Abstract
Background
Common genetic variations influence rejection, infection, drug metabolism, and side effect profiles after pediatric heart transplantation. Reports in adults suggest that genetic background may influence post-transplant renal function. In this multicenter study we investigated the association of genetic polymorphisms (GP) in a panel of candidate genes on renal function in 453 pediatric heart transplant recipients.
Methods
We performed genotyping for functional GPs in 19 candidate genes. Renal function was determined annually after transplantation by calculation of glomerular filtration rate (eGFR). Mixed effects and Cox proportional hazard models were used to assess recipient characteristics and the effect of GPs on longitudinal eGFR and time to eGFR <60 mL/min/1.73m2.
Results
Mean age at transplantation was 6.2 ± 6.1 years and mean follow-up was 5.1 ± 2.5 years. Older age at transplant and black race were independently associated with post-transplant renal dysfunction. In univariate analyses, FASL (C-843T) T allele (p=0.014) and HO-1 (A326G) G allele (p=0.0017) were associated with decreased renal function. After adjusting for age and race, these associations were attenuated [FASL (p=0.075), HO-1 (p=0.053)]. We found no associations of other GPs, including GPs in TGFβ1, CYP3A5, ABCB1, and ACE, with post-transplant renal function.
Conclusions
In this multicenter, large sample of pediatric heart transplant recipients we found no strong associations between GPs in 19 candidate genes and post-transplant renal function. Our findings contradict reported associations of CYP3A5 and TGFβ1 with renal function and suggest that genotyping for these GPs will not facilitate individualized immunosuppression for the purpose of protecting renal function after pediatric heart transplantation.
INTRODUCTION
Renal dysfunction occurs commonly after solid organ transplantation [1–4] and severe renal disease has been associated with increased risks of death in both pediatric and adult heart transplant recipients [5–6]. Because of their well-established acute and chronic nephrotoxic effects, calcineurin inhibitors (CNI) have long been considered the primary culprit in post-transplant renal dysfunction [7–9]. However, despite the use of common immunosuppression protocols, not all recipients show deterioration of renal function at the same pace. Several other factors such as age at transplantation, hypertension, diabetes mellitus, and hyperlipidemia have been shown to contribute to renal dysfunction after heart transplantation [6], but these factors do not explain all the observed variation in renal function. To what extent genetic polymorphisms (GPs) account for some of the variability in renal outcomes after solid organ transplantation is unclear. In our previous work, we have shown that GPs influence the risks of acute rejection, rejection with hemodynamic compromise, infection, and immunosuppressant-related adverse effects after pediatric heart transplantation [10–13]. Associations of post-transplant renal function with polymorphisms of genes involved in inflammation, drug absorption, distribution and metabolism, and regulation of the renin-angiotensin-aldosterone system have been reported, mainly in adult transplant recipients [14–21]. In this multi-center prospective study we sought to explore associations between GPs in a panel of candidate genes and renal function after pediatric heart transplantation.
METHODS
Participants
This cohort consisted of pediatric heart transplant recipients from the six participating centers of the National Heart, Lung and Blood Institute-sponsored Specialized Centers for Clinically Oriented Research (SCCOR) program for “Optimizing Outcomes after Pediatric Heart Transplantation,” comprising the University of Pittsburgh, Stanford University, Loma Linda University, Washington University, Columbia University, and University of Alabama at Birmingham. All patients in the study also participated in the Pediatric Heart Transplantation Study (PHTS), an international, prospective, event-driven database of risk factors for outcomes after listing for heart transplantation [22]. Patients were ≤18 years of age at transplantation, received an allograft between January 1993 and December 2009, and had at least one annual post-transplant serum creatinine recorded for use in calculation of estimated glomerular filtration rate (eGFR). Between 2003 and 2008, past and current transplant recipients were enrolled in the SCCOR study to collect blood for genetic analysis after obtaining approval of the Institutional Review Boards of the participating centers and informed consent from parents or guardians. All patients were maintained on immunosuppressive regimens according to individual institutional protocols.
Data collection
All clinical data were prospectively collected and entered in the PHTS database. Patient characteristics (gender, race/ethnicity, age at transplantation, reason for transplant) were collected at enrollment. Clinical data (including serum creatinine and height) were recorded at transplantation and approximately annually thereafter. Detailed event data were collected at the time of pre-specified serious adverse events including rejection, infection, retransplantation and death. Due to sample size and observation time limitations, post-transplant follow-up of eGFR was censored at the 8-year follow-up for this analysis.
