Abstract
Sarcoidosis is a systemic inflammatory disease characterized by the formation of granulomas in affected organs. Genome-wide association studies (GWASs) of this disease have been conducted only in European population. We present the first sarcoidosis GWAS in African Americans (AAs, 818 cases and 1,088 related controls) followed by replication in independent sets of AAs (455 cases and 557 controls) and European Americans (EAs, 442 cases and 2,284 controls). We evaluated >6 million SNPs either genotyped using the Illumina Omni1-Quad array or imputed from the 1000 Genomes Project data. We identified a novel sarcoidosis-associated locus, NOTCH4, that reached genome-wide significance in the combined AA samples (rs715299, P AA-meta = 6.51×10−10) and demonstrated the independence of this locus from others in the MHC region in the same sample. We replicated previous European GWAS associations within HLA-DRA, HLA-DRB5, HLA-DRB1, BTNL2, and ANXA11 in both our AA and EA datasets. We also confirmed significant associations to the previously reported HLA-C and HLA-B regions in the EA but not AA samples. We further identified suggestive associations with several other genes previously reported in lung or inflammatory diseases.
Introduction
Sarcoidosis is a systemic disease characterized by granulomatous inflammation that primarily affects the lungs, but can affect any organ [1], [2], [3]. While the etiology of this disease remains elusive, the pathophysiology likely involves a dysregulated immune response to environmental agents in a genetically susceptible host. Several environmental exposures have been associated with sarcoidosis including mold, inorganic particles, and insecticides [4], [5], [6]. A significant genetic component to sarcoidosis susceptibility is supported by a 2.5 fold elevated disease risk in siblings and parents of cases [7] as well as potential disease susceptibility loci identified from both linkage and association studies [8], [9], [10], [11], [12].
Sarcoidosis impacts individuals of all races, ages and genders [13], but in the U.S. is most frequent in AAs [14], [15], with disease onset peaking between the ages of 20 and 39 years [16]. The AA population is more commonly affected than EAs [16], [17], [18], [19], with a three-fold higher lifetime risk (2.4%) and age-adjusted annual incidence (35.5 per 100,000) compared to EAs (0.85% and 10.9 per 100,000, respectively). AA patients have higher disease severity and more extra-thoracic involvement than EA patients and are less likely to have disease that resolves [20]. Ethnicity specific prevalence and severity support the involvement of genes and further suggest ethnicity-specific genetic risk profiles.
Genetic associations with specific HLA alleles and sarcoidosis have repeatedly been reported [21], [22], [23], [24]. Heterogeneity of these HLA effects in sarcoidosis across ancestries was observed in the ACCESS study [23] suggesting that while the HLA-DRB1*1101 allele was associated with sarcoidosis in AAs and EAs, the HLA-DRB1*1501 allele was associated with sarcoidosis only in EAs [23]. Recent studies have reported additional susceptibility loci including BTNL2 [9], [25], [26] in both EAs and AAs, and ANXA11 [11] and RAB23 [27] in Germans. The first genome-wide linkage study of AA sarcoidosis families performed by our group found prominent linkage signals on chromosome 5, at 5q11.2, 5p13, and 5q31 [10]. Our admixture study confirmed the latter two of these effects and found regions on chromosomes 6p22.3 and 17p13.3–17p13.1 associated with increased African ancestry [28]. Based on clear evidence of the involvement of genes in the onset and manifestation of sarcoidosis, we sought to confirm sarcoidosis genetic risk loci reported in association scans of European populations and to identify novel risk loci by conducting the first genome-wide association study (GWAS) of sarcoidosis in an American population. We present results from a family-based discovery cohort of AAs as well as two independent replication sets of AA cases and controls and EA cases and controls.
