Abstract
Background
Effective gene transfection without serum deprivation is a prerequisite for successful stem cell-based gene therapy. Polyethylenimine (PEI) is an efficient nonviral gene vector, but its application has been hindered by serum sensitivity and severe cytotoxicity.
Methods
To solve this problem, a new family of lipopolyplexes was developed by coating PEI/DNA polyplexes with three serum-resistant cationic lipids, namely, lysinylated, histidylated, and arginylated cholesterol. The physical properties, transfection efficiency, cellular uptake, subcellular distribution, and cytotoxicity of the lipopolyplexes was investigated.
Results
The outer coat composed of lysinylated or histidylated cholesterol remarkably improved the transfection efficiency of the polyplex with a low PEI/DNA ratio of 2 in the presence of serum. The resulting lysinylated and histidylated cholesterol lipopolyplexes were even more efficient than the best performing polyplex with a high PEI/DNA ratio of 10. Results from cellular uptake and subcellular distribution studies suggest that their higher transfection efficiency may result from accelerated DNA nuclear localization. The superiority of the lipopolyplexes over the best performing polyplex was also confirmed by delivering the therapeutic gene, hVEGF165. Equally importantly, the lipid coating removed the necessity of introducing excess free PEI chains into the transfection solution for higher efficiency, generating lipopolyplexes with no signs of cytotoxicity.
Conclusion
Noncovalent modification of polyplexes with lysinylated and histidylated cholesterol lipids can simultaneously improve efficiency and reduce the toxicity of gene delivery under serum conditions, showing great promise for genetic modification of bone marrow stem cells.
Keywords: gene delivery, nonviral vectors, cationic lipids, polyethylenimine, lipopolyplex, bone marrow stem cells
Introduction
Stem cell-mediated gene therapy has emerged as an attractive strategy for the treatment of cardiovascular diseases, skeletal disorders, ischemic damage, and cancer.1 One of several types of stem cells, mesenchymal stem cells derived from bone marrow (BMSCs) have been extensively investigated because of their attractive characteristics of easy accessibility, low immunogenicity, capacity for self-renewal, and freedom from ethical issues.2
The first step in BMSC-mediated gene therapy is genetic modification of cells, which requires a gene delivery vector to achieve introduction of foreign DNA. Nonviral vectors, such as cationic polymers and cationic lipids, have become a promising alternative to viral vectors by offering a better safety profile, a larger cargo capacity, and easier handling.3–5 Among the cationic polymers, polyethylenimine (PEI) is one of the most successful and widely studied gene carriers. PEI can effectively condense DNA to form nanoparticles6 and promote endosomal escape through a so-called “proton sponge” effect.7,8 Moreover, compared with other commercially available transfection vectors, PEI is very stable, easy to manipulate, and inexpensive.9 However, use of PEI for gene delivery has encountered an efficiency-cytotoxicity dilemma. Optimal PEI performance often requires the presence of excessive PEI chains in the transfection solution, together with serum-free conditions to avoid destabilization of the polyplexes by negatively charged serum proteins.10,11 However, a large amount of uncomplexed PEI chains can cause significant cytotoxicity due to cellular membrane damage.12–14 In addition, serum deprivation is known to induce apoptosis of BMSCs,15 which will hamper their therapeutic applications in vivo. In this context, chemical modifications, such as grafting PEI with polyethylene glycol,16 hyaluronic acid,17 and alginate18 have been investigated to improve gene delivery into BMSCs. However, these modifications have potential disadvantages in terms of technical complexity and poor availability to BMSC-based gene therapy.
Noncovalent modification of PEI/DNA polyplexes with liposomes to form lipopolyplexes has been proposed as a simple and useful strategy to overcome some of the drawbacks of PEI. PEI/DNA polyplexes in combination with liposomes containing DOCSPER,19 DOSPER,19 DOTAP, 20 DOTMA,21 DPPC,22 or DPPG 22 have been shown to improve the transfection of immortal cell lines under serum conditions. Several parameters, in particular the lipid structure and vector/DNA molar ratio, were suggested to be factors influencing the transfection behavior of the lipopolyplexes. Synergism between PEI and lipids in gene delivery has been proposed, whereby PEI improves intracellular trafficking of DNA while lipids promote cellular uptake of the complexes.20,23,24 Despite these positive results, there have been few studies of lipopolyplex-mediated gene delivery into BMSCs, which are inherently more difficult to transfect than immortal cell lines.
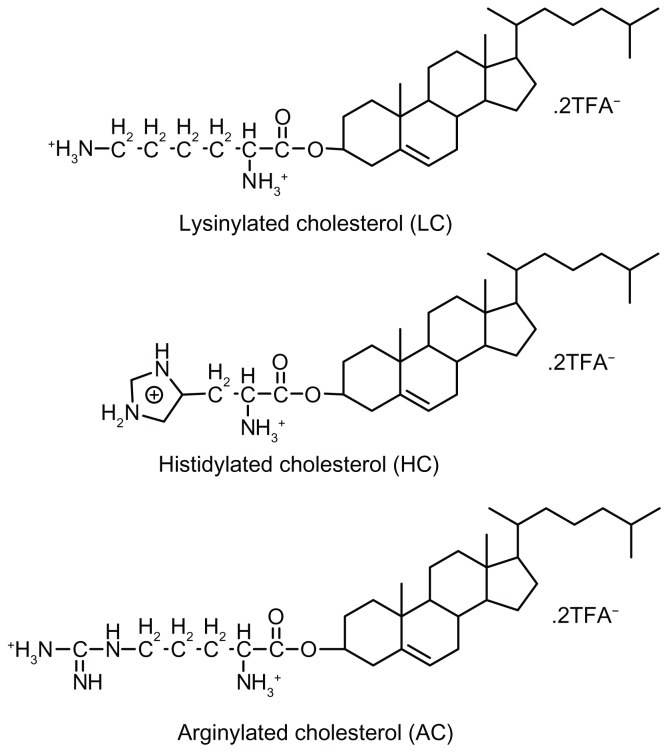
In previous studies, we have reported the synthesis and characterization of three alkaline amino acid-based cationic lipids, ie, lysinylated cholesterol (LC), histidylated cholesterol (HC), and arginylated cholesterol (AC) (Figure 1).25,26 Cationic lipoplexes prepared from these lipids, especially LC and AC, have shown high levels of serum resistance and low toxicity in the transfection of 293T, HeLa, and NIH3T3 cell lines. The present study investigated whether modification of PEI/DNA polyplexes with these lipids could simultaneously improve the efficiency and safety of gene delivery into BMSCs under serum conditions. Parameters including lipid structure and lipid/DNA and PEI/DNA molar ratios were evaluated to determine the optimal conditions for lipopolyplex-mediated gene delivery. Cellular uptake and subcellular localization studies were conducted to explore possible mechanisms underlying the superior performance of certain lipopolyplexes. These lipopolyplexes were further evaluated for cytotoxicity and potential for vascular endothelial growth factor (VEGF) gene delivery.
Figure 1.
Chemical structures of amino acid-based cationic lipids used in this study.
Materials and methods
Chemicals
Amino acid-based lipids including LC, HC, and AC were synthesized as described previously.26 Branched PEI (25 kDa) was purchased from Sigma (St Louis, MO) and was dissolved in 5% glucose at a concentration of 1 mg/mL (stock solution). 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) ammonium salt (Rhod- DOPE) were purchased from Avanti Polar Lipids (Alabaster, AL). Fluorescein isothiocyanate was obtained from Merck (Darmstadt, Germany). The fluorescent dyes, YOYO-1 and TOTO-3, were obtained from Invitrogen (Carlsbad, CA).
