Abstract
Multiplex Automated Genome Engineering (MAGE) employs short oligonucleotides to scarlessly modify genomes. However, insertions of >10 bases are still inefficient, but can be improved substantially by selection of highly modified chromosomes. Here, we describe Co-Selection MAGE (CoS-MAGE) to optimize biosynthesis of aromatic amino acid derivatives by combinatorially inserting multiple T7 promoters simultaneously into 12 genomic operons. Promoter libraries can be quickly generated to study gain-of-function epistatic interactions in gene networks.
The ability to directly manipulate and add new genetic regulatory elements to the chromosome is highly desirable as it bypasses the need to resort to plasmid-based systems where copy number variation can substantially affect the dynamics and robustness of synthetic networks1. Previously, we developed the Multiplex Automated Genome Engineering (MAGE) method to combinatorially introduce sequence changes to many sites in the Escherichia coli genome using libraries of 90-base synthetic oligonucleotides2, 3. While the efficiency for making small modifications was high (>30% for <4bp)2, 4, we observed that the insertion efficiency per MAGE cycle dropped substantially for inserts of larger sizes (<2% for >20bp), thus limiting its utility for integration of regulatory elements or larger sequence motifs. However, we unexpectedly found that a small fraction of isolates contained more than one mutated site in each cycle of MAGE. We hypothesized that the isolation of clones containing a mutation at one site may increase the likelihood that other mutated sites are also found because a certain cell subpopulation may be more electrocompetent or more available to perform oligo-based allelic replacement. We call the process of population enrichment for highly mutated genomes Co-Selection (CoS) MAGE, which we demonstrate here as an enhanced scarless genome engineering approach to efficiently introduce larger regulatory elements such as promoters into targeted regions on the chromosome.
The 20 bp T7-phage promoter (TAATACGACTCACTATAGGG) was chosen for insertion upstream of 12 genes or operons in the E. coli genome that are associated with the biosynthesis of aromatic amino acids (Fig. 1 and Supplementary Fig. 1). In the presence of T7 RNA polymerase, transcription from the T7 promoter (T7p) is strong and orthogonal to endogenous promoters. In E. coli, the industrially relevant blue dye, indigo, and anti-leukemia compound5, indirubin, can be produced from the tryptophan (Trp) intermediate, indole, by heterologous expression of a Methylophaga sp. flavin-containing monooxygenase, bfmo6. Using the MAGE-competent EcNR2 strain2, we first removed known feedback regulation and allosteric inhibition associated with Trp biosynthesis7–10, which involved nonsense inactivation of trpR, P148L mutation in aroF, M293T mutation in trpE, and D146N mutation in aroG. We also inactivated the galK, malK, cat, and bla genes on the chromosome by introducing a revertible premature stop codon in each gene, which was also done at high-efficiency by MAGE using 90-bp oligos to generate the strain EcHW47 (see Online Methods).
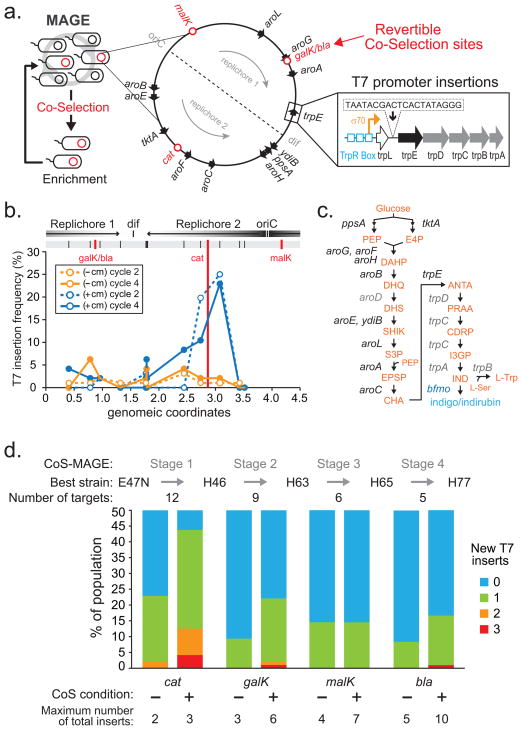
Fig 1. Co-selection MAGE strategy for enriching highly modified genomes.