Results
Genome-wide Association Scan of AA Discovery Set
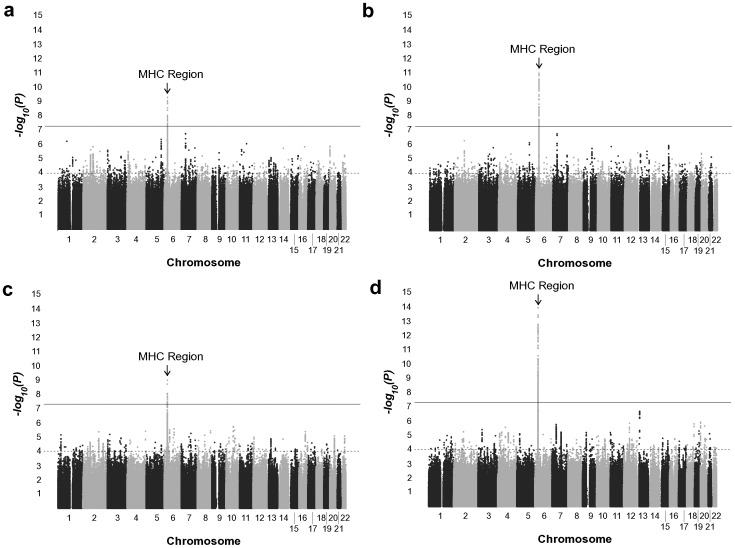
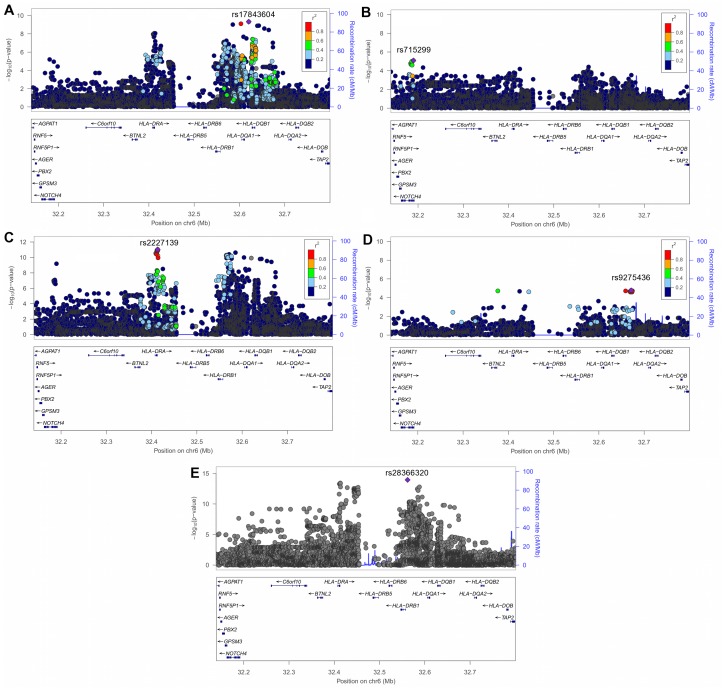
A total of 864,829 single-nucleotide polymorphisms (SNPs) in our AA discovery set passed quality control assessment (Materials and Methods, Figure 1, Table 1). To increase the density of SNPs to be tested for association, we performed genotype imputation across the genome with the 1000 Genomes Project Phase I haplotypes as reference (Materials and Methods). The GWAS of the AA discovery set demonstrated no evidence for inflation of the test statistics (genomic control inflation factor [λGC] = 0.980) after comparing the observed and expected distributions of the SNP-sarcoidosis association P-values calculated using EMMAX (Figure S1, Materials and Methods). This suggests our regression model was able to account for population stratification in this dataset. The quantile-quantile plot revealed the presence of significant genetic effects associated with sarcoidosis (Figure S1). This dataset had good statistical power (at α = 5×10−8) to detect associations from common alleles with odds ratios ≥1.5 (Figure S2). We only found variants within previously reported MHC Class II genes [11], [22] exceeding genome-wide significance in this dataset (Figure 2A, Figure 3A, Table S2); HLA-DRA with the peak signals at multiple SNPs in perfect linkage disequilibrium (LD) with each other (r 2 = 1) including a missense SNP rs7192 (P AA-Disc = 8.73×10−9), HLA-DQA1 (peak signal at rs17843604, P AA-Disc = 4.77×10−10), and HLA-DQB1 (peak signal at rs149288329, P AA-Disc = 1.27×10−9) (Table S2). These SNPs were not LD with each other (r 2≤0.054).
