% of CDH cases. This suggests that deleterious changes in either SOX7 or GATA4 alone are likely to contribute to only a small portion of CDH cases.
Sox7+/Δex2 mice demonstrate strain-dependent variation in CDH penetrance
Only a portion of Sox7+/Δex2 mice develop CDH. Incomplete penetrance and discordance for CDH among monozygotic twins have also been reported in association with recurrent 8p23.1 microdeletions (16). This suggests that other genetic, environmental and/or stochastic factors influence the development of diaphragmatic defects in Sox7+/Δex2 mice and in humans with 8p23.1 deletions. The influence of other genetic factors on the penetrance of CDH is clearly demonstrated by the higher rate of CDH seen in Sox7+/Δex2 mice on a mixed C57BL6/SV129 background compared with the rate observed on a pure C57BL6 background.
This strain-dependent variation in CDH penetrance could prove useful in future studies involving Sox7+/Δex2 mice aimed at identifying modifying genes which contribute to the development of CDH. At the same time, the presence of strongly modifying genes also poses a significant challenge to the use of mouse models as a primary means of identifying genes that cause CDH since it is difficult to predict, a priori, which background will allow the clearest demonstration of a gene's effect on diaphragm development. Indeed, it is likely that the most sensitive background will not only vary based on hernia type but may be specific to the pathway(s) being disrupted. Studies focused on increasing our understanding of the histopathologic and molecular mechanisms that underlie CDH and the genetic properties of common mouse strains may ultimately lead to an improved ability to make such predictions.
Despite the present challenges arising from strain-dependent variability, the increasing availability of mouse knock-out resources—combined with the limitations encountered in screening affected cohorts—makes it likely that mouse knock-out studies will continue to be one of the most effective and efficient means of evaluating the role of candidate genes in the development of CDH and other sporadic birth defects.
The expression of GATA4 and SOX7 in the developing diaphragm and its relationship with CDH
The anterior CDH seen in Sox7+/Δex2 and Gata4+/− mice arises in the ventral midline directly behind the sternum in a region of the anterior diaphragm that would typically be muscularized. Previous studies have shown that GATA4 expression is restricted to the non-muscular, mesenchymal component of the PPF, and the MyoD-negative mesenchymal cells of the diaphragm at E16.5 (17,40). In our immunohistochemical studies, we did not detect GATA4 expression in the nuclei of diaphragmatic muscle cells at P1. These data suggest that the anterior CDH seen in GATA4-deficient mice is unlikely to be due to a primary defect in muscle precursor cells or in the mature muscle cells of the diaphragm.
In contrast to GATA4, a subtle blush of SOX7 nuclear staining was seen in some muscle cells of the anterior and posterior diaphragm at P1 suggesting that SOX7 may be expressed at a low level within the diaphragmatic musculature. It is possible, therefore, that a primary defect in the muscle cells of the diaphragm may contribute to the development of CDH in Sox7+/Δex2 mice. If such a defect exists, it could predispose Sox7+/Δex2 mice to the development of CDH by causing a transient or permanent weakness in the muscular portion of the diaphragm. While a lack of hernias or eventrations in other regions of the muscularized diaphragm argues against a strong, generalized effect on the diaphragmatic musculature, hernias caused by a mild effect on the musculature could arise preferentially in the anterior portion of the diaphragm since it is the last region to be muscularized.
In the developing diaphragm, strong SOX7 expression appears to be limited to the endothelial cells of the diaphragmatic vasculature. This leads us to also consider how decreased SOX7 expression in endothelial cells might contribute to the development of these hernias. One possibility is that decreased SOX7 expression in endothelial cells leads to abnormal development of the vascular network which directly compromises the development or the integrity of the anterior diaphragm due to an inadequate blood supply. However, severe abnormalities in vascular development typically result in early embryonic lethality, and it is difficult to postulate why the anterior diaphragm would be particularly sensitive to this type of defect. The anterior diaphragmatic vasculatures of Sox7+/Δex2 and wild-type mice also appeared to be comparable in our studies.