a, Cells undergoing cycles of MAGE were enriched by a co-selection stage through phenotypic selection of a revertible genomic locus (i.e. malK, galK, cat or bla) and used for subsequent cycles of MAGE in an iterative fashion. Twelve genomic operons were targeted for insertion of the T7 promoter sequence upstream of the first relevant open reading frame (ORF). b, Enhanced T7 insertion frequency near the cat locus was observed only when the population was co-selected by chloramphenicol for cat (blue lines) in comparison to without co-selection (orange lines) after two (dotted lines) and four (solid lines) MAGE cycles as determined by MASC-PCR (Online Methods). c, Biosynthesis of tryptophan and indole derivatives indigo and indirubin. Intermediate products are shown in orange and the first gene in each relevant operon shown in black. Targeted genes that are not first in the operon are shown in gray. d, Each stage of CoS-MAGE involves 4 cycles of MAGE followed by screening of 96 colonies to find the most modified mutant. Population frequency of colonies with 0 to 3 new T7p insertions for each co-selection stage are shown for the corresponding conditions. Co-selection enriches for highly modified genomes, which are used in the subsequent CoS-MAGE stages with a reduced oligo pool to target the remaining sites. The most enriched strains at the end of each CoS stage are noted (H46, H63, H65, H77).
To perform CoS-MAGE, the mixed oligo pool that targets all 12 insertion sites was spiked at a molar ratio of 50:1 with a small amount of an oligo (called CoS oligo) that reverted the function of an inactivated genomic selectable marker (i.e. galK(−), malK(−), cat(−), or bla(−)). Restoration to galK(+) or malK(+) enabled growth on M9 minimal media supplemented with galactose or maltose, respectively, as the sole carbon source. Restoration to cat(+) or bla(+) conferred antibiotic resistance to chloramphenicol (cm) or carbenicillin (carb) respectively. In the first CoS stage, we performed 4 MAGE cycles targeting 12 sites and cat(−). At the end of the CoS stage, 96 colonies plated with and without chloramphenicol were isolated and genotyped by Multiplex-Allele Specific Colony (MASC) PCR3 to query for the presence of T7p insertions at the 12 target sites (see Online Methods). We found that among the isolated clones, target sites near the CoS marker were highly enriched for T7p insertions only when the chloramphenicol CoS was applied (Fig. 1b). At these sites, the frequency of T7p integration was as high as 25% with CoS in comparison to 2–3% without. This increased frequency of integration could be seen after only 2 MAGE cycles (Fig. 1b). We hypothesize that oligo-mediated genomic modifications near the CoS marker were enhanced by capturing cells whose replication forks were transiently in the open state and available for allelic replacement11.
By selecting against cells that do not generate oligo-mediated allelic replacements, we can substantially enrich the population for individual clones with multiple T7p insertions. This co-selection effect can be further enhanced through multiple MAGE cycles. Without CoS enrichment, 10 to 25% of the population contained cells with one T7p after 4 MAGE cycles. With CoS, more than 40% of cells had at least one T7p insertion, and 5% had three insertions (Fig. 1d). Using clone H46 that contained the most T7p insertions (3 of 12), we performed a second stage of CoS-MAGE by targeting the remaining 9 sites using another CoS marker, galK. This stage produced clones with up to 3 more T7p insertions. Two more stages of CoS-MAGE generated a clone with 10 T7p insertions (Fig. 1d). In comparison, we could only generate up to 5 insertions in a single clone in the absence of CoS. Co-selection by malK during the third stage did not yield mutliple additional mutants, likely due to the fact that malK is far from any target site (>500 kb, see Supplementary Table 1), providing support for the replication-dependency hypothesis in which co-selection affects neighboring chromosomal regions. These results demonstrate that CoS-MAGE can substantially enhance the generation of genomic insertions (Supplementary Note). We inserted T7p into the two remaining sites by CoS-MAGE to produce a clone that contains all12 T7 promoters for orthogonally driving gene expression.
One important feature of CoS-MAGE is the capacity to produce combinatorial libraries of variants. In the process of generating the 12-target strain, we were able to isolate intermediate strains with fewer T7p insertions but in different combinations (Fig. 2). We recovered 80 unique variants with 1 to 12 T7p insertions including all 12 single inserts and two complete panels of 11 double inserts (aroG or trpE with all others). Combinatorial T7 promoter expression was used to probe the tunable parameters within the biosynthesis network. We could indirectly measure the level of tryptophan produced through expression of bfmo from each strain in the library when driven by a plasmid-encoded T7 polymerase (pN249). Indigo and indirubin pigments, the major products of the bfmo monooxygenase6, were readily extracted by DMSO treatment (Online Methods).