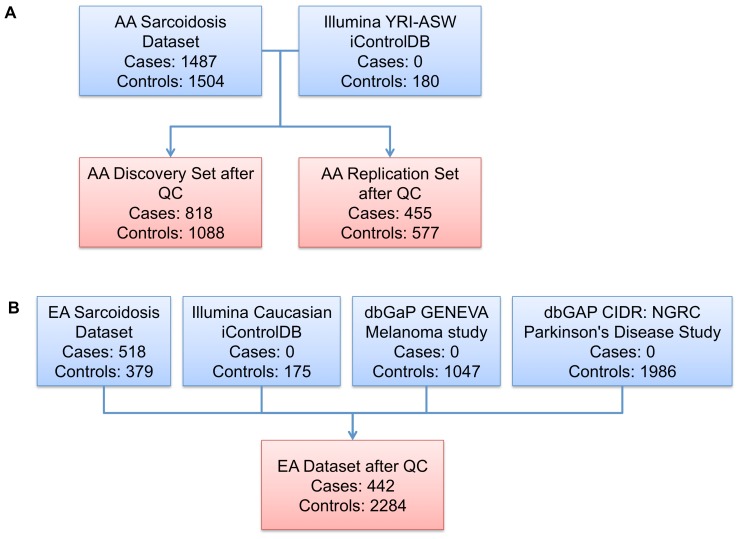
Figure 1. A graphical overview of the GWAS datasets.
(A–B) Summary of the AA (A) and EA (B) datasets.
Table 1. Sample summary before and after quality control (QC).
| African American | European American | |||||
| Characteristic | All samplesbefore QC | Discovery setafter QC | Replication setafter QC | All Samples after QC | Replication set before QC | Replication set after QC |
| Cases | 1487 | 818 | 455 | 1273 | 518 | 442 |
| Controls | 1504 | 908 | 577 | 1465 | 379 | 339 |
| External Controls | 180a | 180 | 0 | 180 | 3208b | 1945 |
| Unknown Affection Status | 2 | 0 | 0 | 0 | 0 | 0 |
| Male | 889 | 575 | 244 | 819 | 1847 | 1173 |
| Female | 2264 | 1331 | 768 | 2099 | 2247 | 1553 |
| Unknown Gender | 20 | 0 | 0 | 0 | 11 | 0 |
| Total | 3173 | 1906 | 1012 | 2918 | 4105 | 2726 |
Taken from the Illumina YRI-ASW iControlDB;
175 Caucasian healthy controls from the Illumina iControlDB, 1047 controls from the dbGaP GENEVA Melanoma study, and 1986 controls from the dbGAP CIDR: NGRC Parkinson’s Disease Study.
Figure 2. Manhattan plots of SNP-sarcoidosis association test results.
(A–D) Association results in the AA discovery set (A), a meta-analysis between the AA discovery and AA replication sets (B), the EA dataset (C), and a meta-analysis of the AA discovery, AA replication and EA datasets (D). The black horizontal line represents the threshold for genome-wide significance (P<5×10−8) and the gray line is the suggestive evidence of association threshold (P<1×10−4).
Figure 3. Regional association plots of SNP-sarcoidosis association test results within the MHC Class II region.
(A–D) Association results in the AA discovery set (A), AA replication set (B), a meta-analysis between the AA discovery and AA replication sets (C), the EA dataset (D), and a meta-analysis of the AA discovery, AA replication and EA datasets (E). Each SNP is colored according to its LD (r 2) with the top SNP, except for (E) since the meta-analysis was performed on two different populations. The recombination rate is denoted by the blue solid line. Plots were drawn using LocusZoom [100].