Alternatively, decreased expression of SOX7 may result in abnormal endothelial signaling. Endothelial cells from various sources have been shown to express a variety of signaling molecules including members of the FGF, TGF-beta, Wnt and PDGF families, as well as Notch ligands, neurotrophins and components of the basement membrane (43). Endothelial signaling has also been shown to play an important role in the development of other organs such as the liver, pancreas and kidney (43,44). As a transcription factor, SOX7 may regulate the expression of signaling proteins causing abnormal or delayed diaphragm development.
Further studies will be needed to determine whether decreased SOX7 expression in the muscle cells or in the vascular endothelium of the diaphragm is sufficient to cause anterior CDH, and to elucidate the molecular mechanisms by which SOX7 regulates diaphragm development. However, since the patterns of SOX7 and GATA4 expression in the diaphragm are different, it is unlikely that SOX7 directly regulates GATA4 expression during diaphragm development.
Sox7 is required for normal cardiovascular development
It is possible that haploinsufficiency of SOX7 may also contribute to the development of cardiac defects in patients with 8p23.1 (16). The majority of Sox7Δex2/Δex2 embryos demonstrates evidence of cardiac failure—dilated pericardial sacs and failure of yolk sac vascular remodeling—at E10.5 and likely die shortly thereafter. This early lethality precludes their use in studies of diaphragm development but suggests that SOX7 plays a critical role in the development of the cardiovascular system. We are presently investigating the cause of the cardiovascular failure in Sox7Δex2/Δex2 embryos which may be due to a primary cardiac defect, a defect in vascular development or a combination of these factors.
MATERIALS AND METHODS
Patient accrual and DNA preparation
Informed consent was obtained from individuals with both isolated and non-isolated CDH and their parents in accordance with internal review board-approved protocols. DNA extracted from whole blood, fibroblast cultures or EBV-immortalized lymphoblastic cells was used for sequencing studies. In cases where the amount of genomic DNA available was limited, ∼10 ng of DNA was amplified using a GenomiPhi DNA amplification kit (GE Healthcare Life Sciences, NJ, USA) according to the manufacturer's instructions. This amplified DNA was then used as a template for PCR amplification and sequencing.
Sequencing of GATA4 and SOX7
PCR primers were designed to amplify the coding sequence and the intron–exon boundaries of SOX7 from genomic DNA. Sequence changes in PCR-amplified products were identified by comparison with control DNA sequences using Sequencher 4.7 software (Gene Codes Corporation, Ann Arbor, MI, USA). A similar procedure was used to screen for mutations in the coding sequence and intervening intron/exon boundaries of GATA4 using primers previously described by Okubo et al. (20).
Generation and genotyping of Sox7 transgenic mice
All mouse studies were performed in accordance with protocols approved by the internal review board of Baylor College of Medicine and conform to relevant regulatory standards.
A Sox7 targeting vector was created using a recombineering strategy (Fig. 1) (45). Briefly, genomic DNA containing both exons of Sox7 was retrieved into plasmid PL253 from bacterial artificial chromosome clone bMQ124L22. A neomycin cassette flanked by loxP sites was then targeted into the first intron after which the neomycin gene was removed by induction of Cre activity in EL350 cells incubated in 0.1% arabinose. This left a single loxP site and a new EcoRI restriction site upstream of exon 2. A second neomycin cassette flanked by Frt sites and bearing a 3′ loxP site was targeted upstream of the second exon of Sox7 creating a new HindIII restriction site downstream of exon 2.