Fig 2. Indigo production from synthetic T7p regulated operons.
Strain E7N is an E2N-derivative that contains the bfmo-encoding pJ401 plasmid. Strain E47N is an E7N derivative with inactivated trpR repression and absent allosteric feedback through aroF(P148L), trpE(M293T), and aroG(D146N). Strains H1-H80 are T7p derivatives of E47N. The top bar graph shows relative change in growth rate for each strain normalized to E7N, Normalized percent values for each strain are calculated by (y−x)/y where x is the doubling time of each strain and y is doubling time of the E7N wild-type. n = 3 for each strain, error bars, s.e.m. The bottom bar graph shows indigo production based on measured indirubin levels (Online Methods). Color coding reflects grouping by the number of T7 insertions. Genotypes for each clone are marked by black boxes and ordered by their relative location along the biosynthesis pathway (ppsA at the top is proximal and trpE at the bottom is distal). The gray box of strain H70 corresponds to an insert in aroH with a mutated T7 sequence (TAATACACTCAGTATGGG).
Within our library, strains with more than 4-fold improvement in indigo production over the E2N ancestor (expressing T7RNAP) were found (Fig. 2) and could be visually appreciated (Supplementary Fig. 2). Based on calibration with commercially available indigo, we estimate that our best variants (H33, H76) produced >8.6 mg g−1 dry cell weight of indigo. Interestingly, we observed synergistic epistasis between certain T7p insert combinations. For example, aroC (H11) and trpE (H12) did not significantly improve production individually relative to the E47N ancestral strain (P = 0.29 and 0.11 respectively; Student’s t test here and below), but in combination (H33) produced 62% more than E47N (P = 8.4 × 10−10). Many other pairs also showed synergetic interactions, such as aroG+ydiB (H18), aroA+aroH (H35), aroA+aroF (H36), aroA+ydiB (H37) and aroA+aroC (H39).
Analysis of higher-order mutants revealed more unexpected results. Strain H77, with 10 of 12 inserts, showed a 20% improvement over E47N (P = 2.5 × 10−3), but the addition of aroB in H78 significantly reduced the yield to 20% below E47N (P = 4.2 × 10−3). Addition of the final tktA insert to this strain in the 12-promoter H80 strain returned yield to 29% above E47N (P = 2.5 × 10−3). Such large and opposite effects of aroB insertion and subsequent tktA insertion suggest that interactions in this gene network are unexpectedly complex, nonlinear, and epistatic. Growth rate measurements of each strain under bmfo-induced conditions also revealed changes ranging from −30% to +30% relative to the ancestral strain (Fig. 2). However, no notable correlation could be extracted between growth rate and indigo levels (Supplementary Fig. 3). We hypothesize that these types of complex epistatic interactions are more prevalent in endogenous networks than previously expected, especially in the context of large-scale modulation of gene expression using synthetic regulatory modules.
Using CoS-MAGE, we can probe epistasis in endogenous and engineered genetic pathways. With a fully synthetic regulon such as the 12 promoter-bearing H80 strain, a reverse optimization can be done by CoS-MAGE using degenerate oligos to fine-tune the expression of each T7 promoter in the pathway with orthogonal T7 polymerases12. Switchable CoS markers (antibiotic resistance genes, fluorescent proteins, or metabolic genes) can be placed anywhere on the genome and can be easily activated or inactivated with individual oligos. We mapped a list of 21 useful genomic markers (Supplementary Fig. 4) with their relevant switching oligos and selection schemes (Supplementary Table 2). Longer oligos can be used in MAGE, with increased homology arm length correlating with increased integration efficiency (Supplementary Fig. 5). We speculate that the use of longer oligonucleotides (100–200 bases) may enable the integration of even larger regulatory sequences for efficient CoS-MAGE. The efficiency improvements of our co-selection method over other MAGE implementations (Supplementary Fig. 6 and Supplementary Table 3) make it more feasible to perform the method in the absence of automated instrumentation. The CoS strategy can be applied to improve engineering of other organisms where oligo-mediated recombineering may be less efficient13. Furthermore, plasmid engineering can benefit from co-selection to combinatorially generate vector libraries of synthetic circuits14. Multiplexed modification of endogenous transcription, translation, and protein-coding sequences will enable larger genome-scale engineering efforts to further push the limits of engineered biological systems15.