Detection of genetic polymorphisms
A sample of 3 to 6 ml anti-coagulated venous blood was obtained from each patient enrolled in the SCCOR study. DNA was extracted from peripheral blood mononuclear cells by a standard phenol/chloroform procedure for genotype analysis of 19 genetic polymorphisms (GPs) in 14 candidate genes that have either been associated with renal dysfunction after transplantation or are theorized to play a role in renal response to injury (Table 1). Genotypes were assessed by single specific primer-polymerase chain reaction (PCR) and/or sequencing, as previously described [23]. PCRs were performed by a 7900HT v Fast Real-Time PCR System instrument (Applied Biosystems, Foster City, CA), and data was processed by SDS 2.2.2 software (Applied Biosystems).
Table 1.
Candidate genes and polymorphisms explored for association with renal dysfunction after pediatric heart transplantation.
| Class | Gene | Polymorphism | SNP ID |
|---|---|---|---|
| Pharmacogenomic | |||
| CYP3A5 | *1/*3 | rs776746 | |
| ABCB1 | 2677 G>T(A) | rs2032582 | |
| 3435 C>T | rs1045642 | ||
| Cytokines, cytokine receptors and growth factors | |||
| TNFα | −308 G>A | rs1800629 | |
| TGFβ1 | +869 T>C | rs1800471 | |
| +915 C>G | rs1800471 | ||
| IL-6 | −174 G>C | rs1800795 | |
| IL-4 | −590 C>T | rs2243250 | |
| IL-1RN | VNTRs in intron 2 | rs380092 | |
| IL-10 | −1082 G>A | rs1800896 | |
| IL15RA | +21G>A | rs7913599 | |
| +5165T>A | rs3736863 | ||
| Other molecules | |||
| ACE | 287-bp insertion/deletion (intron 16) | rs4646994 | |
| HO-1 | −489 A>T | rs2071746 | |
| 326 A>G | rs17879828 | ||
| NOS2 | Ser608Leu | rs2297518 | |
| +38 C>G | rs10459953 | ||
| FAS | −670 A>G | rs71800682 | |
| FASL | −844 C>T | rs763110 | |
Primary outcome measure and statistical analysis
The primary outcome measure was change in eGFR after transplantation, which was calculated using the modified Schwartz formula [24]. In 10–20 instances at each annual follow-up time where height data was missing, an imputed height based on the cohort’s annual change in height was used.
Assessment of Hardy-Weinberg Equilibrium of genotypes was performed using the chi-square test (1 degree of freedom). Two statistical approaches were used to assess renal function after heart transplantation. Multivariable longitudinal linear mixed models were used to analyze the repeated annual eGFR outcome measures adjusting for post-transplant follow-up year and accounting for within-person correlation. Cox proportional hazard models were used to analyze the time to onset of renal dysfunction, defined as eGFR <60mL/min/1.73m2. This corresponds to the National Kidney Foundation’s chronic kidney disease stages III – V and is an indication to treat in an attempt to slow progression of renal disease, regardless of cause [25]. Because renal function at the time of heart transplantation is often abnormal as a result of low cardiac output, only patients with eGFR ≥60mL/min/1.73m2 at the one-year post-transplant assessment were included in the Cox models of post-transplant renal dysfunction. The effects of clinical characteristics (recipient sex, recipient race, age at transplant, CNI use) on renal function were examined. For each GP, models without and with adjustment for recipient sex, race, and age at transplant were created to estimate the effect of the level of exposure for each variant allele (absent, heterozygous, homozygous) on longitudinal eGFR values and time to renal dysfunction after HTx. In addition to assessments based on the ‘allelic content’ of the 19 GPs, we also tested pre-specified genotypes and haplotypes that have been reported to be associated with renal disease [19–20, 25–27] for association with post-transplant renal dysfunction. The genotypes and haplotypes are: IL1-RN allele 2 homozygote, TGFβ1 high producer (TGFβ1 codon 10/25 alleles TC/GG, TT/GG), high IL6/low IL4 producers (IL6/IL4 -590C>T alleles GG/CC), and high TNFα/low IL6/low IL4 producers (TNFα/IL6/IL4 -590C>T alleles AA/CC/CC, AA/CG/CC, GA/CC/CC, GA/CG/CC). Adjusted beta-coefficients for longitudinal eGFR and hazard ratios (HR) for renal dysfunction with 95% confidence intervals are reported.