The targeting vector was linearized and electroporated into 129SvEv embryonic stem (ES) cells and proper integration of the 5′ loxP site was confirmed by detection of a 6504 bp EcoRI digestion product (wild-type allele 8785 bp) using an 850 bp Southern probe generated using primers CKO-5′-South 1R (5′-AAGCAACTTTCCCAACCTGTATTC-3′) and CKO-5′-South 1L (5′-CTCCCAAGACCTTTCCTAATCC-3′). Integration of the 3′ neomycin cassette was confirmed by detection of a 13 530 bp HindIII digestion product (wild-type allele 17 372 bp) using a 953 bp Southern probe generated using primers CKO-3′-South 1R (5′-GTGGGGGCACACAGCACGTAAG-3′) and CKO-3′-South 1L (5′-GGAAGGGGCGAGGTATGTGA-3′).
Properly targeted embryonic stem cells were injected into C57 albino blastocysts. Chimeric mice were identified by eye and coat color and were mated to C57 albino females. Agouti progeny bearing the targeted Sox7 allele were identified by PCR analysis of tail DNA using probes Sox7Junct 5R (5′-CTTCGATTCTACACATTAGTGC-3′), Sox7Junct 5L (5′-CTAAAGCGCATGCTCCAGAC-3′) and Sox7Junct 6L (5′-CATGAATGCTCCCAATGAATGC-3′) which amplified a 419 bp product from the targeted allele and a 660 bp product from the wild-type allele.
The second exon and 3′ untranslated region of Sox7 were excised in vivo by mating these mice with EIIa-cre mice that express Cre recombinase in the female germline. The Sox7Δex2 allele was detected by PCR analysis of tail DNA using probes Sox7 Del 1F (5′-AAACTGTCCTGCTATGGTCAGAAAGTCCTA-3′) and Sox7 Del 1R (5′-AGTGCTACTGAATAATGGGTGTGGGTTATG-3′) which yields a 258 bp product.
Histology and immunohistochemistry
Hernial sacs and portions of the liver were dissected free from Sox7+/Δex2 mice and placed in a solution of 10% formaldehyde at room temperature for ∼48 h. The specimens were trimmed, washed in phosphate-buffered saline (PBS), dehydrated in ethanol, embedded in paraffin and sectioned coronally. Representative sections were then stained with hematoxylin and eosin.
For routine histological analyses, embryos were fixed in a solution of 10% formaldehyde overnight, washed in PBS, dehydrated in ethanol, embedded in paraffin and sectioned at 8 µm. Representative sections were then stained with hematoxylin and eosin.
For immunohistochemistry, embryos were fixed in a solution of 10% formaldehyde overnight, embedded in paraffin and sectioned at 8 µm. Sections were mounted on glass slides, dewaxed in xylene and rehydrated using a graded series of alcohol washes. For antigen retrieval, slides were immersed in 250 ml of Vector Antigen Unmasking Solution (Vector Laboratories, Burlingame, CA, USA) and placed in a 1200 watt microwave for 3 to 7 min at 70% power and 10 min at 10% power. Slides were then cooled to room temperature and washed in PBS.
For GATA4 immunohistochemistry, slides were incubated in 0.3% H2O2 for 10 min to quench endogenous peroxidases. GATA4 was then detected using Vector M.O.M. Immunodetection kit (Vector Laboratories) in conjunction with a 1:50 dilution of GATA4 mouse primary monoclonal antibody Sc-25310 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) according to manufacturer's instructions.
For SOX7 immunohistochemistry, slides were blocked for 1 h in a PBS-based solution containing 0.5% bovine serum albumin and 5% normal donkey serum followed by an overnight exposure to a 1:200 dilution of SOX7 goat polyclonal antibody AF2766 (R& D Systems, Minneapolis, MN, USA). Slides were washed in PBS and exposed to a 1:200 dilution of donkey anti-goat IgG 705-065-003 (Jackson ImmunoResearch Laboratories, West Grove, PA, USA) for 1 h at room temperature. Slides were then washed in PBS and sections were incubated in ABC solution (Vector Laboratories) for 30 min before being washed again in PBS.