Plasmid construction and preparation
The firefly luciferase (Luc) reporter gene from pGL3 (Promega, Madison, WI) was cloned into pcDNA3.1 (Invitrogen) between the NotI and XbaI sites, generating pcDNA-Luc. Plasmid pcDNA-hVEGF165 was constructed by inserting the full length human VEGF165 cDNA (GenBank AB021221.1) into pcDNA3.1 between the NheI and EcoRI sites. The nucleotide sequence was confirmed by DNA sequencing. Plasmids were amplified in Escherichia coli DH5α and extracted using an endotoxin-free plasmid purification kit (Qiagen, Hilden, Germany). The DNA concentration was measured at an absorbance of 260 nm and adjusted to 1 mg/mL in 5% glucose. In some experiments, pcDNA-Luc was labeled with YOYO-1 or TOTO-3 according to the manufacturer’s protocol.
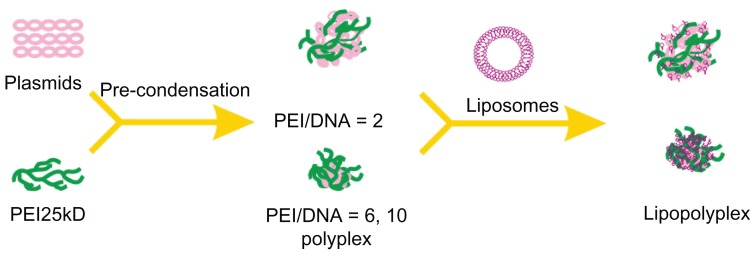
Preparation of polyplexes, lipoplexes, and lipopolyplexes
Polyplexes were prepared by adding DNA solutions to PEI solutions at PEI nitrogen/DNA phosphate ratios (hereinafter referred to as the PEI/DNA ratio) of 2, 6, and 10, respectively. The mixtures were incubated for 15 minutes at room temperature before use. LC, HC, and AC liposomes were prepared as described previously.26 Lipoplexes were obtained by gently mixing liposome solutions and DNA solutions at cationic lipid nitrogen/DNA phosphate ratios (hereinafter referred to as the lipid/DNA ratio) of 2.7 and 5.3, respectively. To obtain lipopolyplexes, polyplexes with various PEI/DNA ratios were first prepared and incubated as described above. The resultant polyplex solutions were then pipetted into each of the three liposome solutions at lipid/DNA ratios of 2.7 and 5.3, respectively. The ternary complexes were incubated for a further 15 minutes at room temperature before use.
Size and zeta potential measurements
The average size and zeta potential of the complexes was measured by dynamic light scattering using a Zetasizer NS (Malvern Instruments, Worcestershire, UK) at 25°C. Next, 50 μL of various complex solutions (containing 1 μg of pcDNA-Luc) were prepared. After 15 minutes of incubation, the complex solutions were diluted with double-distilled water to a final volume of 1 mL and measured. To determine the average size and zeta potential in the presence of 10% serum, the complex solutions were mixed with an equal volume of prefiltered (0.22 μm) Alpha Minimal Essential Medium (α-MEM; Gibco-Invitrogen, Carlsbad, CA) containing 20% fetal bovine serum (Hyclone, Logan, UT). The mixture was incubated for another 15 minutes, and diluted with double-distilled water to a final volume of 1 mL and measured.
Transmission electron microscopy
A 20 μL drop of freshly prepared lipopolyplexes was placed on a carbon-coated copper grid and air-dried for 5 minutes. The rest of the sample on the grid was absorbed with paper and dried by air. After staining with phosphotungstic acid, samples were visualized with a JEM-100SX electron microscope (JEOL, Akishima, Japan) operating at 100 kV.
Atomic force microscopy
The samples were prepared by placing 10 μL of lipopolyplex solution on freshly cleaved mica. After 5 minutes, the mica disk was gently rinsed by dipping once into double-distilled water and the excess water was removed using filter paper. After drying at room temperature, the samples were visualized by atomic force microscopy (Asylum Research MFP-3D-Bio, Santa Barbara, CA).
Isolation and expansion of BMSCs
Animal care and experimental procedures were performed according to the Guidelines for Animal Experimentation in China. The BMSCs were isolated and cultured as previously described.27 Briefly, bone marrow cavities from Sprague-Dawley rats (6 weeks old, West China Animal Culture Center of Sichuan University) were flushed with growth medium composed of α-MEM plus 10% fetal bovine serum, 2 mM L-glutamine, and 1% penicillin-streptomycin (Gibco-Invitrogen). Cells were passed through a 70 μm strainer and then plated in 100 mm culture dishes with growth medium (passage 0). After 3 days of culture, nonadherent cells were removed by a change of medium, and thereafter the medium was changed on alternate days. Cells were subcultured at approximately 80% confluence by trypsinization and those at passage 2–3 were used for this study.
Transfection experiments with luciferase-encoding plasmids
BMSCs were seeded into a 24-well plate at a density of 1 × 104 cells/cm2. After 24 hours of incubation, the cells were transfected with various complexes (0.8 μg of DNA per well). The cells were harvested in lysis buffer 48 hours after transfection. Luciferase activity was quantified using a luciferase assay kit (Promega) according to the given protocol. Briefly, the cell lysate was first centrifuged at 12,000 × g for 5 minutes at 4°C. Then, 100 μL of luciferase substrate was added to 20 μL of lysate supernatant and light emission was measured for 10 seconds using a Varioskan Flash microplate reader (Thermo Fisher Scientific, Waltham, MA). The protein concentration of the supernatant was measured using a BCA protein assay kit (Pierce, Rockford, IL). Luciferase activity was expressed as relative light units per mg of protein.
Cellular uptake assays
Cellular uptake of DNA was investigated by flow cytometry. Briefly, complexes containing YOYO-1-labeled DNA were added to subconfluent cultures of BMSCs in 100 mm plates (18 μg of DNA per plate). After incubation for 4 hours, the cells were trypsinized and washed with phosphate-buffered solution three times to remove nonspecific extracellular fluorescent probes. The cells were then suspended in phosphate- buffered solution and analyzed for fluorescence on a FACSAria flow cytometer (BD Biosciences, San Jose, CA) with 488 nm excitation. The fluorescence of 10,000 single cells was measured for each sample.
Confocal laser scanning microscopy
DNA, PEI, and lipids were respectively labeled with TOTO-3, fluorescein isothiocyanate,28 and Rhod-DOPE (1% molar ratio in liposome formulation). BMSCs grown in 35 mm glass-bottomed dishes were treated with various complexes (3 μg of DNA per dish). After incubation for 4 hours at 37°C, the treatment medium was removed, and the cells were washed three times with phosphate-buffered solution and incubated with Hoechst 33258 (Beyotime Institute of Biotechnology, Jiangsu China) for 30 minutes to stain nuclei. Images were obtained using a confocal laser scanning microscope (TCP SP5, Leica, Solms, Germany).
Cytotoxicity assays
Cytotoxicity was evaluated using a Cell Counting Kit-8 (CCK-8, Dojindo, Kumamoto, Japan). BMSCs were seeded at a density of 1 × 104 cells/cm2 in 96-well plates with growth medium 24 hours prior to treatment. The complexes (0.2 μg of DNA per well) with various lipid/PEI/DNA ratios were added into the wells. After 4 hours, the transfection medium was replaced with fresh growth medium and the cells were cultured for another 48 hours. Then, 10 μL of CCK-8 solution was added to each well and the plates were incubated at 37°C for 2 hours. Absorbance was measured at a wavelength of 450 nm and a reference wavelength of 630 nm, using a Varioskan Flash microplate reader.