All analyses were conducted based on a two-sided alpha of 0.05 and were performed using SAS 9.1 software (SAS Institute, Cary, NC). Based on our sample size and the overall observed event rate in the Cox models, we had 80% power to detect a hazard ratio of 1.52 for renal dysfunction for a polymorphism with 25% of patients in one group and 75% in the other. For the longitudinal analyses we had 80% power to detect an absolute difference of 10.8 ml/min/1.73m2 between gene polymorphism groups assuming the same genotype frequency (i.e. 25%:75%).
RESULTS
A total of 453 patients were included in the longitudinal analysis of eGFR following transplantation. Mean post-transplant follow-up was 5.1 ± 2.5 years and 135 of these patients had 8 year follow-up data. Mean age at HTx was 6.2 ± 6.1 years and 33% were <1 year old at HTx. The cohort was 58% male, 60% White, 13% Black, and 23% Hispanic. The majority of listing diagnoses were cardiomyopathy (50%) and congenital heart disease (45%). At each annual follow-up time nearly all patients (>98%)were taking a CNI. At 1 year post HTx, 56% of patients were taking cyclosporine and 43% tacrolimus and at 7 years post HTx this distribution was 57% and 43%.
Three hundred two patients were also evaluated using the survival analysis approach. Except for the IL4 -590 C>T polymorphism, there were no statistical differences in the level of exposure for each variant allele between patients included and excluded from Cox modeling (IL4 -590 CC genotype prevalence of included patients 58% vs. excluded 47%, p=0.026). Despite this, there were no differences in the level of exposure for any of the IL4 containing, pre-specified haplotypes between patients included and excluded from Cox modeling.
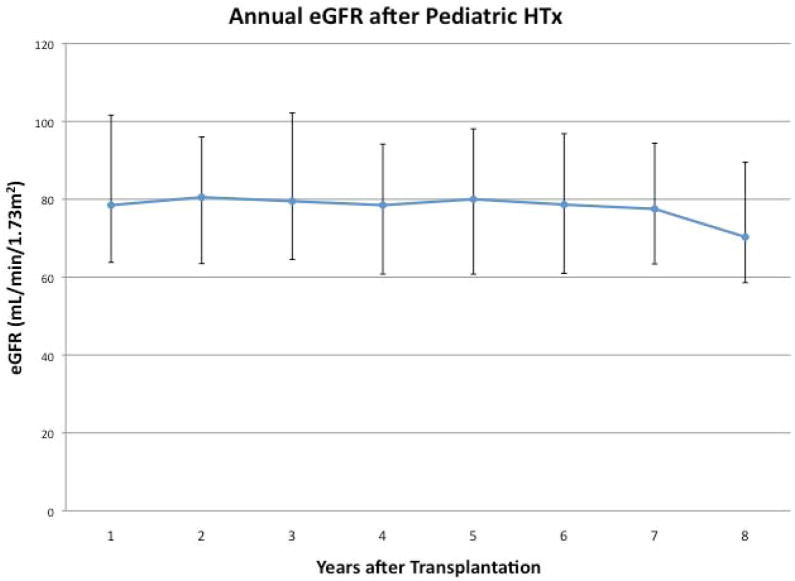
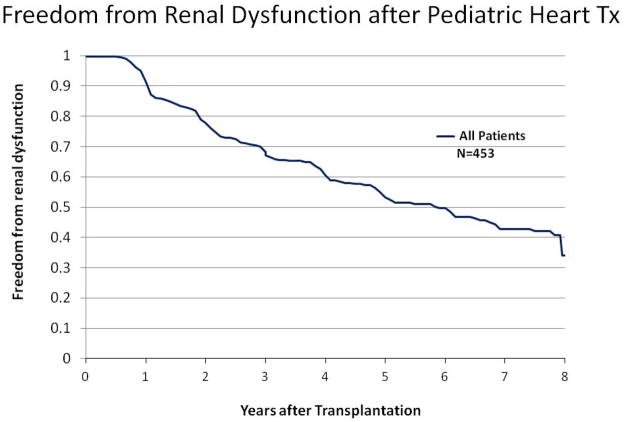
Figure 1 shows the mean eGFR annually post-HTx and freedom from eGFR <60 ml/min/1.73m2 among all patients, regardless of eGFR at HTx. Among clinical characteristics assessed, older age at transplant (p=0.02) and recipient Black race (p=0.038) were independently associated with lower eGFR following HTx).
