Abstract
During the course of HIV infection, some HIV-1 viruses switch from using the CCR5 (R5) coreceptor to using CXCR4 (X4). Here, we describe two subtype C isolates from a Zimbabwean patient that switched from using R5 to using both R5 and X4 with an accompanying addition of five amino acids to the V3 loop region of envelope. The insert appears to be derived from the human genome rather than a duplication within HIV-1.
Initial infection with HIV-1 occurs most often with viruses that utilize the chemokine coreceptor CCR5 (R5). However, in up to 50% of HIV-1 subtype B infections, envelope glycoproteins acquire amino acid changes that lead to using the coreceptor, CXCR4 (X4); this switch precedes progression to AIDS [1,2]. The prevalence of X4 usage in subtype C HIV-1 viruses has been thought to be lower compared to subtype B HIV-1, but studies of X4-using subtype C viruses have become more common [3–6]. The mechanism of coreceptor switch is unknown. Here, we report on subtype C virus isolates from an individual in Zimbabwe where, over 2 years, there was a switch from an R5 to X4 using virus that occurred concurrent with an unusual insert within the V3 loop of envelope.
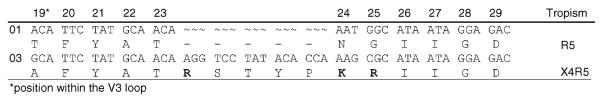
A blood sample was first collected from a 36-year-old patient, TC02, in 2001 in Harare, Zimbabwe. Peripheral blood mononuclear cells (PBMCs) were cultivated with donor PBMCs and the virus isolate was characterized as using R5 through both an MT-2 assay and GHOST assay using fluorescence-activated cell sorting to identify infected cells [6]. A second blood sample was collected in 2003 and virus was grown from the PBMCs. This second virus isolate grew on MT-2 cells, forming syncytia, and used both R5 and X4 coreceptors in the GHOST assay. Over the 2 years between isolates, the CD4+ cells increased from 9 to 81 and the viral load decreased from 5.09 log10 copies RNA/ml to 3.94 log10 copies RNA/ml on a nonsuppressive antiretroviral regimen, consisting of lamivudine, stavudine, and saquinavir. There was evidence for evolution of HIV-associated drug resistance in the second sample, with development of an L90M mutation in the viral protease and M184V in the viral reverse transcriptase compared to wild-type virus at baseline. Upon sequencing the C2-V5 region of both samples, we discovered that the second sample contained an unusual five amino acid insert within the V3 loop (Fig. 1). This insertion was observed in the primary isolate from 2003 as well as three biological clones obtained by limiting dilution of PBMCs infected with the initial isolate. The five amino acid insert, RSYTP, is located between amino acids 23 and 24, just after the crown region.
Fig. 1. Comparison of TC02 isolate V3 loop envelope sequences in 2001 and 2003.
The env C2-V5 region was sequenced after reverse transcription and amplification of viral RNA from virus stocks isolated in 2001 and 2003 from cultured peripheral blood mononuclear cells. Sequences were manually aligned in Bioedit. Amino acids 19–29 of the V3 loop as well as their corresponding nucleotides are shown and positively-charged amino acids are displayed in bold. Accession numbers for C2-V5 env sequences: AY265929 (2001), FJ445693 (2003).
Insertions and deletions within the envelope V3 loop are common, especially among X4-using viruses. These insertions are usually one to three amino acids, and they often occur just before the crown region. Insertions have been reported in other HIV-1 genes and are frequently the result of duplications from other parts of the HIV-1 genome. For example, a five amino acid insert within reverse transcriptase was attributed to a duplication from the beginning of envelope [7]. However, a BLAST search of the HIV-1 Los Alamos database showed that the 15 nucleotides that comprise RSYTP are not found anywhere within the HIV-1 genome. A search of Genbank revealed that the sequence is found in the human genome. This sequence of 15 nucleotides is present in the transcription factor AP-2 on chromosome 6, as well as within chromosome 10. Sixteen nucleotides (the 15 and the next A nucleotide) are found in chromosome 16. Shorter 13 and 14 base fragments are found throughout the human genome.