ONLINE METHODS
Media, chemicals, and reagents
All strains were grown in rich LB-min media containing tryptone (10 g l−1), yeast extract (5 g l−1), and NaCl (5 g l−1), and buffered to pH 7.45 with NaOH. Chloramphenicol (cm) at 20 μg ml−1, kanamycin (kan) at 30 μg ml−1, carbenicillin (carb) at 50 μg ml−1, or spectinomycin (spec) at 100 μg ml−1 were supplemented to liquid LB-min media or LB-min agar plates (LB-min with 15 g l−1 agar) for selection. Isopropyl β-D-1-thiogalactopyranoside (IPTG) was used at 0.25 mM to induce the Plac promoter. MacConkey-Galactose (Mac-Gal) or MacConkey-Maltose (Mac-Mal) agar plates were made by supplementing Difco MacConkey Agar Base (40 g l−1) which contains peptone (17.0 g l−1), proteose peptone (3.0 g l−1), bile salts no. 3 (1.5 g l−1), NaCl (5.0 g l−1), agar (13.5 g l−1), neutral red (30 mg l−1), and crystal violet (1 mg l−1), with D-(+)-galactose or D-(+)maltose monohydrate at 10 g l−1 respectively. M9 minimal media was made by adding 5X M9 base (64 g l−1 Na2HPO4-7H2O or 30 g l−1 Na2HPO4, 15 g l−1 KH2PO4, 2.5 g l−1 NaCl 5.0 g l−1 NH4Cl) with MgSO4-7dH2O (1 mM), vitamin B1 (5×10−5 %), D-biotin (0.2 μg l−1) and 0.2% D-(+)-galactose or D-(+)-maltose monohydrate. Multiplex PCR kits were purchased from Qiagen (Cat #206143). Standard 96-well format agarose gel electrophoresis system (Bio-Rad Sub-Cell Model 96 Cell) and reagents were used.
Oligonucleotides (Oligos)
All primers and oligonucleotides were obtained from Integrated DNA Technologies with standard purification. MAGE oligos were designed to insert T7 promoters 35 bp upstream of the start codon of the protein coding sequence of 12 gene/operons (see Supplementary Fig. 1). These oligos had minimized secondary structure (>−12 kcal/mol) and targeted the replicating lagging strand. Additionally, each oligo contained four phosphorothioate linkages at the 5′ terminus. Sequences of all MAGE oligos and primers used for sequencing and MASC-PCR are provided in Supplementary Table 4.
Co-Selection MAGE Cycling Process
MAGE cycling has been previously described3. In brief, cells were grown in 3 mL volume with proper antibiotics in a 30 °C rotating incubator. At OD600 of 0.6, cells were heat-shocked in a 42 °C shaking water-bath for 15 min. One mL of cells was made electrocompetent per reaction at 4 °C by several buffer exchanges with sterile distilled water through centrifugation and resuspension. Oligos were added to concentrated cells for each 50 ul electroporation reaction. For multiplex reactions, targeting oligos were mixed in equal molar to reach a final total oligo concentration of ~12 μM. Co-selection oligos (e.g. cat_restore_oligo, galK_restore_oligo, malK_restore_oligo, bla_restore_oligo), which would restore the inactivated nonsense mutation to functional wild-type sequence, were added to the oligo pool at a molar ratio of 1:50 (1 CoS oligo per 50 targeting oligo). Electroporation was carried out in 1-mm gap cuvettes with conditions 1.8 kV, 200 Ω, 25 μF, and cells were recovered into fresh LB media for subsequent MAGE cycle. Each stage of Co-Selection was done at the end of 4 cycles of MAGE by plating on the appropriate selective condition (LB-cm for cat(+), LB-carb for bla(+), M9-Gal for galK(+), Mac-Gal for red galK(+), M9-Mal for malK(+), or Mac-Mal for red malK(+)). Isolating and MASC-PCR genotyping 48–96 colonies resulted in numerous positive clones that were subsequently sequence verified. A streamlined Co-Selection MAGE protocol is described here. Each cycle of MAGE takes ~2 hrs, and 4 cycles can be easily done in a single day, ending with plating on the selection media plates (for the co-selection step). Overnight selection on plates will yield colonies in the morning, which can be picked directly into MASC-PCR reactions to identify the best clone (2 hr turnaround). At the same time, each colony can be grown up in 96-well format in-parallel to the PCR genotyping step. The best clone (taken from the already 2-hr grown 96-well plate) can then be used for the next 4 cycles of MAGE in the same day (Day 2) and iterated in subsequent days in the same manner. For selection that required a minimal media growth step (M9-gal, M9-mal), an additional day is needed to allow the colonies to form for each Co-Selection step, thus doubling the total time needed. We would like to note that 4 cycles of MAGE is not needed each day before the co-selection enrichment is applied (2 or even 1 cycle may be sufficient). We performed 4 cycles per day because it was practically as easy as 2 with low sample numbers and some minor benefit can be gained from additional rounds of standard MAGE. This co-selection method is also a practical way of implementing MAGE in the absence of an automated instrument to perform MAGE, while still achieving a high frequency of site-directed mutagenesis.