Genome-wide Meta-Analysis of the AA Discovery and Replication Sets
After assessing association between SNPs and sarcoidosis using logistic regression in the AA replication set (Materials and Methods, Figure 1, Table 1), we found little evidence for inflation of the test statistics in this dataset (λGC = 1.030, Figure S1). A meta-analysis of the AA discovery and replication sets yielded additional MHC SNPs that surpassed genome-wide significance in the meta-analysis results not present in either set alone. These included a genotyped SNP in the previously unreported neurogenic locus notch homolog protein 4 (NOTCH4) gene (rs715299, P AA-meta = 6.51×10−10) and other SNPs within the MHC Class II genes (Figure 1B, Figure 3C, Table 2, Table S2).
Table 2. Regions of association meeting genome-wide significance and their most significant SNPs grouped by sample.
| CHR | BP(hg 19) | SNP | Gene | Alleles1 | African Americans | European Americans | PAll-Meta | Heterogeneity Test | |||||||||
| MAFAA-Disc 2 | ORAA-Disc 3 | P AA-Disc | MAFAA-Rep 2 | ORAA-Rep 3 | P AA-Rep | PAA-Meta | MAFEA 2 | OREA 3 | P EA | Q | I 2 (%) | ||||||
| 6 | 32,411,646 | rs7192 | HLA-DRA a | G/T | 0.424 | 1.66 | 8.73E-09 | 0.445 | 1.40 | 3.44E-04 | 1.40E-11 | 0.395 | 1.35 | 1.26E-04 | 5.28E-14 | 0.304 | 16 |
| 6 | 32,620,283 | rs17843604 | HLA-DQA1 a | C/T | 0.402 | 0.63 | 4.77E-10 | 0.378 | 0.80 | 1.70E-02 | 1.21E-10 | 0.56 | 0.91 | 1.81E-01 | 2.73E-08 | 5.33E-05 | 89.8 |
| 6 | 32,642,794 | rs149288329 | HLA-DQB1 a | T/C | 0.025 | 1.92 | 1.27E-09 | 0.038 | 1.87 | 1.15E-02 | 1.55E-10 | NA | NA | NA | NA | NA | NA |
| 6 | 32,189,841 | rs715299 | NOTCH4 b | T/G | 0.454 | 1.30 | 1.12E-05 | 0.480 | 1.52 | 8.14E-06 | 6.51E-10 | 0.324 | 1.14 | 9.58E-02 | 2.15E-08 | 0.064 | 63.6 |
| 6 | 31,272,612 | rs6457375 | HLA-C c | A/G | 0.423 | 0.88 | 4.24E-01 | 0.403 | 1.17 | 9.06E-02 | 7.26E-01 | 0.49 | 1.58 | 1.98E-09 | 9.80E-06 | 1.84E-05 | 90.8 |
| 6 | 31,326,324 | rs2596475 | HLA-B c | T/C | 0.287 | 0.90 | 5.27E-01 | 0.263 | 1.00 | 9.84E-01 | 6.01E-01 | 0.386 | 1.52 | 3.82E-08 | 2.72E-05 | 7.45E-05 | 89.5 |
| 6 | 32,446,853 | rs17203612 | HLA-DRB5 c | T/C | 0.270 | 0.64 | 2.66E-05 | 0.243 | 0.79 | 2.42E-02 | 2.33E-06 | 0.438 | 0.63 | 1.82E-08 | 2.80E-13 | 0.209 | 36.1 |
Major/minor allele of AAs as the reference;
Minor allele frequency;
The odds ratio (OR) was calculated with respect to the minor allele of AAs.
Previously reported sarcoidosis loci meeting genome-wide significance in the AA discovery set.