Transfection experiments with hVEGF165-encoding plasmids
BMSCs were plated at a density of 1.0 × 104 cells/cm2 in 12-well plates and treated with the complexes (1.6 μg of DNA per well) 24 hours after plating. After incubation for 4 hours, the medium was replaced with 1 mL of fresh growth medium and the cells were cultured for another 48 hours. Total RNA was extracted from the cells using Trizol reagent ( Invitrogen) and treated with RNase-free Dnase I (MBI-Fermentas, St Leon-Rot, Germany) prior to cDNA synthesis to eliminate any residual plasmid or genomic DNA. Quantity and quality of RNA samples were evaluated by spectrophotometry and agarose electrophoresis. cDNA was synthesized from 2 μg of pure total RNA using RevertAid™ M-MuLV reverse transcriptase (MBI-Fermentas). Real-time polymerase chain reaction (PCR) was performed on a C1000™ Thermal Cycler (CFX 96™ Real-Time System, BioRad, Munich, Germany), using SYBR Premix Ex Taq™ II (Takara, Dalian, China). The sequences of the primers were as follows: hVEGF165, 5′-AGGAGGGCAGAATCATCACGAA-3′ (forward) and 5′-TGGTGATGTTGGACTCCTCAGT-3′ (reverse); β-actin, 5′-CTCCTCCCTGGAGAAGAGCTA-3′ (forward) and 5′-CCTTCTGCATCCTGTCGGCAA-3′ (reverse). The specificity of amplification was verified by melt-curve analysis and agarose gel electrophoresis. Relative gene expressions were calculated using the 2−Δ(ΔCt) method, where ΔCt = Ct (hVEGF165) − Ct (β-actin) and Δ(ΔCt) = ΔCt (sample) − ΔCt (control).29
To assess VEGF protein secretion, the culture medium was collected from each well 48 hours after transfection and diluted with α-MEM to a final volume of 2 mL. The medium was then centrifuged to remove debris and analyzed in triplicate for quantification of VEGF content using a RayBio® human VEGF enzyme-linked immunosorbent assay kit (RayBiotech, Norcross, GA), according to the manufacturer’s instructions. Enzyme-linked immunosorbent assay data were obtained using the Varioskan Flash microplate reader at a wavelength of 450 nm. Results were then normalized for the number of cells and time in culture.30
Statistical analysis
Quantitative data were expressed as the mean ± standard deviation. Comparisons among groups were made using the Student’s t-test assuming two-tailed distribution and unequal variances. A P value < 0.05 was considered to be statistically significant.
Results
Physicochemical properties of polyplexes and lipopolyplexes
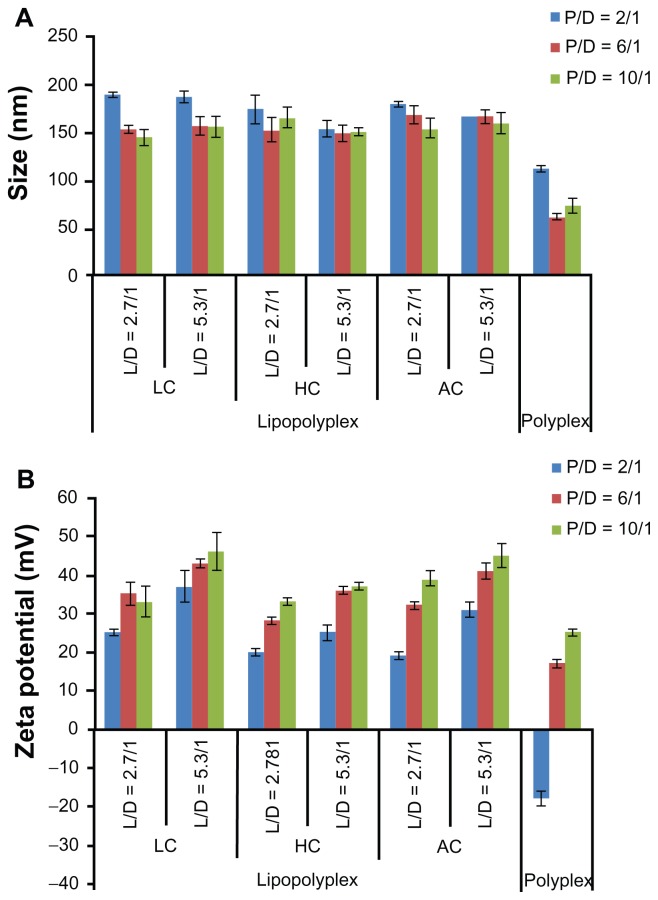
The physical properties of the polyplexes and lipopolyplexes were evaluated by dynamic light scattering. The particle size and zeta potential of polyplexes prepared at a PEI/DNA ratio of 2 were 112 ± 3 nm (Figure 2A) and −18 ± 2 mV (Figure 2B), respectively. The negative zeta potential suggested that the DNA molecules were insufficiently condensed by PEI, with part of them still exposed on the surface of the complexes. With the increase in the PEI/DNA ratio, the size of the polyplexes decreased to around 70 nm and the zeta potential increased to 25 mV, indicating complete DNA complexation.
Figure 2.
Average particle size (A) and zeta potential (B) of various lipopolyplexes and corresponding polyplexes in the absence of serum.
Note: Data are expressed as the mean ± standard deviation (n = 3).
Abbreviations: LC, lysinylated cholesterol; HC, histidylated cholesterol; AC, arginylated cholesterol; L, lipid; P, polyethylenimine; D, DNA.
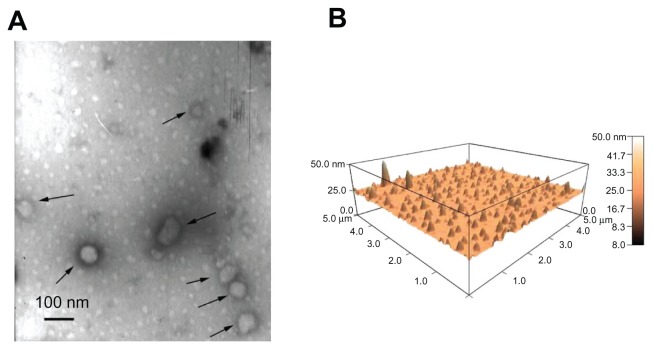
The diameter of the three lipopolyplexes was in a narrow range of 145–190 nm (Figure 2A), which was 1.5-fold to 2.5-fold larger than that of the corresponding polyplexes. The zeta potential of the lipopolyplexes generally increased with increasing lipid/DNA and PEI/DNA ratios, and was higher than that of the corresponding polyplexes (Figure 2B). Moreover, it was found that HC lipopolyplexes had a lower zeta potential than the LC and AC lipopolyplexes, possibly due to the weak alkaline nature of the imidazole group on HC. Images obtained by transmission electron microscopy (Figure 3A) and atomic force microscopy (Figure 3B) showed that all the lipopolyplexes were well dispersed spherical vehicles. As a result of sample shrinkage during drying, the diameter (around 100 nm) was relatively smaller than that obtained by dynamic light scattering.
Figure 3.
Morphology of lipopolyplexes. Representative images of lipopolyplexes visualized by transmission electron microscopy (A) (black arrows) and atomic force microscopy (B) in the absence of serum.