Figure 1.
Figure 1a: Median and interquartile range eGFR for the cohort annually after transplantation.
Figure 1b: Time is measured as years after transplantation among all patients, including those who have eGFR<60 mL/min/1.73m2 on first post-transplant assessment.
The GPs of CYP3A5, IL4, IL6, and HO-1(326A>G) were found to have statistically significantly deviation from Hardy-Weinberg Equilibrium (p<0.05). Among the 19 GPs assessed, only FASL (-844C>T) T allele (log rank p=0.033, unadjusted beta for repeated measures annual eGFR p=0.014) and HO-1 (326A>G) G allele (log rank p=0.004, unadjusted beta p=0.169) were associated with decreased renal function after heart transplantation. Both alleles were observed more frequently in Blacks (p<0.0001). After adjusting for age at HTx, race, and sex, the associations with renal function were no longer significant (FASL adjusted HR=1.09 per T allele, 95% CI [0.80 – 1.48], p=0.58; adjusted beta for repeated measures eGFR p=0.075 and HO-1 adjusted HR=1.67 per G allele, 95% CI [1.00 – 2.82], p=0.053; adjusted beta for repeated measures eGFR p=0.754). We also found no associations between renal function and any of the pre-specified genotypes or haplotypes.
DISCUSSION
The development of chronic renal insufficiency is a significant event after transplantation that is associated with increased risk of death in adult and pediatric transplant recipients [5–6]. Because not everyone is at equal risk, the ability to identify risk factors would be of benefit by allowing for targeted interventions to minimize these adverse outcomes. Though clinical risk factors have been identified, they do not fully account for the variability observed in the development of renal dysfunction after solid organ transplantation. The discovery of genetically based risk factors should enable further explanation of the variability in risk and, more importantly, may allow for alterations in standard post-transplant care tailored to individual patients. This is the promise of personalized medicine and was the primary rationale for undertaking the current study.
Unfortunately our study did not find any strong genetic markers of post-transplant renal dysfunction among the GPs and pre-selected genotypes and haplotypes we examined. We did find weak associations of FASL -843C>T and HO-1 326A>G with renal dysfunction that did not persist in multivariable analysis. To our knowledge these associations have not previously been reported in any population, post-transplant or otherwise. Both FASL and HO-1 are expressed in the kidney and have been implicated in various experimental models of renal dysfunction [26–31]. However, no role or mechanism in post-transplant renal dysfunction has been described. It must also be considered that these associations may be spurious, arising as a result of the many univariate statistical comparisons made.
A handful of studies have looked at GPs as risk factors for renal dysfunction after solid organ transplantation. With regard to GPs in CYP3A5, our findings are consistent with those of Klauke [14] and contradict the reported associations by de Denus [21]. Our findings with regard to TGFβ1 also agree with Klauke [14] yet contradict those of others, including our own group’s single center study in which TGFB1 codon 10 and 25 polymorphisms were found to be associated with renal function after pediatric heart transplantation [15–17, 19]. While each of these studies differs to some extent methodologically or in sample size, our study’s findings are important because they represent the first multi-center, ethnically diverse study of this topic and the largest to date.
There are several limitations to our study. For the most part our analysis was performed on the basis of the level of exposure to each variant allele. Thus we would not uncover associations of haplotypes aside from the pre-specified haplotypes that we explored. Also four GPs were found to have statistically significantly deviation from Hardy-Weinberg equilibrium. This implies some ‘missingness’ of genotypes in our population due to chance, genotyping error, or ascertainment bias [32]. Of these reasons, ascertainment bias is most likely due possibly to the relatively small numbers of subjects in our study. Though we included all enrolled patients with appropriate data, our sample size was fixed by the study’s design - SCCOR cohort with accompanying PHTS clinical data. Our study is the largest of its kind in the transplant literature; however the fixed sample size may have lead us to falsely reject causality between GPs and post transplant renal function when in fact causality exists (i.e. type II error) [32].