Potentially novel region meeting genome-wide significance after the meta-analysis of AA datasets.
Previously reported sarcoidosis loci meeting genome-wide significance in the EA dataset.
Stepwise Conditional Association of the MHC Region in Combined AA Dataset
Since the MHC region is known for its extensive regions of high LD [29], we sought to assess whether the novel AA association signal within NOTCH4 was independent of the signals within the MHC Class II genes. We performed stepwise conditional association analyses (Materials and Methods) among variants with P AA-meta <5×10−8 in the MHC region in the combined AA set and at step one used the most significant SNP (rs2227139, HLA-DRA) as the covariate. After adjusting for this HLA-DRA SNP, we observed significant residual associations in several other regions; the most significant of which was at rs146146117 (HLA-DQA1, P conditional = 6.81×10−8, Table S3). Significant residual associations remained after the next step of adjusting for HLA-DRA and HLA-DQA1 SNPs; the most significant residual association was within HLA-DRB1 (rs9461776, P conditional = 1.45×10−7, Table S3). We continued to step three by adding this HLA-DRB1 SNP into the regression and found the most significant residual signals at NOTCH4 (rs715299, P conditional = 1.74×10−6) and HLA-DQA1 (rs9272320, P conditional = 7.04×10−6) (Table S3). The subsequent (and final) step adding this HLA-DQA1 SNP (rs9272320) as a covariate resulted in diminished association signals for the remaining significant SNPs within the MHC class II genes (P conditional ≥0.014), whereas NOTCH4 remained significant (rs715299, P conditional = 8.85×10−5) (Table S3). While the P-value for NOTCH4 did not retain the GWAS threshold of 5×10−8 after rigorous conditioning, it remains the only significant effect well exceeding the suggestive level of association. It suggests that the observed signal within NOTCH4 is independent of the evaluated SNPs within the MHC Class II genes. These analyses also showed the existence of multiple independent signals within this MHC region (Table 2).
Confirmation of Previously Reported SNPs Associated with Sarcoidosis in the Combined AA Datasets
Three significant SNPs reported in the previous German GWAS in the MHC region (P<1×10−6) [11] were also replicated in our combined AA datasets (rs7194 [in perfect LD with rs7192], HLA-DRA, P AA-meta = 1.40×10−11; rs9268853, HLA-DRB5, P AA-meta = 7.40×10−4; and rs615672, HLA-DRB1, P AA-meta = 2.60×10−9, Table 3). The previously reported peak SNP within BTNL2 (rs2076530) [9], [11], [25] was not strongly associated with sarcoidosis in our AA datasets (P AA-meta = 0.024, Table 3). However, a SNP with 4 kb upstream of rs2076530, rs9268482, was suggestive of association (P AA-meta = 6.32×10−6, Table 3). Interestingly, we also identified a suggestive association at a BTNL2 coding-synonymous SNP, rs9268480 (P AA-meta = 1.03×10−5), only 28 bp upstream of rs2076530 and in high LD with rs9268482 (r 2 = 0.996). Since BNTL2 is only 170 kb apart from NOTCH4, we sought to assess whether the signal within NOTCH4 is independent of the signal within BTNL2 using conditional association analyses. When adjusting for one of those associated BTNL2 SNPs (rs9268482), we found NOTCH4 remained significant (rs715299, P conditional = 2.86×10−8). On the other hand, after adjusting for the NOTCH4 SNP, we still observed a significant residual signal at the BTNL2 SNP (rs9268482, P conditional = 1.26×10−4). These indicated the signal within NOTCH4 is also independent of the BTNL2 signal.