Slides were stained by exposing sections to Nova Red substrate (Vector Laboratories) for a duration between 30 s and 2 min after which slides were washed in water and counterstained with methyl green (Vector Laboratories).
Visualization of diaphragmatic vasculature
Juvenile and adult mice between the ages of 3.5 and 8.5 weeks were euthanized by exposure to CO2. Carcasses were stored at room temperature for ∼1.5 to 3 h before dissection. After removal of the skin, a cut was made through the thorax. The contents of the abdominal and thoracic cavities were removed and the remaining tissues were trimmed leaving the intact diaphragm supported by a ring of tissue. The diaphragm was washed with PBS and photographed from the abdominal side.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health (grant numbers T32-GM007330-33S1 to M.J.W, F30-HL099469 to M.J.W., KO8-HD050583 to D.A.S. and R01-HD065667 to D.A.S.) and the Sophia Foundation for Scientific Research (grant number SSWO551 to D.V.).
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank the families who participated in this research study, Elda Munivez for her technical help, and Dr Eric N. Olson who kindly provided the Gata4+/− mice used in this study. The authors have no conflicts of interest to declare.
REFERENCES
- 1.Skari H., Bjornland K., Haugen G., Egeland T., Emblem R. Congenital diaphragmatic hernia: a meta-analysis of mortality factors. J. Pediatr. Surg. 2000;35:1187–1197. doi: 10.1053/jpsu.2000.8725. [DOI] [PubMed] [Google Scholar]
- 2.Bollmann R., Kalache K., Mau H., Chaoui R., Tennstedt C. Associated malformations and chromosomal defects in congenital diaphragmatic hernia. Fetal Diagn. Ther. 1995;10:52–59. doi: 10.1159/000264193. [DOI] [PubMed] [Google Scholar]
- 3.Tibboel D., Gaag A.V. Etiologic and genetic factors in congenital diaphragmatic hernia. Clin. Perinatol. 1996;23:689–699. [PubMed] [Google Scholar]
- 4.Lin A.E., Pober B.R., Adatia I. Congenital diaphragmatic hernia and associated cardiovascular malformations: type, frequency, and impact on management. Am. J. Med. Genet. C Semin. Med. Genet., 145C. 2007:201–216. doi: 10.1002/ajmg.c.30131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holder A.M., Klaassens M., Tibboel D., de Klein A., Lee B., Scott D.A. Genetic factors in congenital diaphragmatic hernia. Am. J. Hum. Genet. 2007;80:825–845. doi: 10.1086/513442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Esch H., Backx L., Pijkels E., Fryns J.P. Congenital diaphragmatic hernia is part of the new 15q24 microdeletion syndrome. Eur. J. Med. Genet. 2009;52:153–156. doi: 10.1016/j.ejmg.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Wat M.J., Enciso V.B., Wiszniewski W., Resnick T., Bader P., Roeder E.R., Freedenberg D., Brown C., Stankiewicz P., Cheung S.W., et al. Recurrent microdeletions of 15q25.2 are associated with increased risk of congenital diaphragmatic hernia, cognitive deficits and possibly Diamond–Blackfan anaemia. J. Med. Genet. 2010;47:777–781. doi: 10.1136/jmg.2009.075903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ackerman K.G., Herron B.J., Vargas S.O., Huang H., Tevosian S.G., Kochilas L., Rao C., Pober B.R., Babiuk R.P., Epstein J.A., et al. Fog2 is required for normal diaphragm and lung development in mice and humans. PLoS Genet. 