Multiplex Allele-specific Colony PCR (MASC-PCR)
Multiplex allele-specific colony PCR (MASC-PCR) was done as previously described3 as a fast, reproducible and cost effective way to query the genotype of clones at up to 12 loci per reaction. In brief, two sets of primers (wild-type forward/reverse, designated as f/r, and mutant forward and reverse, designated as fm/r) were used to query each genomic locus with (f/r) corresponding to the wild-type allele and (fm/r) corresponding to the mutant allele. The forward primers (f and fm) were identical except at the 3′ terminus, which corresponded to either the wild-type (f) or mutant (fm) allele. The reverse primer was the same for both alleles. If a clone has the wild-type allele, only primer set f/r should produce a product and not primer set fm/r. Conversely, if a clone has the mutant allele, only primer set fm/r should produce a PCR product and not f/r. This allele specific PCR reaction can be multiplexed to amplify amplicons of different lengths (100, 150, 200, 250, 300, 400, 500, 600, 700, 850, 1000, 1200 bp) corresponding to each of the 12 loci in the MASC-PCR reaction. Primers were generally designed for a target Tm of 62 °C, but a gradient PCR was done to experimentally determine the optimal Tm to improve specificity of distinguishing wild-type and mutant alleles. The Multiplex PCR kit from Qiagen (Cat #206143) was used in 20 μL reactions with 1 μL of a 1 in 100 water dilution of a picked colony and final primer concentration of 0.2 μM. PCR cycles were as follows: Step 1 - 15 min at 95 °C; Step 2 - 30 sec at 94 °C; Step 3 - 30 sec at optimal Tm; Step 4 - 80 sec at 72°C; Step 5 - repeat Steps 2–4 for 26 times; and Step 6 - 5 min at 72°C. Electrophoresis was done on a 1.5% ethidium bromide agarose gel and visualized under UV.
Base Strain Construction
The MAGE-competent ΔmutS::cat base strain EcNR2 has been previously described2. EcNR2 contained a heat-shock inducible genomic λ-Red system that required growth at 30–32°C, with the bla gene replacing the bioA locus. Transformation of EcNR2 with pJ401 (courtesy of Howard Salis), which expressed bfmo by 0.25 mM IPTG induction, and selection on LB-kan plates yielded EcHW7. The TrpR repressor was inactivated in EcHW7 by MAGE using trpR_KO_Oligo. MASC-PCR screening of 48 colonies (using primer set trpR_KO_f/fm/r) yielded several positive clones, one of which was designated EcHW44. To remove tryptophan-associated allosteric feedback, single nucleotide mutations were introduced to aroF, trpE, and aroG of EcHW44 using aroF(P148L)_oligo, trpE(M293T)_oligo, aroG(D146N)_oligo by applying 4 cycles of MAGE along with oligo galK_mut45_oligo. Following the 4 MAGE cycles, cells were plated on Mac-Gal+kan plates and 48 overnight galK(−) white colonies were picked, pooled, and regrown together for another 2 cycles of MAGE with aroF(P148L)_oligo, trpE(M293T)_oligo, aroG(D146N)_oligo and galK_mut45_oligo (total oligo concentration of 10 μM). Following the 2 MAGE cycles, cells were plated on Mac-Gal+kan plates and 32 overnight malK(−) white colonies were picked and genotyped for the aroF, trpE, and aroG mutations by MASC-PCR using primer sets aroF(P148L)_f/fm/r, trpE(M293T)_f/fm/r, and aroG(D146N)_f/fm/r to identify a positive clone EcHW45, which had all the mutations. EcHW45 was used to generate the inactivated cat(−) genotype using cat_mut45_oligo and subsequently made bla(−) with bla_mut45_oligo to generate EcHW47. Screening of cat(−) and bla(−) was easily done by replica plating from LB plates onto LB-cm+carb or more quickly done by picking ~30 colonies into LB and LB-cm+carb media to identify cm- or carb-sensitive strains. EcHW47 contained aroF(P148L), trpE(M293T), aroG(D146N) and revertible nonsense mutations in the inactivated galK(−), malK(−), cat(−) and bla(−) loci. Transformation of the IPTG-inducible T7 RNA polymerase gene on pN249 (courtesy of Karsten Temme) into EcNR2, EcHW7 and EcHW47 and selection on LB-spec yielded E2N, E7N and E47N respectively.