The use of estimated rather than serially measured GFR may have underestimated the degree of renal dysfunction in this population [33]. While this could have hindered our ability to find associations based on the survival analysis approach, it should not have affected our ability to detect associations using the repeated measures approach. Finally it is important to note that only a few of the GPs we explored have been associated with renal dysfunction after solid organ transplantation (TGFβ1, CYP3A5, ABCB1, ACE). While many of the other GPs we explored have been associated with end-stage renal disease in non-transplant recipients (TNFα, IL-6, IL-4, IL-1RN, IL-10) or implicated to play a role in kidney injury (HO-1, NOS2, FAS, FASL, IL15RA), the lack of associations for these GPs in this exploratory analysis is not necessarily unexpected.
In summary, among 19 candidate GPs assessed in this large, multi-center, pediatric cohort, we found no strong associations with renal function after heart transplantation. Previously reported associations of TGFβ1 and CYP3A5 with post-transplant renal function were not observed. These data suggest that genotyping for these GPs is unlikely to facilitate individualized immunosuppression for the purpose of preserving renal function after pediatric heart transplantation.
Footnotes
DISCLOSURES
Dr. Feingold’s effort on this project was supported by the National Institutes of Health through Grant Numbers KL2 RR024154 and KL2TR000146. This project was also supported by the National Heart Lung Blood Institute, National Institutes of Health (5P50 HL 07432-05). The content is solely the responsibility of the authors and does not necessarily represent the views of the National Heart Lung Blood Institute or the National Institutes of Health.
No author has a financial interest or other potential conflict of interest related to subject matter or materials mentioned in the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wilkinson AH, Cohen DJ. Renal failure in the recipients of nonrenal solid organ transplants. J Am Soc Nephrol. 1999 May;10(5):1136–44. doi: 10.1681/ASN.V1051136. [DOI] [PubMed] [Google Scholar]
- 2.Filler G, Sharma AP. High prevalence of chronic kidney disease in pediatric solid organ transplantation. Pediatr Transplant. 2009 Feb;13(1):7–10. doi: 10.1111/j.1399-3046.2008.01077.x. [DOI] [PubMed] [Google Scholar]
- 3.Feingold B, Zheng J, Law YM, Morrow WR, Hoffman TM, Schechtman KB, Dipchand AI, Canter CE. Risk Factors for Late Renal Dysfunction after Pediatric Heart Transplantation: A Multi-institutional Study. Pediatr Transplant. 2011;15:699–705. doi: 10.1111/j.1399-3046.2011.01564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fisher NC, Nightingale PG, Gunson BK, Lipkin GW, Neuberger JM. Chronic renal failure following liver transplantation: a retrospective analysis. Transplantation. 1998;66:59–66. doi: 10.1097/00007890-199807150-00010. [DOI] [PubMed] [Google Scholar]
- 5.Lee Ck, Christensen LL, Magee JC, Ojo AO, Harmon WE, Bridges ND. Pre-transplant Risk Factors for Chronic Renal Dysfunction After Pediatric Heart Transplantation: A 10-Year National Cohort Study. J Heart Lung Transplant. 2007;26:458–65. doi: 10.1016/j.healun.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 6.Ojo AO, Held PJ, Port FK, Wolfe RA, Leichtman AB, Young EW, Arndorfer J, Christensen L, Merion RM. Chronic Renal Failure after Transplantation of a Nonrenal Organ. N Engl J Med. 2003;349:931–940. doi: 10.1056/NEJMoa021744. [DOI] [PubMed] [Google Scholar]