2005;1:58–65. doi: 10.1371/journal.pgen.0010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wat M.J., Veenma D., Hogue J., Holder A.M., Yu Z., Wat J.J., Hanchard N., Shchelochkov O.A., Fernandes C.J., Johnson A., et al. Genomic alterations that contribute to the development of isolated and non-isolated congenital diaphragmatic hernia. J. Med. Genet. 2011;48:299–307. doi: 10.1136/jmg.2011.089680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott D.A., Cooper M.L., Stankiewicz P., Patel A., Potocki L., Cheung S.W. Congenital diaphragmatic hernia in WAGR syndrome. Am. J. Med. Genet. A. 2005;134:430–433. doi: 10.1002/ajmg.a.30654. [DOI] [PubMed] [Google Scholar]
- 11.Antonius T., van Bon B., Eggink A., van der Burgt I., Noordam K., van Heijst A. Denys–Drash syndrome and congenital diaphragmatic hernia: another case with the 1097G > A(Arg366His) mutation. Am. J. Med. Genet. A., 146A. 2008:496–499. doi: 10.1002/ajmg.a.32168. [DOI] [PubMed] [Google Scholar]
- 12.Devriendt K., Deloof E., Moerman P., Legius E., Vanhole C., de Zegher F., Proesmans W., Devlieger H. Diaphragmatic hernia in Denys–Drash syndrome. Am. J. Med. Genet. A. 1995;57:97–101. doi: 10.1002/ajmg.1320570120. [DOI] [PubMed] [Google Scholar]
- 13.Kreidberg J.A., Sariola H., Loring J.M., Maeda M., Pelletier J., Housman D., Jaenisch R. WT-1 is required for early kidney development. Cell. 1993;74:679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- 14.You L.R., Takamoto N., Yu C.T., Tanaka T., Kodama T., Demayo F.J., Tsai S.Y., Tsai M.J. Mouse lacking COUP-TFII as an animal model of Bochdalek-type congenital diaphragmatic hernia. Proc. Natl. Acad. Sci. U.S.A. 2005;102:16351–16356. doi: 10.1073/pnas.0507832102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scott D.A., Klaassens M., Holder A.M., Lally K.P., Fernandes C.J., Galjaard R.J., Tibboel D., de Klein A., Lee B. Genome-wide oligonucleotide-based array comparative genome hybridization analysis of non-isolated congenital diaphragmatic hernia. Hum. Mol. Genet. 2007;16:424–430. doi: 10.1093/hmg/ddl475. [DOI] [PubMed] [Google Scholar]
- 16.Wat M.J., Shchelochkov O.A., Holder A.M., Breman A.M., Dagli A., Bacino C., Scaglia F., Zori R.T., Cheung S.W., Scott D.A., et al. Chromosome 8p23.1 deletions as a cause of complex congenital heart defects and diaphragmatic hernia. Am. J. Med. Genet. A. 2009;149A:1661–1677. doi: 10.1002/ajmg.a.32896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jay P.Y., Bielinska M., Erlich J.M., Mannisto S., Pu W.T., Heikinheimo M., Wilson D.B. Impaired mesenchymal cell function in Gata4 mutant mice leads to diaphragmatic hernias and primary lung defects. Dev. Biol. 2007;301:602–614. doi: 10.1016/j.ydbio.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garg V., Kathiriya I.S., Barnes R., Schluterman M.K., King I.N., Butler C.A., Rothrock C.R., Eapen R.S., Hirayama-Yamada K., Joo K., et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424:443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- 19.Hirayama-Yamada K., Kamisago M., Akimoto K., Aotsuka H., Nakamura Y., Tomita H., Furutani M., Imamura S., Takao A., Nakazawa M., et al. Phenotypes with GATA4 or NKX2.5 mutations in familial atrial septal defect. Am. J. Med. Genet. A. 2005;135:47–52. doi: 10.1002/ajmg.a.30684. [DOI] [PubMed] [Google Scholar]