T7 Promoter Insertions by CoS-MAGE
The E47N strain was used to insert T7 promoters into 12 genomic loci 35 bp upstream of aroL, aroG, aroA, trpE, ydiB, ppsA, aroH, aroC, aroF, tktA, aroE, and aroB. For the first stage of CoS-MAGE, 12 oligos were pooled together in equal molar ratio for a final concentration of 1 μM per oligo. The cat_restore_oligo (CoS oligo at final concentration of 0.02 μM) was added to this pool. The E47N strain was cycled 4 times by MAGE using the oligo pool over the course of 8 hours (a typical workday). In general, the number of cat(+) cells in the population was ~1 in 2000 so dilutions were made accordingly when plating. The cell population was diluted and spread on LB+cm (for co-selection for cat) or on LB (without co-selection) overnight. We isolated and genotyped 96 colonies from each plate by MASC-PCR for presence of the T7 promoter sequences at each of the 12 target sites. Of the co-selected colonies assayed, clone H46 had the most T7 inserts (aroH, aroC and aroF) so it was used for the second stage of CoS-MAGE. In the second stage, 9 oligos corresponding to the remaining targets were pooled together in equal molar ratio as well as the galK_restore_oligo (CoS oligo at final per oligo concentration of 1 μM). We increased the CoS oligo to target oligo ratio because at the original (1:50 ratio) we expected to find 1 red galK(+) colony in ~2000 colonies on MAC-Gal, which would be infeasible for clonal isolation. Boosting the CoS oligo concentration can increase the number of galK(+) colonies to levels screenable by plating. Alternatively, plating on M9-Gal for galK(+) colonies was also feasible and would not need higher galK CoS oligo concentration, but at a longer delay due to slower colony growth on minimal media (~2.5X of wild-type). The 9 target plus galK CoS oligo pool was used in the second CoS stage through 4 cycles of MAGE on H46, and the cell population was plated on Mac-Gal. Isolation and MASC-PCR genotyping of 96 red galK(+) colonies grown overnight resulted many additional T7 inserts. The best clone (H63) had 6 inserts in total (aroF, aroC, aroH, ppsA, ydiB and aroL). The third CoS stage was done on the remaining 6 targets similarly as described above using malK_restore_oligo as the CoS oligo and plating on Mac-Mal to isolate red malK(+) colonies. The best clone (H65) containing 7 inserts (aroE aroF, aroC, aroH, ppsA, ydiB, aroL) was isolated. The fourth CoS stage was done on the remaining 5 targets using bla_restore_oligo. Selection on LB-carb and isolation of 96 colonies resulted in the best clone H77 that contained 10 inserts (without tktA and aroB). We reiterated CoS-MAGE on the remaining targets to generate the H80 clone, which contained all 12 desired T7 promoter insertions as well as other clones containing unique insertions through MASC-PCR every 2 MAGE cycles. A summary of genomic distance of each locus to the relevant Co-selectable marker is shown in Supplementary Table 1. In total, 80 genotypically unique strains (H1-H80) were isolated containing 1 to 12 T7 promoter insertions. We transformed the T7 polymerase-encoding pN249 plasmid into each strain and selected on LB-spectinomycin for the presence of the plasmid.