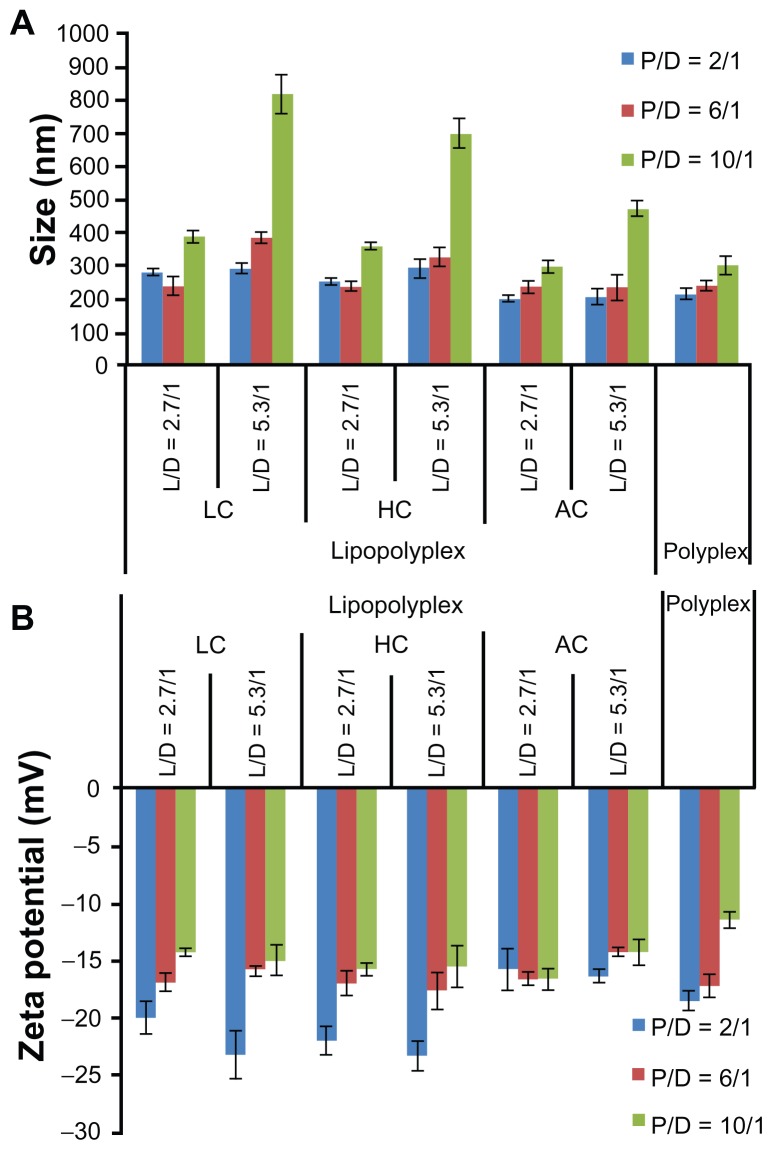
After incubation with 10% fetal bovine serum, most of the lipopolyplexes showed only moderately increased particle size (200–400 nm, Figure 4A) and a reversed zeta potential (Figure 4B). These results suggest that the lipopolyplexes attracted negatively charged proteins onto their surface, while being stable against aggregation under serum conditions. The particle size of the lipopolyplexes with a lipid/PEI/DNA ratio of 5.3/10/1 and polyplexes with a PEI/DNA ratio of 6 and 10 increased by approximately 3.0–5.5-fold, indicating some degree of particle aggregation induced by serum exposure.
Figure 4.
Average particle size (A) and zeta potential (B) of various lipopolyplexes and the corresponding polyplexes after incubation with serum.
Notes: Particle size and zeta potential of the complexes was determined by dynamic light scattering after incubation in 10% fetal bovine serum for 15 minutes. Data are expressed as the mean ± standard deviation (n = 3).
Abbreviations: LC, lysinylated cholesterol; HC, histidylated cholesterol; AC, arginylated cholesterol; L, lipid; P, polyethylenimine; D, DNA.
Transfection studies with plasmids encoding luciferase
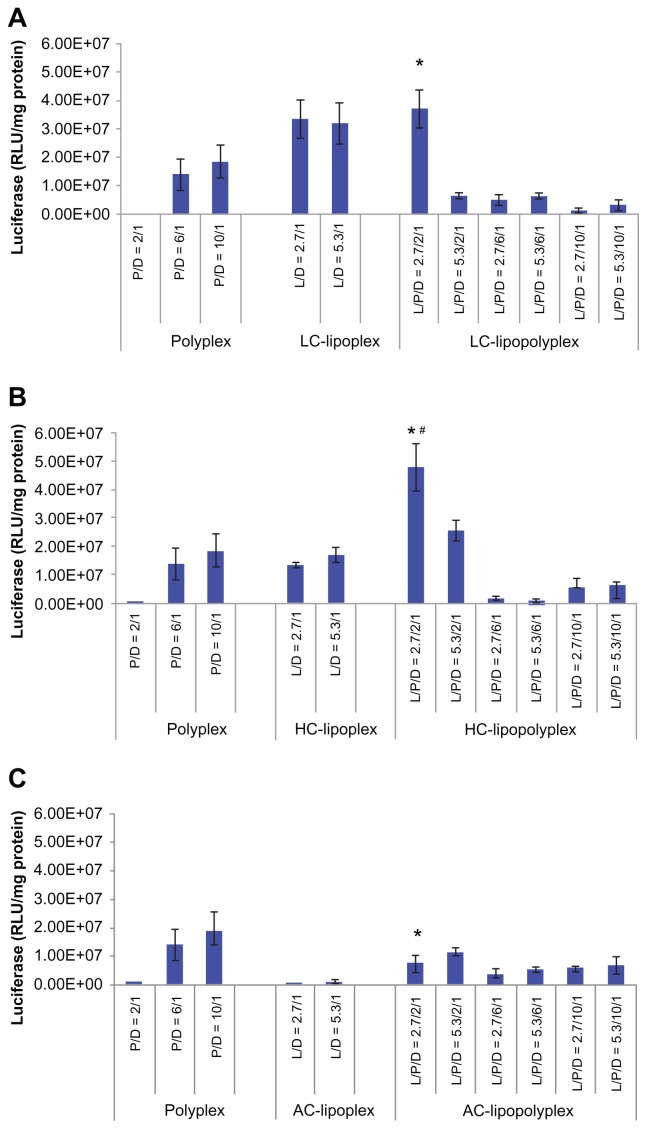
Transfection of BMSCs with pcDNA-Luc was carried out in culture medium containing 10% fetal bovine serum to determine the transfection efficiency of the various complexes. The negatively charged low PEI/DNA ratio polyplex (PEI/DNA = 2) showed almost no transfection activity (Figure 5). By combining it with LC or HC at a lipid/DNA ratio of 2.7, the transfection efficiency was significantly improved by three orders of magnitude (Figure 5A and B), generating LC and HC lipopolyplexes that were 2.0 and 2.6 times more efficient than the commonly used high PEI/DNA ratio polyplex (PEI/DNA = 10, P <0.05), respectively. The LC lipopolyplex was slight more efficient than the corresponding LC lipoplex, but the difference was not statistically significant (P > 0.05). In contrast, the transfection efficiency of the HC lipopolyplex was higher than the combined value of the corresponding polyplex and lipoplex (P < 0.05), indicating that HC acted in synergy with PEI in facilitating transfection at the lipid/PEI/DNA ratio of 2.7/2/1. However, it is worth noting that the transfection activity of both the LC and HC lipopolyplexes dropped dramatically when the PEI/DNA ratio was higher than 2 or the lipid/DNA ratio was higher than 2.7. Although a synergistic effect on gene delivery was also observed between PEI and AC at the lipid/PEI/DNA ratio of 2.7/2/1, the AC lipopolyplex showed significantly lower transfection efficiency than the high PEI/DNA ratio polyplex (P < 0.05). This result suggests that lipid chemistry exerted a significant influence on gene delivery into BMSCs by lipopolyplexes.
Figure 5.
Transfection efficiencies of LC lipopolyplexes (A), HC lipopolyplexes (B), and AC lipopolyplexes (C), and the corresponding lipoplexes and polyplexes in bone marrow stem cells.
Notes: Complexes were prepared at the indicated PEI/DNA and lipid/DNA ratios. Bone marrow stem cells were incubated for 4 hours with complexes in medium containing 10% fetal bovine serum. Forty-eight hours after transfection, the cells were lysed for quantification of luciferase activity. Data are shown as the mean ± standard deviation (n = 3, *P < 0.05 versus polyplex with a PEI/DNA ratio of 10; #P < 0.05 versus HC lipoplex with the lipid/DNA ratio of 2.7).
Abbreviations: LC, lysinylated cholesterol; HC, histidylated cholesterol; AC, arginylated cholesterol; L, lipid; P, polyethylenimine; D, DNA.