Table 3. Replication of previously reported SNPs associated with sarcoidosis [9], [11], [25], [27].
| CHR | BP(hg 19) | SNP | Gene | Alleles1 | African Americans | European Americans | PAll-Meta | Heterogeneity Test | |||||||||
| MAFAA-Disc 2 | ORAA-Disc | P AA-Disc | MAFAA-Rep 2 | ORAA-Rep | P AA-Rep | PAA-Meta | MAFEA 2 | OREA | P EA | Q | I 2 (%) | ||||||
| 6 | 32,363,816 | rs2076530 | BTNL2 | T/C | 0.309 | 0.84 | 2.50E-01 | 0.312 | 0.80 | 2.46E-02 | 2.42E-02 | 0.434 | 0.70 | 4.19E-06 | 1.44E-06 | 0.324 | 11.3 |
| 6 | 32,412,480 | rs7194 | HLA-DRA | A/G | 0.424 | 1.66 | 8.73E-09 | 0.445 | 1.40 | 3.44E-04 | 1.40E-11 | 0.395 | 1.35 | 1.26E-04 | 5.28E-14 | 0.304 | 16 |
| 6 | 32,429,643 | rs9268853 | HLA-DRB5 | T/C | 0.214 | 0.72 | 1.16E-03 | 0.197 | 0.86 | 2.03E-01 | 7.40E-04 | 0.331 | 0.76 | 9.79E-04 | 2.39E-06 | 0.544 | 0 |
| 6 | 32,574,171 | rs615672 | HLA-DRB1 | C/G | 0.449 | 0.64 | 1.23E-06 | 0.438 | 0.72 | 5.50E-04 | 2.60E-09 | 0.643 | 0.81 | 8.00E-03 | 9.97E-10 | 2.00E-07 | 93.5 |
| 6 | 57055354 | rs1040461 | RAB23 | C/T | 0.158 | 1.13 | 3.24E-02 | 0.177 | 1.21 | 1.18E-01 | 8.04E-03 | 0.079 | 0.89 | 4.18E-01 | 1.80E-01 | 0.257 | 26.4 |
| 10 | 81,926,702 | rs1049550 | ANXA11 | G/A | 0.185 | 0.68 | 7.91E-04 | 0.187 | 0.88 | 2.89E-01 | 8.46E-04 | 0.409 | 0.81 | 8.33E-03 | 2.30E-05 | 0.356 | 3.2 |
Major/minor allele of AAs as the reference;
Minor allele frequency;
The odds ratio (OR) was calculated with respect to the minor allele of AAs.
We saw modest association with two other previously reported susceptibility genes: ANXA11 [11] and RAB23 [27]. A non-synonymous SNP within ANXA11, rs1049550, was associated with sarcoidosis in our combined AA datasets at P AA-meta = 8.46×10−4 (Table 3). A similar modest association was seen with a non-synonymous SNP within RAB23 (rs1040461, P AA-meta = 8.04×10−3, Table 3). We did find suggestive evidence of association on 5q11.2 (peak signal at rs116137605 within a region between SNX18 and ESM1, P AA-meta = 3.09×10−5) a region identified in our previous linkage and fine-mapping studies [10], [28], [30].
Genome-wide Association Scan of EA Dataset
We found 682,921 genotyped SNPs passed quality control measures in our EA dataset (Materials and Methods, Figure 1, Table 1). After performing imputation with the 1000 Genomes Project haplotypes, the SNP-sarcoidosis association calculated using logistic regression of the EA dataset showed little evidence for inflation of the test statistics (λGC = 1.027, Figure S1). This dataset also had good statistical power (at α = 5×10−8) to detect associations from common alleles with odds ratios ≥1.5 (Figure S2). We observed genome-wide significance SNPs within previously reported MHC genes [9], [11], [24] including HLA-C (peak signal at rs6457375, P EA = 1.98×10−9), HLA-B (peak signal at rs2596475, P EA = 3.82×10−8), and HLA-DRB5 (peak signal at rs17203612, P EA = 1.82×10−8) (Figure 2C, Figure 3D, Table 2, Table S2). However, we did not find any variant within NOTCH4 passed genome-wide significance in this dataset (Figure S3). Stepwise conditional association analyses further demonstrated two independent signals exist within this region tagged by rs6457375 (HLA-C) and rs17203612 (HLA-DRB5) (Table S4).