Sequencing Verification
We sequenced all 12 target sites for the expected T7 sequence in each of the 80 strains (H1-H80). The PCR primers amplified ~150–180 bp around each T7 insertion locus by a standard protocol. All sequenced sites were verified to contain the correct T7 sequence (TAATACGACTCACTATAGGG ) except site aroH of strain H70 where the T7 sequence was TAATACACTCAGTATGGG.
Pigment Extraction and Quantification
To quantify the amount of up-regulation in the modified biosynthesis pathway, we measured indigo/indirubin production in H1-H80 and ancestral strains E2N, E7N and E47N following previously described methods3. Each strain was grown in 10 mL of LB-kan+spec+IPTG for 24 hours at 30 °C prior to extraction. E2N was grown in LB-spec+IPTG only. For pigment extraction, 1 mL of each strain was concentrated by centrifugation in a deep-wall 96-well plate. Excess media was discarded and each cell pellet was resuspended in 1 mL of DMSO. The samples were sonicated for 10 seconds and centrifuged again to concentrate the cell debris, which appeared white. The blue supernatant was removed and stored at room temperature in the dark to allow accurate measurement of indigo and indirubin levels. After 3 days, 200 μL of each supernatant was transferred to a 96-well plate and absorbance at 550 nm was measured on a Molecular Devices spectrophotometer to quantify relative indirubin levels. Indigo at 620 nm was also measured. However, the absorbance level decayed over time (Supplementary Fig. 2) likely due to indigo instability. Thus to determine absolute indigo production levels, we took the relative indirubin levels against strain H33 (best producer) and normalized against the measured indigo production level (at Ab620) of an immediately extracted H33 sample. Indigo levels were determined by mapping the Ab620 to a calibration curve generated using dilutions of commercially obtained indigo powder. Five ml of cells were dried and weighed to derive the dry cell weight. Pigment extractions were repeated four separate times on different days for each strain to ensure statistical confidence.
Statistical Analysis
The P-values described in the paper were determined by performing the Student’s t-test between the two sample groups to derive the t-score, which is then used to compute the p-value.
Supplementary Material
Supplementary Figure 1. Insertion design used to introduce T7 promoters upstream of genes/operons.
Supplementary Figure 2. Pigment quantification.
Supplementary Figure 3. Correlation of indigo production level and growth rate
Supplementary Figure 4. Genomic map and list of 21 available Co-Selection markers that can be used to enhance MAGE efficiency.
Supplementary Figure 5. Characterization of the insertion efficiency of various lengths
Supplementary Figure 6. Comparison of the length of time in experimental hours needed for different MAGE strategies to generate clones with 20bp T7 promoter insertions.
Supplementary Table 1. Summary of stages of CoS-MAGE to generate the H77 strain.
Supplementary Table 2. Summary of CoS Markers available in EcNR2 (MG1655 derivative)
Supplementary Table 3. Side-by-side comparison of various genome engineering strategies to make a strain that contains 20bp T7 insertion mutation across 10 genomic loci.
Supplementary Table 4. Oligo and primer sequences used in the study.
Supplementary Note. Co-selection mechanism and enhancement analysis
Acknowledgments
We thank J. Keasling, A. Juminaga, P. Carr, S. Kosuri, J. Aach, and T. Gianoulis for helpful discussions and data interpretation. We thank H. Salis (Penn State) for generously providing pJ401 and C. Voigt (MIT) for generously providing pN249. This work was funded by multiple programs from the National Science Foundation (SynBERC, Grant #SA5283-11210), the Department of Energy (Genome to Life Center, Grant #DE-FG02-03ER6344) and the Wyss Institute for Biologically Inspired Engineering. Additional funding support comes from the Next-Generation BioGreen 21 Program (SSAC, Grant #PJ008109), Rural Development Administration, and from the Intelligent Synthetic Biology Center of Global Frontier Project funded by the Ministry of Education, Science and Technology (2011-0031956), Korea. Harris Wang is supported by the Wyss Institute Technology Development Fellowship and the National Institutes of Health Director’s Early Independence Award (Grant Number DP5OD009172). Jaehwan Jeong is supported by the National Junior research fellowship from the National Research Foundation of Korea (2012-0000391).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
STATEMENT OF RESPONSIBILITY
H.H.W., H.B.K., D.H.B. and G.M.C. designed the study. H.H.W,, H.B.K., L.C. and J.H.J performed the experiments. H.H.W., H.B.K. and D.H.B. analyzed the data and prepared the initial manuscript. All authors helped edit and revise the final manuscript.