Cellular uptake and subcellular distribution of DNA
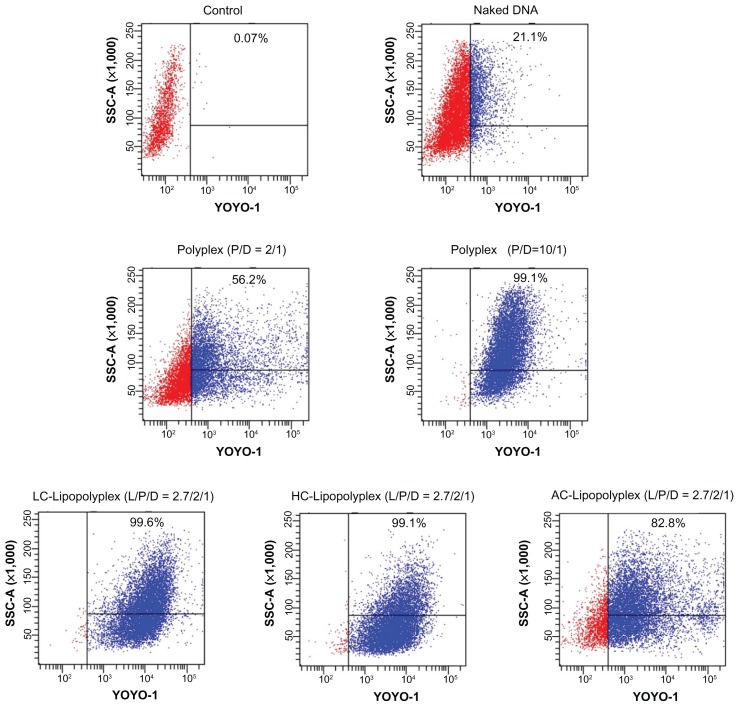
As described above, LC and HC lipopolyplexes with a lipid/PEI/DNA ratio of 2.7/2/1 were more efficient than the low and high PEI/DNA ratio polyplexes, while the corresponding AC lipopolyplex was ineffective. To gain insight into the mechanisms involved in their different transfection profiles, the cellular uptake of DNA induced by these complexes was analyzed by flow cytometry. The data are presented as the percentage of cells with internalized YOYO-DNA. As shown in Figure 6, three lipopolyplexes offered a much higher percentage of positive cells than the corresponding polyplexes, demonstrating that addition of cationic lipids to the low PEI/DNA ratio polyplex facilitated cellular uptake. Notably, despite a significant difference in transfection efficiency (Figure 5), LC and HC lipopolyplexes induced the same level of cellular uptake (more than 99% of the cells were positive) as did the high PEI/DNA ratio polyplex. Therefore, it is speculated that the higher efficiency of the LC and HC lipopolyplexes was derived from an improvement in intracellular trafficking of DNA.
Figure 6.
Representative flow cytometry pictures of DNA uptake by bone marrow stem cells.
Notes: Bone marrow stem cells were treated with various complexes in the presence of 10% fetal bovine serum for 4 hours. The percentage of cells containing YOYO-labeled DNA was measured by flow cytometry. Untreated cells were used as the negative control.
Abbreviations: LC, lysinylated cholesterol; HC, histidylated cholesterol; AC, arginylated cholesterol; L, lipid; P, polyethylenimine; D, DNA.
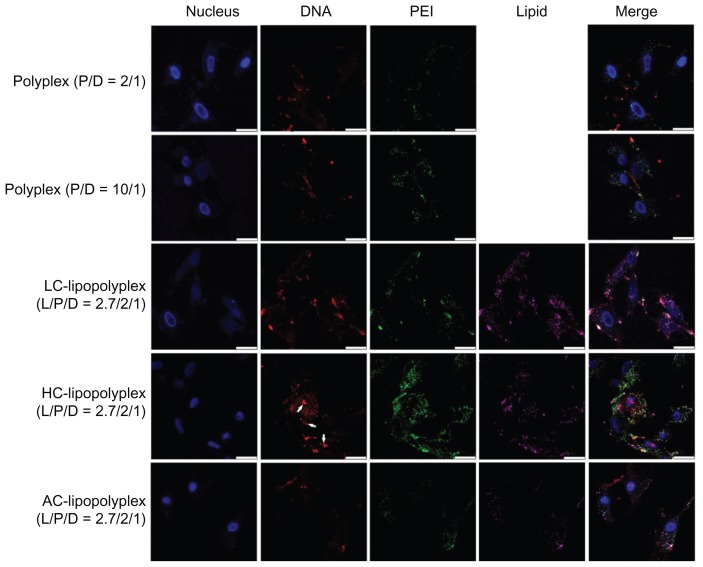
Subcellular distribution of the complexes was visualized by confocal laser scanning microscopy. As shown in Figure 7, only weak DNA (red) and PEI (green) signals were observed in cells treated with the low PEI/DNA ratio polyplex, confirming poor cellular uptake, whereas much higher levels of cellular uptake were detected for LC and HC lipopolyplexes. These observations were consistent with the data obtained from flow cytometry analysis. For the high PEI/DNA ratio polyplex, DNA was localized mainly around the cell membrane and in the cytoplasm, but not in the nucleus (blue). In contrast, accumulation of DNA in the perinuclear region was readily observed in cells treated with LC and HC lipopolyplexes. In particular, nuclear localization of free DNA (white arrows) could be detected for the HC lipopolyplex. Given that nuclear trafficking of DNA is a prerequisite for gene expression,31 these observations suggested an underlying reason for the higher transfection activity of LC and HC lipopolyplexes. Compared with LC and HC lipopolyplexes, the AC lipopolyplex induced a significantly smaller degree of DNA uptake (Figure 6), although DNA was occasionally found in the nuclei. Confocal laser scanning microscopic analysis also revealed a lower degree of cellular internalization of the AC lipopolyplex components (Figure 7), which was in line with the lower transfection efficiency of the complex.
Figure 7.
Representative confocal laser scanning microscopy fluorescence images of subcellular localization of various complexes.
Notes: Fluorescein isothiocyanate-labeled PEI is shown in green, rhodamine-labeled lipids are shown in purple, TOTO-labeled DNA is shown in red, and Hoechst-stained nuclei are shown in blue. Bone marrow stem cells were treated for 4 hours with the various complexes formed at the indicated PEI/DNA and lipid/DNA ratios in medium containing 10% fetal bovine serum and observed under confocal laser scanning microscopy (white arrows, free DNA; scale bar 25 μm).
Abbreviations: LC, lysinylated cholesterol; HC, histidylated cholesterol; AC, arginylated cholesterol; L, lipid; P, polyethylenimine; D, DNA.
Cytotoxicity
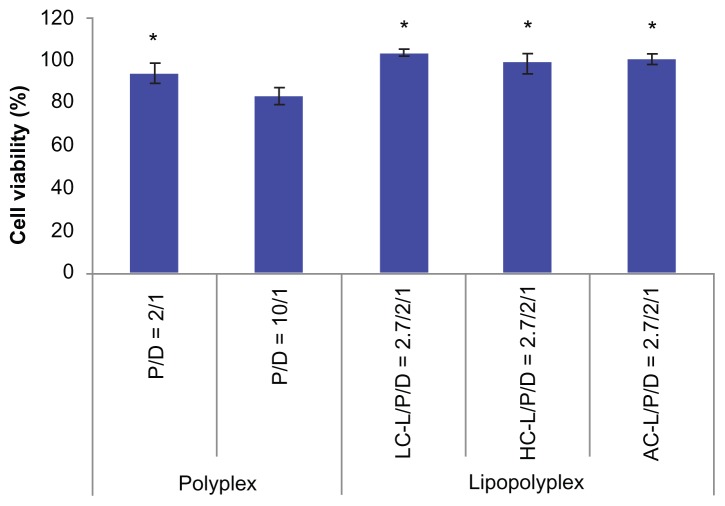
The cytotoxicity of the various complexes was evaluated by determining the percentage of metabolic activity of the transfected cells in relation to the unexposed cells using the CCK-8 assay. In view of the consistency of the incubation time used in gene transfection tests, we adopted 4 hours as the incubation time for this study. The high PEI/DNA ratio polyplex showed apparent cytotoxicity, as reflected by 81% cell viability (Figure 8). In contrast, the low PEI/DNA ratio polyplex and three lipopolyplexes were almost nontoxic, suggesting that addition of lipids did not add to the toxicity of the complexes.