- 20.Okubo A., Miyoshi O., Baba K., Takagi M., Tsukamoto K., Kinoshita A., Yoshiura K., Kishino T., Ohta T., Niikawa N., et al. A novel GATA4 mutation completely segregated with atrial septal defect in a large Japanese family. J. Med. Genet. 2004;41:e97. doi: 10.1136/jmg.2004.018895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarkozy A., Conti E., Neri C., D'Agostino R., Digilio M.C., Esposito G., Toscano A., Marino B., Pizzuti A., Dallapiccola B. Spectrum of atrial septal defects associated with mutations of NKX2.5 and GATA4 transcription factors. J. Med. Genet. 2005;42:e16. doi: 10.1136/jmg.2004.026740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nemer G., Fadlalah F., Usta J., Nemer M., Dbaibo G., Obeid M., Bitar F. A novel mutation in the GATA4 gene in patients with tetralogy of fallot. Hum. Mutat. 2006;27:293–294. doi: 10.1002/humu.9410. [DOI] [PubMed] [Google Scholar]
- 23.Reamon-Buettner S.M., Borlak J. GATA4 zinc finger mutations as a molecular rationale for septation defects of the human heart. J. Med. Genet. 2005;42:e32. doi: 10.1136/jmg.2004.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haqq C.M., King C.Y., Ukiyama E., Falsafi S., Haqq T.N., Donahoe P.K., Weiss M.A. Molecular basis of mammalian sexual determination: activation of Mullerian inhibiting substance gene expression by SRY. Science. 1994;266:1494–1500. doi: 10.1126/science.7985018. [DOI] [PubMed] [Google Scholar]
- 25.Avilion A.A., Nicolis S.K., Pevny L.H., Perez L., Vivian N., Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seguin C.A., Draper J.S., Nagy A., Rossant J. Establishment of endoderm progenitors by SOX transcription factor expression in human embryonic stem cells. Cell Stem Cell. 2008;3:182–195. doi: 10.1016/j.stem.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 27.Futaki S., Hayashi Y., Emoto T., Weber C.N., Sekiguchi K. Sox7 plays crucial roles in parietal endoderm differentiation in F9 embryonal carcinoma cells through regulating Gata-4 and Gata-6 expression. Mol. Cell. Biol. 2004;24:10492–10503. doi: 10.1128/MCB.24.23.10492-10503.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taniguchi K., Hiraoka Y., Ogawa M., Sakai Y., Kido S., Aiso S. Isolation and characterization of a mouse SRY-related cDNA, mSox7. Biochim. Biophys. Acta. 1999;1445:225–231. doi: 10.1016/s0167-4781(99)00047-0. [DOI] [PubMed] [Google Scholar]
- 29.Takash W., Canizares J., Bonneaud N., Poulat F., Mattei M.G., Jay P., Berta P. SOX7 transcription factor: sequence, chromosomal localisation, expression, transactivation and interference with Wnt signalling. Nucleic Acids Res. 2001;29:4274–4283. doi: 10.1093/nar/29.21.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gandillet A., Serrano A.G., Pearson S., Lie A.L.M., Lacaud G., Kouskoff V. Sox7-sustained expression alters the balance between proliferation and differentiation of hematopoietic progenitors at the onset of blood specification. Blood. 2009;114:4813–4822. doi: 10.1182/blood-2009-06-226290. [DOI] [PubMed] [Google Scholar]
- 31.Hosking B., Francois M., Wilhelm D., Orsenigo F., Caprini A., Svingen T., Tutt D., Davidson T., Browne C., Dejana E., et al. Sox7 and Sox17 are strain-specific modifiers of the lymphangiogenic defects caused by Sox18 dysfunction in mice. Development. 2009;136:2385–2391. doi: 10.1242/dev.034827. [DOI] [PubMed] [Google Scholar]