Note: Supplementary information is available on the Nature Methods website.
References
- 1.Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467:167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang HH, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang HH, Church GM. Multiplexed genome engineering and genotyping methods applications for synthetic biology and metabolic engineering. Methods in enzymology. 2011;498:409–426. doi: 10.1016/B978-0-12-385120-8.00018-8. [DOI] [PubMed] [Google Scholar]
- 4.Costantino N, Court DL. Enhanced levels of lambda Red-mediated recombinants in mismatch repair mutants. Proc Natl Acad Sci U S A. 2003;100:15748–15753. doi: 10.1073/pnas.2434959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoessel R, et al. Indirubin, the active constituent of a Chinese antileukaemia medicine, inhibits cyclin-dependent kinases. Nat Cell Biol. 1999;1:60–67. doi: 10.1038/9035. [DOI] [PubMed] [Google Scholar]
- 6.Choi HS, et al. A novel flavin-containing monooxygenase from Methylophaga sp strain SK1 and its indigo synthesis in Escherichia coli. Biochem Biophys Res Commun. 2003;306:930–936. doi: 10.1016/s0006-291x(03)01087-8. [DOI] [PubMed] [Google Scholar]
- 7.Caligiuri MG, Bauerle R. Identification of amino acid residues involved in feedback regulation of the anthranilate synthase complex from Salmonella typhimurium. Evidence for an amino-terminal regulatory site. J Biol Chem. 1991;266:8328–8335. [PubMed] [Google Scholar]
- 8.Kikuchi Y, Tsujimoto K, Kurahashi O. Mutational analysis of the feedback sites of phenylalanine-sensitive 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase of Escherichia coli. Appl Environ Microbiol. 1997;63:761–762. doi: 10.1128/aem.63.2.761-762.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weaver LM, Herrmann KM. Cloning of an aroF allele encoding a tyrosine-insensitive 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase. J Bacteriol. 1990;172:6581–6584. doi: 10.1128/jb.172.11.6581-6584.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bongaerts J, Kramer M, Muller U, Raeven L, Wubbolts M. Metabolic engineering for microbial production of aromatic amino acids and derived compounds. Metab Eng. 2001;3:289–300. doi: 10.1006/mben.2001.0196. [DOI] [PubMed] [Google Scholar]
- 11.Carr P, Wang HH, Church GM. Co-selection of Oligonucleotides Accelerate Genome-scale Engineering. in revision 2012. [Google Scholar]
- 12.Temme K, Hill R, Segall-Shapiro TH, Voigt CA. Modular Control of Multiple Pathways using Engineered Orthogonal T7 Polymerases. Nucleic Acids Res. doi: 10.1093/nar/gks597. (submitted 2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swingle B, et al. Oligonucleotide recombination in Gram-negative bacteria. Mol Microbiol. 2010;75:138–148. doi: 10.1111/j.1365-2958.2009.06976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yokobayashi Y, Weiss R, Arnold FH. Directed evolution of a genetic circuit. Proc Natl Acad Sci U S A. 2002;99:16587–16591. doi: 10.1073/pnas.252535999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isaacs FJ, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Insertion design used to introduce T7 promoters upstream of genes/operons.
Supplementary Figure 2. Pigment quantification.
Supplementary Figure 3. Correlation of indigo production level and growth rate
Supplementary Figure 4. Genomic map and list of 21 available Co-Selection markers that can be used to enhance MAGE efficiency.
Supplementary Figure 5. Characterization of the insertion efficiency of various lengths
Supplementary Figure 6. Comparison of the length of time in experimental hours needed for different MAGE strategies to generate clones with 20bp T7 promoter insertions.
Supplementary Table 1. Summary of stages of CoS-MAGE to generate the H77 strain.
Supplementary Table 2. Summary of CoS Markers available in EcNR2 (MG1655 derivative)
Supplementary Table 3. Side-by-side comparison of various genome engineering strategies to make a strain that contains 20bp T7 insertion mutation across 10 genomic loci.
Supplementary Table 4. Oligo and primer sequences used in the study.
Supplementary Note. Co-selection mechanism and enhancement analysis