- 7.Naesens M, Kuypers DRJ, Sarwal M. Calcineurin Inhibitor Nephrotoxicity. Clin J Am Soc Nephrol. 2009;4:481–508. doi: 10.2215/CJN.04800908. doi: 10.2215. [DOI] [PubMed] [Google Scholar]
- 8.Myers B, Ross J, Newton L, Luetscher J, Perlroth M. Cyclosporine associated chronic nephropathy. N Engl J Med. 1984;311:699–705. doi: 10.1056/NEJM198409133111103. [DOI] [PubMed] [Google Scholar]
- 9.Ader J-L, Rostaing L. Cyclosporin nephrotoxicity: pathophysiology and comparison with FK-506. Curr Opin Nephrol Hypertens. 1998;7:539–45. doi: 10.1097/00041552-199809000-00009. [DOI] [PubMed] [Google Scholar]
- 10.Girnita DM, Brooks MM, Webber SA, et al. Genetic polymorphisms impact the risk of acute rejection in pediatric heart transplantation: a multi-institutional study. Transplantation. 2008;85:1632. doi: 10.1097/TP.0b013e3181722edc. [DOI] [PubMed] [Google Scholar]
- 11.Ohmann EL, Burckart GJ, Brooks MM, Chen Y, Pravica V, Girnita DM, Zeevi A, Webber SA. Genetic polymorphisms influence mycophenonlate mofetil–related adverse events in pediatric heart transplant patients. J Heart Lung Transplant. 2010;29:509–516. doi: 10.1016/j.healun.2009.11.602. [DOI] [PubMed] [Google Scholar]
- 12.Ohmann EL, Brooks MM, Webber SA, Girnita DM, Ferrell RE, Burckart GJ, Chinnock R, Canter C, Addonizio L, Bernstein D, Kirklin JK, Naftel DC, Zeevi A. Association of genetic polymorphisms and risk of late post-transplantation infection in pediatric heart recipients. J Heart Lung Transplant. 2010;29:1342–51. doi: 10.1016/j.healun.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 13.Girnita DM, Ohmann EL, Brooks MM, Webber SA, Burckart GJ, Ferrell RE, Ranganathan S, Chinnock R, Canter C, Addonizio L, Bernstein D, Kirklin JK, Naftel DC, Zeevi A. Gene Polymorphisms Impact the Risk of Rejection With Hemodynamic Compromise: A Multicenter Study. Transplantation. 2011;91(12):1326–32. doi: 10.1097/TP.0b013e31821c1e10. [DOI] [PubMed] [Google Scholar]
- 14.Klauke B, Wirth A, Zittermann A, et al. No association between single nucleotide polymorphisms and the development of nephrotoxicity after orthotopic heart transplantation. J Heart Lung Transplant. 2008;27:741–5. doi: 10.1016/j.healun.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Lacha J, Hubacek JA, Viklicky O, Malek I, Hutchinson I, Vitko S. TGF- β1 gene polymorphism is a risk factor for renal dysfunction in heart transplant recipients. Transplant Proc. 2001;33:1567–9. doi: 10.1016/s0041-1345(00)02596-3. [DOI] [PubMed] [Google Scholar]
- 16.Baan CC, Balk AHMM, Holweg CTJ, Van Riemsdijk IC, Maat LP, Vantrimpont PJ, Niesters HG, Weimar W. Renal failure after clinical heart transplantation is associated with the TGF-β1 codon 10 gene polymorphism. J Heart Lung Transplant. 2000;19:866–72. doi: 10.1016/s1053-2498(00)00155-8. [DOI] [PubMed] [Google Scholar]
- 17.van de Wetering J, Weimar CHE, Balk AHMM, Roodnat JI, Holweg CTJ, Baan CC, van Domburg RT, Weimar W. The Impact of Transforming Growth Factor- β1 Gene Polymorphism on End-Stage Renal Failure After Heart Transplantation. Transplantation. 2006;82:1744–8. doi: 10.1097/01.tp.0000250360.78553.5e. [DOI] [PubMed] [Google Scholar]
- 18.Zhang W, Chen B, Zhang W, Chen N, Yu Z, Cai W. Effect of MDR1 gene polymorphism on progression of end-stage renal disease. Acta Pharmacol Sin. 2007;28(4):579–583. doi: 10.1111/j.1745-7254.2007.00517.x. [DOI] [PubMed] [Google Scholar]