Figure 8.
Cytotoxicity of various complexes determined by Cell Counting Kit-8 assay.
Notes: The results show the relative cell viability of bone marrow stem cells exposed for 4 hours to complexes formed at the indicated PEI/DNA and lipid/DNA ratios in the presence of 10% fetal bovine serum. The viability of untreated cells was taken as 100%. Values are represented the mean ± standard deviation (n = 3; *P < 0.05 versus polyplex with a PEI/DNA ratio of 10).
Abbreviations: LC, lysinylated cholesterol; HC, histidylated cholesterol; AC, arginylated cholesterol; L, lipid; P, polyethylenimine; D, DNA.
Transfection studies with plasmids encoding hVEGF165
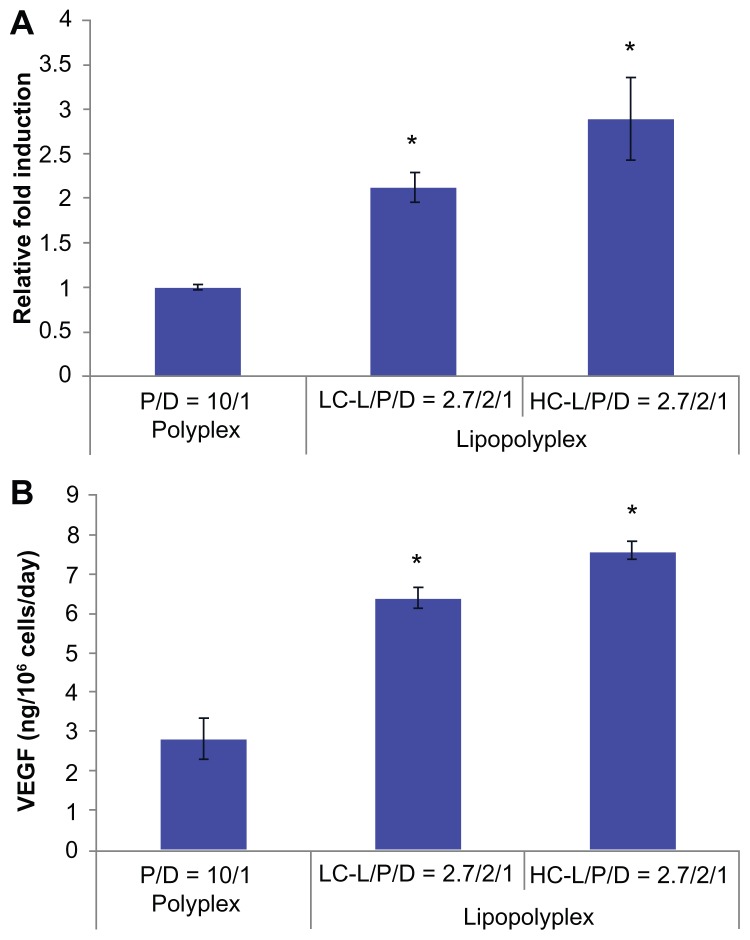
The potential application of the optimized lipopolyplexes for cell-based gene therapy was also evaluated by delivering the therapeutic gene hVEGF165. The transfection activity was evaluated at both the mRNA and protein levels. The results from real-time PCR demonstrated that the LC and HC lipopolyplexes conferred significantly higher hVEGF165 mRNA levels than the high PEI/DNA ratio polyplex (P < 0.05, Figure 9A), which was in good agreement with the results obtained with the luciferase-encoding plasmids. The results were also confirmed by measuring hVEGF165 protein secretion in the culture medium using an enzyme-linked immunosorbent assay (Figure 9B). Calculations based on cell number and culture time revealed that LC and HC lipopolyplexes provided hVEGF165 expression levels ranging from 6.2 to 7.8 ng/106 cells/day.
Figure 9.
hVEGF165 mRNA (A) and protein (B) expression by transfected bone marrow stem cells.
Notes: Bone marrow stem cells were transfected with complexes prepared at the indicated lipid/DNA and PEI/DNA ratios in the presence of 10% fetal bovine serum. Cells and the culture supernatant were harvested 48 hours after transfection. hVEGF165 mRNA levels in bone marrow stem cells were quantified by real-time polymerase chain reaction. Secretion of vascular endothelial growth factor protein in the culture supernatant was quantified by enzyme-linked immunosorbent assay. Results are normalized for the number of cells and duration of culture, and are expressed as the mean ± standard deviation (n = 3; *P < 0.05 versus the polyplex).
Abbreviations: LC, lysinylated cholesterol; HC, histidylated cholesterol; L, lipid; P, polyethylenimine; D, DNA.
Discussion
The clinical application of BMSC-mediated gene therapy relies heavily on the development of gene carriers that can effectively deliver genes into cells without affecting their viability and fate. However, application of most existing nonviral vectors, including the “golden standard” polymer PEI, has been limited by a correlation between efficiency and cytotoxicity. The current study sought to solve this dilemma by noncovalent modification of PEI/DNA polyplexes with amino acid-based cationic lipids which had been proven to be serum-resistant and nontoxic.
In the absence of serum, all lipopolyplexes were well dispersed nanoparticles, giving a larger particle size and higher zeta potential than the corresponding polyplexes (Figures 2 and 3). This suggests that cationic lipids formed an outer coat around the preformed polyplexes, in agreement with the “core-shell” structure of lipopolyplexes proposed by earlier studies.21,22 Nevertheless, it should be noted that the surface charge of lipopolyplexes also increased with increasing PEI/DNA ratio. Thus, it is possible that the polyplex core was not entirely wrapped by the lipids, as shown in Scheme 1. After exposure to serum, most of the lipopolyplexes did not aggregate (Figure 4). However, absorption of serum proteins inevitably altered the size and surface charge of the nanoparticles.
Scheme 1.
Schematic illustration depicting formation of lipopolyplexes by combining preformed polyplex core with cationic lipids.
Abbreviation: PEI, polyethylenimine.
Complete complexation of DNA with 25 kDa branched PEI is known to occur when the PEI/DNA ratio reaches 3.32 However, PEI/DNA polyplexes usually give optimal transfection at a higher ratio of 10,32 where a considerable number of PEI chains remain free in solution.33,34 The excessive PEI chains can significantly increase transfection, presumably via their ability to induce membrane destabilization and disruption.35 Paradoxically, it is these free PEI chains that are held responsible for the immediate cytotoxicity of the complexes.36,37 The low PEI/DNA ratio polyplex of 2 prepared in this study was loosely compacted, conferred poor cellular uptake of DNA (Figures 6 and 7), and had little transfection activity (Figure 5). Modification of this low PEI/DNA ratio polyplex with LC or HC at a lipid/DNA ratio of 2.7 remarkably increased DNA uptake and transfection efficiency. It is most likely that the lipid coating protected the fragile polyplex core from dissociation or aggregation caused by serum proteins. In support of this hypothesis, the resulting lipopolyplexes (Supplementary Figure S1B–D) had a more homogeneous size distribution than the polyplex (Supplementary Figure S1A) after exposure to 10% fetal bovine serum. Complexation of DNA with excessive PEI chains (PEI/DNA ratio of 10) was found to be equally effective in promoting cellular uptake (Figure 6). Nevertheless, the transfection efficiency was several-fold lower than that of the best LC and HC lipopolyplexes (Figure 5). Confocal laser scanning microscopic observations suggest that nuclear localization of DNA was relatively hindered (Figure 7), presumably due to inhibition of the endosome lytic power of PEI by serum proteins.38,39 The lipid coating of the lipopolyplexes may help to overcome this obstacle by destabilizing the endosomal membranes.40 Equally importantly, in contrast with the high PEI/DNA ratio polyplex, LC and HC lipopolyplexes avoided introduction of excessive PEI chains and were completely nontoxic (Figure 8), conferring an additional advantage for the genetic modification of BMSCs.