- 32.Kanai-Azuma M., Kanai Y., Gad J.M., Tajima Y., Taya C., Kurohmaru M., Sanai Y., Yonekawa H., Yazaki K., Tam P.P., et al. Depletion of definitive gut endoderm in Sox17-null mutant mice. Development. 2002;129:2367–2379. doi: 10.1242/dev.129.10.2367. [DOI] [PubMed] [Google Scholar]
- 33.Katoh M. Expression of human SOX7 in normal tissues and tumors. Int. J. Mol. Med. 2002;9:363–368. [PubMed] [Google Scholar]
- 34.Lioubinski O., Muller M., Wegner M., Sander M. Expression of Sox transcription factors in the developing mouse pancreas. Dev. Dyn. 2003;227:402–408. doi: 10.1002/dvdy.10311. [DOI] [PubMed] [Google Scholar]
- 35.Matsui T., Kanai-Azuma M., Hara K., Matoba S., Hiramatsu R., Kawakami H., Kurohmaru M., Koopman P., Kanai Y. Redundant roles of Sox17 and Sox18 in postnatal angiogenesis in mice. J. Cell Sci. 2006;119:3513–3526. doi: 10.1242/jcs.03081. [DOI] [PubMed] [Google Scholar]
- 36.Sakamoto Y., Hara K., Kanai-Azuma M., Matsui T., Miura Y., Tsunekawa N., Kurohmaru M., Saijoh Y., Koopman P., Kanai Y. Redundant roles of Sox17 and Sox18 in early cardiovascular development of mouse embryos. Biochem. Biophys. Res. Commun. 2007;360:539–544. doi: 10.1016/j.bbrc.2007.06.093. [DOI] [PubMed] [Google Scholar]
- 37.Tam P.P., Khoo P.L., Wong N., Tsang T.E., Behringer R.R. Regionalization of cell fates and cell movement in the endoderm of the mouse gastrula and the impact of loss of Lhx1(Lim1) function. Dev. Biol. 2004;274:171–187. doi: 10.1016/j.ydbio.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 38.Young N., Hahn C.N., Poh A., Dong C., Wilhelm D., Olsson J., Muscat G.E., Parsons P., Gamble J.R., Koopman P. Effect of disrupted SOX18 transcription factor function on tumor growth, vascularization, and endothelial development. J. Natl. Cancer Inst. 2006;98:1060–1067. doi: 10.1093/jnci/djj299. [DOI] [PubMed] [Google Scholar]
- 39.Molkentin J.D., Lin Q., Duncan S.A., Olson E.N. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 40.Clugston R.D., Zhang W., Greer J.J. Gene expression in the developing diaphragm: significance for congenital diaphragmatic hernia. Am. J. Physiol. Lung Cell Mol. Physiol. 2008;294:L665–L675. doi: 10.1152/ajplung.00027.2008. [DOI] [PubMed] [Google Scholar]
- 41.Shimokawa O., Miyake N., Yoshimura T., Sosonkina N., Harada N., Mizuguchi T., Kondoh S., Kishino T., Ohta T., Remco V., et al. Molecular characterization of del(8)(p23.1p23.1) in a case of congenital diaphragmatic hernia. Am. J. Med. Genet. A. 2005;136:49–51. doi: 10.1002/ajmg.a.30778. [DOI] [PubMed] [Google Scholar]
- 42.Slavotinek A., Lee S.S., Davis R., Shrit A., Leppig K.A., Rhim J., Jasnosz K., Albertson D., Pinkel D. Fryns syndrome phenotype caused by chromosome microdeletions at 15q26.2 and 8p23.1. J. Med. Genet. 2005;42:730–736. doi: 10.1136/jmg.2004.028787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lammert E., Cleaver O., Melton D. Role of endothelial cells in early pancreas and liver development. Mech. Dev. 2003;120:59–64. doi: 10.1016/s0925-4773(02)00332-5. [DOI] [PubMed] [Google Scholar]
- 44.Crivellato E. The role of angiogenic growth factors in organogenesis. Int. J. Dev. Biol. 2011;55:365–375. doi: 10.1387/ijdb.103214ec. [DOI] [PubMed] [Google Scholar]
- 45.Liu P., Jenkins N.A., Copeland N.G. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.