Another explanation for the change in tropism may be the switch to lysine (K) and arginine (R) at positions 24 and 25, immediately 3′ to the insert. The 2001 isolate contained neutral amino acids asperagine (N) and glycine (G) at these two positions. The insert increases the V3 loop length from 35 to 40 amino acids and increases the net charge. The K and R after the insert contribute to the increase in charge as well, resulting in an increase in net charge from +5 to +8. Analysis of the V3 loop amino acid sequence with the insert removed using WebPSSM (http://indra.mullins.microbiol.washington.edu/pssm/) predicts that the virus would be more likely to use R5 for entry, whereas the presence of positive amino acids at positions 24 and 25 is consistent with a virus that would also use X4 as a coreceptor.
These sequential isolates and the characterization of tropism in biologically cloned virus provides an example of an increase in V3 loop length and charge coinciding with a change in coreceptor usage, from an exclusively R5-using virus to a population capable of using both R5 and X4 for viral entry. The five amino acid insert adds an additional charged residue (R) and a second proline. Because of their unique R-group, prolines contribute to loop structures and increase flexibility in the secondary structures of peptides. This unique insert provides a further demonstration of the relationship between charged residues within the V3 loop, the increased flexibility of a ‘promiscuous’ envelope protein and the emergence of X4 and R5/X4 usage. The presence of the insert in the V3 loop of this virus increased both charge and flexibility, which suggests that the acquisition of host (human) sequences through recombination may lead to changes in tropism. However, more evidence is needed to prove the origin of this insert.
References
- 1.Connor RI, Sheridan KE, Ceradini D, Choe S, Landau NR. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koot M, van Leeuwen R, de Goede RE, Keet IP, Danner S, Schattenkerk JK Eeftinck, et al. Conversion rate towards a syncytium-inducing (SI) phenotype during different stages of human immunodeficiency virus type 1 infection and prognostic value of SI phenotype for survival after AIDS diagnosis. J Infect Dis. 1999;179:254–258. doi: 10.1086/314539. [DOI] [PubMed] [Google Scholar]
- 3.Batra M, Tien PC, Shafer RW, Contag CH, Katzenstein DA. HIV type 1 envelope subtype C sequences from recent seroconverters in Zimbabwe. AIDS Res Hum Retroviruses. 2000;16:973–979. doi: 10.1089/08892220050058399. [DOI] [PubMed] [Google Scholar]
- 4.Cilliers T, Nhlapo J, Coetzer M, Orlovic D, Ketas T, Olson WC, et al. The CCR5 and CXCR4 coreceptors are both used by human immunodeficiency virus type 1 primary isolates from subtype C. J Virol. 2003;77:4449–4456. doi: 10.1128/JVI.77.7.4449-4456.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coetzer M, Cilliers T, Papathanasopoulos M, Ramjee G, Karim SA, Williamson C, Morris L. Longitudinal analysis of HIV type 1 subtype C envelope sequences from South Africa. AIDS Res Hum Retroviruses. 2007;23:316–321. doi: 10.1089/aid.2006.0207. [DOI] [PubMed] [Google Scholar]
- 6.Johnston ER, Zijenah LS, Mutetwa S, Kantor R, Kittinunvorakoon C, Katzenstein DA. High frequency of syncytium-inducing and CXCR4-tropic viruses among human immunodeficiency virus type 1 subtype C-infected patients receiving antiretroviral treatment. J Virol. 2003;77:7682–7688. doi: 10.1128/JVI.77.13.7682-7688.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lobato RL, Kim EY, Kagan RM, Merigan TC. Genotypic and phenotypic analysis of a novel 15-base insertion occurring between codons 69 and 70 of HIV type 1 reverse transcriptase. AIDS Res Hum Retroviruses. 2002;18:733–736. doi: 10.1089/088922202760072375. [DOI] [PubMed] [Google Scholar]