It is noteworthy that the transfection activity of the LC and HC lipopolyplexes significantly decreased when the PEI/DNA ratio was higher than 2 or the lipid/DNA ratio was higher than 2.7 (Figure 5). This indicates that the success of lipopolyplexes relies on a loosely condensed polyplex core modified by a moderate amount of lipid. Interestingly, earlier studies have also reported that the highest enhancement of transfection by PEI and DOTAP-Chol or DOTMA was achieved at relatively low PEI/DNA ratios of 4.523 and 2,21 respectively, although the reason for this was unclear. It is possible that the relaxed polyplex core benefited transfection by achieving a balance between DNA protection and subsequent DNA liberation for gene expression. In the event of a higher lipid/DNA ratio, excessive cationic lipids might compete with PEI for DNA molecules, destabilizing the polyplex core. In other words, a supramolecular structure combining the advantages of the polyplex core and lipid shell may rest on relatively low lipid/DNA and PEI/DNA ratios.
It was not expected that the AC lipopolyplex with a lipid/PEI/DNA ratio of 2.7/2/1 resulted in much lower levels of cellular uptake and transfection than the corresponding LC and HC lipopolyplexes, because they had a similar particle size and surface charge in general (Figures 2 and 4). In the meantime, AC lipoplexes were also found to be inefficient, irrespective of the lipid/DNA ratio. This was quite different from the results of our earlier study showing that AC lipoplexes were more efficient than HC lipoplexes for gene delivery into NIH3T3 and HeLa cells in the presence of serum.25 This cell type-dependent transfection efficiency can be explained by the notion that cellular internalization of nonviral vectors is affected by cell membrane composition and physiology.41,42 On the other hand, the surface chemistry of nanoparticles can influence the composition of the protein corona that forms on their surface under serum conditions.43 Accordingly, the lack of efficiency of the AC lipopolyplex might result from unfavorable interactions between the cell membrane of BMSCs and the protein corona on the AC lipopolyplex. It also suggested that the optimal design of lipopolyplexes may vary for different cell types.
VEGF is one of the most potent direct-acting angiogenic proteins identified to date and has been widely used in cell-based gene therapy.44 It has been demonstrated that implantation of mouse myoblasts engineered to secrete a corresponding isoform of VEGF at a level of 5 ng/106 cells/day is sufficient to establish normal homogeneous capillary-like vessels.30 In view of this, the results of this study (Figure 9) suggest that the optimized LC and HC lipopolyplexes were more efficient than the high PEI/DNA ratio polyplex in terms of therapeutic gene delivery into BMSCs, and might be able to transfect BMSCs under serum conditions to express functional levels of VEGF protein. Further in vivo studies are in progress to evaluate the therapeutic effect of BMSCs genetically modified with the lipopolyplexes.
Conclusion
In summary, this study demonstrates that noncovalent modification of PEI/DNA polyplexes with lysine/histidine-based cationic lipids to form lipopolyplexes allows a compromise to achieve higher transfection efficiency under serum conditions while minimizing toxicity. The transfection efficiency of the lipopolyplexes peaked at a relatively low lipid/PEI/DNA ratio. A partially condensed polyplex core modified by an outer shell of lipids contributed to a supramolecular structure that combined the advantages of PEI and lipids for gene delivery. Further insights into structure-function relationships will contribute to the design of future lipopolyplexes for more efficient gene delivery into BMSCs.
Supplementary figure
Representative particle size distributions of various complexes after incubation with serum.
Note: Particle size distributions of the complexes were determined by dynamic light scattering after incubation in 10% fetal bovine serum for 15 minutes.
Abbreviations: LC, lysinylated cholesterol; HC, histidylated cholesterol; AC, arginylated cholesterol; L, lipid; P, polyethylenimine; D, DNA.
Acknowledgments
This research was financially supported by the National Basic Research Program of China (National 973 Program, 2011CB606206) and the National Natural Science Foundation of China (51133004, 50903051, and 30970730).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Asahara T, Murohara T, Sullivan A, et al. Isolation of putative progenitor endothelial cells for angiogenesis. Science. 1997;275(5302):964–967. doi: 10.1126/science.275.5302.964. [DOI] [PubMed] [Google Scholar]
- 2.Si YL, Zhao YL, Hao HJ, Fu XB, Han WD. MSCs: biological characteristics, clinical applications and their outstanding concerns. Ageing Res Rev. 2011;10(1):93–103. doi: 10.1016/j.arr.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Davis ME. Non-viral gene delivery systems. Curr Opin Biotechnol. 2002;13(2):128–131. doi: 10.1016/s0958-1669(02)00294-x. [DOI] [PubMed] [Google Scholar]
- 4.Santos JL, Pandita D, Rodrigues J, Pêgo AP, Granja PL, Tomás H. Non-viral gene delivery to mesenchymal stem cells: methods, strategies and application in bone tissue engineering and regeneration. Curr Gene Ther. 2011;11(1):46–57. doi: 10.2174/156652311794520102. [DOI] [PubMed] [Google Scholar]
- 5.Hu YL, Fu YH, Tabata Y, Gao JQ. Mesenchymal stem cells: a promising targeted-delivery vehicle in cancer gene therapy. J Control Release. 2010;147(2):154–162. doi: 10.1016/j.jconrel.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 6.Godbey WT, Wu KK, Mikos AG. Poly(ethylenimine) and its role in gene delivery. J Control Release. 1999;60(2–3):149–160. doi: 10.1016/s0168-3659(99)00090-5. [DOI] [PubMed] [Google Scholar]
- 7.Godbey WT, Wu KK, Mikos AG. Tracking the intracellular path of poly(ethylenimine)/DNA complexes for gene delivery. Proc Natl Acad Sci U S A. 1999;96(9):5177–5181. doi: 10.1073/pnas.96.9.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clamme JP. Intracellular dynamics of the gene delivery vehicle polyethylenimine during transfection: investigation by two-photon fluorescence correlation spectroscopy. Biochim Biophys Acta. 2003;1617(1–2):52–61. doi: 10.1016/j.bbamem.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 9.Guo W, Lee RJ. Efficient gene delivery via non-covalent complexes of folic acid and polyethylenimine. J Control Release. 2001;77(1–2):131–138. doi: 10.1016/s0168-3659(01)00456-4. [DOI] [PubMed] [Google Scholar]
- 10.Gao X, Kim KS, Liu D. Nonviral gene delivery: what we know and what is next. AAPS J. 2007;9(1):E92–E104. doi: 10.1208/aapsj0901009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merdan T, Kopecek J, Kissel T. Prospects for cationic polymers in gene and oligonucleotide therapy against cancer. Adv Drug Deliv Rev. 2002;54(5):715–758. doi: 10.1016/s0169-409x(02)00046-7. [DOI] [PubMed] [Google Scholar]
- 12.Lv H, Zhang S, Wang B, Cui S, Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J Control Release. 2006;114(1):100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 13.Godbey WT, Barry MA, Saggau P, Wu KK, Mikos AG. Poly(ethylenimine)-mediated transfection: a new paradigm for gene delivery. J Biomed Mater Res A. 2000;51(3):321–328. doi: 10.1002/1097-4636(20000905)51:3<321::aid-jbm5>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 14.Fischer D, Bieber T, Li Y, Elsässer HP, Kissel T. A novel non-viral vector for DNA delivery based on low molecular weight, branched polyethylenimine: effect of molecular weight on transfection efficiency and cytotoxicity. Pharm Res. 1999;16(8):1273–1279. doi: 10.1023/a:1014861900478. [DOI] [PubMed] [Google Scholar]