- 19.Di Filippo S, Zeevi A, McDade KK, Boyle GJ, Miller SA, Gandhi SK, Webber SA. Impact of TGFβ1 Gene Polymorphisms on Late Renal Function in Pediatric Heart Transplantation. Human Immunol. 2005;66:133–9. doi: 10.1016/j.humimm.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 20.Büscher R, Nagel D, Finkelberg I, Büscher AK, Wingen A, Kranz B, Vester U, Hoyer PF. Donor and recipient ACE I/D genotype are associated with loss of renal function in children following renal transplantation. Pediatric Transplant. 2011;15(2):214–20. doi: 10.1111/j.1399-3046.2010.01449.x. [DOI] [PubMed] [Google Scholar]
- 21.de Denus S, Zakrzewski M, Barhdadi A, Leblanc M, Racine N, Bélanger F, Carrier M, Ducharme A, Dubé M, Turgeon J, White M. Association between renal function and CYP3A5 genotype in heart transplant recipients treated with calcineurin inhibitors. J Heart Lung Transplant. 2011;30:326–31. doi: 10.1016/j.healun.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 22.Hsu DT, Naftel DC, Webber SA, et al. Lessons learned from the pediatric heart transplant study. Congenit Heart Dis. 2006;1:54–62. doi: 10.1111/j.1747-0803.2006.00011.x. [DOI] [PubMed] [Google Scholar]
- 23.Girnita DM, Webber SA, Ferrell R, et al. Disparate distribution of 16 candidate single nucleotide polymorphisms among racial and ethnic groups of pediatric heart transplant patients. Transplantation. 2006;82:1774–80. doi: 10.1097/01.tp.0000250656.33731.08. [DOI] [PubMed] [Google Scholar]
- 24.Schwartz GJ, Muñoz A, Schneider MF, et al. New equations to estimate GFR in children with CKD. J Am Soc Nephrol. 2009;20(3):629–37. doi: 10.1681/ASN.2008030287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.National kidney foundation. K/DOQI clinical practice guidelines for chronic kidney disease: Evaluation, classification and stratification. Am J Kidney Dis. 2002;39:S1–S266. [PubMed] [Google Scholar]
- 26.Linkermann A, Himmerkus N, Rölver L, Keyser KA, Steen P, Bräsen JH, Bleich M, Kunzendorf U, Krautwald S. Renal tubular Fas ligand mediates fratricide in cisplatin-induced acute kidney failure. Kidney Int. 2011;79(2):169–78. doi: 10.1038/ki.2010.317. [DOI] [PubMed] [Google Scholar]
- 27.Lorz C, Ortiz A, Justo P, et al. Proapoptotic Fas ligand is expressed by normal kidney tubular epithelium and injured glomeruli. J Am Soc Nephrol. 2000;11:1266–77. doi: 10.1681/ASN.V1171266. [DOI] [PubMed] [Google Scholar]
- 28.Ortiz A, Lorz C, Egido J. The Fas ligand/Fas system in renal injury. Nephrol Dial Transplant. 1999;14:1831–34. doi: 10.1093/ndt/14.8.1831. [DOI] [PubMed] [Google Scholar]
- 29.Agarwal A, Nick HS. Renal Response to Tissue Injury: Lessons from Heme Oxygenase-1 Gene Ablation and Expression. J Am Soc Nephrol. 2000;11:965–73. doi: 10.1681/ASN.V115965. [DOI] [PubMed] [Google Scholar]
- 30.Agarwal A, Balla J, Alam J, Croatt AJ, Nath KA. Induction of heme oxygenase in toxic renal injury: A protective role in cisplatin nephrotoxicity in the rat. Kidney Int. 1995;48:1298–1307. doi: 10.1038/ki.1995.414. [DOI] [PubMed] [Google Scholar]
- 31.Agarwal A, Kim Y, Matas AJ, Alam J, Nath KA. Gas-generating systems in acute renal allograft rejection in the rat: Co-induction of heme oxygenase and nitric oxide synthase. Transplantation. 1996;61:93–8. doi: 10.1097/00007890-199601150-00019. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez S, Gaunt TR, Day INM. Hardy-Weinberg Equilibrium Testing of Biological Ascertainment for Mendelian Randomization Studies. Am J Epidemol. 2009;169:505–514. doi: 10.1093/aje/kwn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bharat W, Manlhiot C, McCrindle BW, Pollock-BarZiv S, Dipchand AI. The profile of renal function over time in a cohort of pediatric heart transplant recipients. Pediatr Transplant. 2009;13:111–18. doi: 10.1111/j.1399-3046.2007.00848.x. [DOI] [PubMed] [Google Scholar]