- 15.Zhu W, Chen J, Cong X, Hu S, Chen X. Hypoxia and serum deprivation-induced apoptosis in mesenchymal stem cells. Stem Cells. 2006;24(2):416–425. doi: 10.1634/stemcells.2005-0121. [DOI] [PubMed] [Google Scholar]
- 16.Chen XA, Zhang LJ, He ZJ, et al. Plasmid-encapsulated polyethylene glycol-grafted polyethylenimine nanoparticles for gene delivery into rat mesenchymal stem cells. Int J Nanomedicine. 2011;6:843–853. doi: 10.2147/IJN.S17155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saraf A, Hacker MC, Sitharaman B, Grande-Allen KJ, Barry MA, Mikos AG. Synthesis and conformational evaluation of a novel gene delivery vector for human mesenchymal stem cells. Biomacromolecules. 2008;9(3):818–827. doi: 10.1021/bm701146f. [DOI] [PubMed] [Google Scholar]
- 18.He W, Guo Z, Wen Y, et al. Alginate-graft-PEI as a gene delivery vector with high efficiency and low cytotoxicity. J Biomater Sci Polym Ed. 2012;23(1–4):315–331. doi: 10.1163/092050610X550359. [DOI] [PubMed] [Google Scholar]
- 19.Pelisek J, Gaedtke L, DeRouchey J, Walker GF, Nikol S, Wagner E. Optimized lipopolyplex formulations for gene transfer to human colon carcinoma cells under in vitro conditions. J Gene Med. 2006;8(2):186–197. doi: 10.1002/jgm.836. [DOI] [PubMed] [Google Scholar]
- 20.García L, Buñuales M, Düzgüneš N, Tros de Ilarduya C. Serum-resistant lipopolyplexes for gene delivery to liver tumour cells. Eur J Pharm Biopharm. 2007;67(1):58–66. doi: 10.1016/j.ejpb.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 21.Matsumoto M, Kishikawa R, Kurosaki T, et al. Hybrid vector including polyethylenimine and cationic lipid, DOTMA, for gene delivery. Int J Pharm. 2008;363(1–2):58–65. doi: 10.1016/j.ijpharm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 22.Schäfer J, Höbel S, Bakowsky U, Aigner A. Liposome-polyethylenimine complexes for enhanced DNA and siRNA delivery. Biomaterials. 2010;31(26):6892–6900. doi: 10.1016/j.biomaterials.2010.05.043. [DOI] [PubMed] [Google Scholar]
- 23.Lee CH, Ni YH, Chen CC, Chou C, Chang FH. Synergistic effect of polyethylenimine and cationic liposomes in nucleic acid delivery to human cancer cells. Biochim Biophys Acta. 2003;1611(1–2):55–62. doi: 10.1016/s0005-2736(03)00027-0. [DOI] [PubMed] [Google Scholar]
- 24.Lampela P, Elomaa M, Ruponen M, Urtti A, Männistö PT, Raasmaja A. Different synergistic roles of small polyethylenimine and Dosper in gene delivery. J Control Release. 2003;88(1):173–183. doi: 10.1016/s0168-3659(02)00489-3. [DOI] [PubMed] [Google Scholar]
- 25.Li L, Song H, Luo K, et al. Gene transfer efficacies of serum-resistant amino acids-based cationic lipids: dependence on headgroup, lipoplex stability and cellular uptake. Int J Pharm. 2011;408(1–2):183–190. doi: 10.1016/j.ijpharm.2011.01.051. [DOI] [PubMed] [Google Scholar]
- 26.Li L, Nie Y, Zhu R, et al. Preparation and gene delivery of alkaline amino acids-based cationic liposomes. Arch Pharm Res. 2008;31(7):924–931. doi: 10.1007/s12272-001-1248-8. [DOI] [PubMed] [Google Scholar]
- 27.Menon LG, Picinich S, Koneru R, et al. Differential gene expression associated with migration of mesenchymal stem cells to conditioned medium from tumor cells or bone marrow cells. Stem Cells. 2007;25(2):520–528. doi: 10.1634/stemcells.2006-0257. [DOI] [PubMed] [Google Scholar]
- 28.Ji J, Fu J, Shen J. Fabrication of a superhydrophobic surface from the amplified exponential growth of a multilayer. Adv Mater. 18(11):1441–1444. [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Ozawa CR, Banfi A, Glazer NL, et al. Microenvironmental VEGF concentration, not total dose, determines a threshold between normal and aberrant angiogenesis. J Clin Invest. 2004;113(4):516–527. doi: 10.1172/JCI18420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaffer DV, Fidelman NA, Dan N, Lauffenburger DA. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol Bioeng. 2000;67(5):598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 32.Yue Y, Jin F, Deng R, et al. Revisit complexation between DNA and polyethylenimine – effect of uncomplexed chains free in the solution mixture on gene transfection. J Control Release. 2011;155(1):67–76. doi: 10.1016/j.jconrel.2010.10.028. [DOI] [PubMed] [Google Scholar]
- 33.Finsinger D, Remy JS, Erbacher P, Koch C, Plank C. Protective copolymers for nonviral gene vectors: synthesis, vector characterization and application in gene delivery. Gene Ther. 2000;7(14):1183–1192. doi: 10.1038/sj.gt.3301227. [DOI] [PubMed] [Google Scholar]
- 34.Clamme JP, Azoulay J, Mély Y. Monitoring of the formation and dissociation of polyethylenimine/DNA complexes by two photon fluorescence correlation spectroscopy. Biophys J. 2003;84(3):1960–1968. doi: 10.1016/S0006-3495(03)75004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yue Y, Jin F, Deng R, et al. Revisit complexation between DNA and polyethylenimine – effect of length of free polycationic chains on gene transfection. J Control Release. 152(1):143–151. doi: 10.1016/j.jconrel.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 36.Boeckle S, von Gersdorff K, van der Piepen S, Culmsee C, Wagner E, Ogris M. Purification of polyethylenimine polyplexes highlights the role of free polycations in gene transfer. J Gene Med. 2004;6(10):1102–1111. doi: 10.1002/jgm.598. [DOI] [PubMed] [Google Scholar]
- 37.Erbacher P, Bettinger T, Brion E, et al. Genuine DNA/polyethylenimine (PEI) complexes improve transfection properties and cell survival. J Drug Target. 2004;12(4):223–236. doi: 10.1080/10611860410001723487. [DOI] [PubMed] [Google Scholar]
- 38.Guo W, Lee RJ. Efficient gene delivery via non-covalent complexes of folic acid and polyethylenimine. J Control Release. 2001;77(1–2):131–138. doi: 10.1016/s0168-3659(01)00456-4. [DOI] [PubMed] [Google Scholar]
- 39.Guo W, Lee RJ. Efficient gene delivery using anionic liposome-complexed polyplexes (LPDII) Biosci Rep. 2000;20(5):419–432. doi: 10.1023/a:1010338219401. [DOI] [PubMed] [Google Scholar]
- 40.Wasungu L, Hoekstra D. Cationic lipids, lipoplexes and intracellular delivery of genes. J Control Release. 2006;116(2):255–264. doi: 10.1016/j.jconrel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 41.Dastan T, Turan K. In vitro characterization and delivery of chitosan-DNA microparticles into mammalian cells. J Pharm Pharm Sci. 2004;7(2):205–214. [PubMed] [Google Scholar]
- 42.Chen J, Yu Z, Chen H, Gao J, Liang W. Transfection efficiency and intracellular fate of polycation liposomes combined with protamine. Biomaterials. 2011;32(5):1412–1418. doi: 10.1016/j.biomaterials.2010.09.074. [DOI] [PubMed] [Google Scholar]
- 43.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc Natl Acad Sci U S A. 2008;105(38):14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferrara N. Vascular endothelial growth factor: molecular and biological aspects. Curr Top Microbiol Immunol. 1999;237:1–30. doi: 10.1007/978-3-642-59953-8_1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative particle size distributions of various complexes after incubation with serum.
Note: Particle size distributions of the complexes were determined by dynamic light scattering after incubation in 10% fetal bovine serum for 15 minutes.
Abbreviations: LC, lysinylated cholesterol; HC, histidylated cholesterol; AC, arginylated cholesterol; L, lipid; P, polyethylenimine; D, DNA.