Confirmation of Previously Identified Loci in EA Dataset
We replicated significant SNPs from the German GWAS [11] in the EA dataset including rs7194 (HLA-DRA, P EA = 1.26×10−4), rs9268853 (HLA-DRB5, P EA = 9.79×10−4), rs615672 (HLA-DRB1, P EA = 8.00×10−3), and rs1049550 (ANXA11, P EA = 8.33×10−3) (Table 3). We also replicated the BTNL2 SNP, rs2076530 [9], [11], [25], in our EA dataset (P EA = 4.19×10−6, Table 3). We did not, however, confirm the RAB23 association [27] in this dataset (rs1040461, P EA = 0.418, Table 3).
Meta-analysis Results of All Datasets
Among regions that met genome-wide significance in the AA meta-analysis, we also found significant associations within HLA-DRA, HLA-DRB1, and HLA-DQA1 in the EA dataset (8.25×10−5≤P EA ≤3.97×10−2, 3.77×10−14≤P All-meta ≤7.23×10−8) (Figure 3E, Table S2). We found a weak association to the NOTCH4 SNP (rs715299) in the EA dataset (P EA = 0.096), perhaps suggesting its ethnicity specific effect (the Cochran’s Q test of heterogeneity P = 0.064 and the inconsistency index I 2 = 63.60%, see Materials and Methods). Conversely, when evaluating regions reaching genome-wide significant in the EA dataset, variants within HLA-DRB5, HLA-DRB1, and HLA-DQA1 were also significant in the AA datasets (1.81×10−7≤P AA-meta ≤1.28×10−5, 1.16×10−14≤P All-meta ≤2.65×10−12, Table S2), whereas HLA-C and HLA-B were not (P AA-meta ≥0.575, Table S2).
Suggestive Association Regions
We observed multiple regions reached suggestive association (P all-meta <1×10−4) in the meta-analysis of all AA and EA datasets. These included variants within TRAK1, SLC44A4, GLI3-C7orf25, ATP8A2, and TGM3 (Tables S5). We observed additional suggestive association regions (P<1×10−4) that were unique to one ethnic group. For example, we identified variants with suggestive association within FHIT, PRDM1, FRMD3, DMBT1 and a region between ZSCAN2 and ALPK3 in the combined AA datasets only (Tables S5). We also observed suggestive association only in the EA dataset within CASP10, RARB, and NCR3 among others (Tables S5). Several of these suggestive effects fall within genes implicated in other lung or inflammatory diseases (Table S6).
Discussion
Previously reported GWASs of sarcoidosis have been limited to European (specifically German) samples. Ours is the first GWAS of sarcoidosis in Americans and, even more importantly, of AAs, the population most commonly and severely affected. Our results, while demonstrating some shared effects across ethnicities, strongly support the presence of ethnic specific genetic effects. We identified significant association between sarcoidosis and a previously unreported locus (NOTCH4) in our AA datasets. This association was determined to be independent of other neighboring MHC genes and is an attractive biological candidate. NOTCH4 encodes a member of the Notch family that is involved in controlling cell fate decisions during developmental processes and regulating the activity of T cell immune responses [31], [32]. The Notch signaling pathway also plays a role in endothelial cell differentiation, apoptosis and proliferation [33], [34], [35], [36]. Further, NOTCH4 is highly expressed in the lung and may play a key role in the lung development and diseases such as asthma and lung arteriovenous shunts [37], [38], [39], [40], [41]. NOTCH4 has also been associated with neonatal lupus [42], multiple sclerosis [43], systemic sclerosis [44], and other immune-related disorders [45], [46], [47], [48]. We also saw evidence of suggestive association of NOTCH4 in our EA dataset. While further studies are needed to define the role of NOTCH4 in the specific pathogenesis of sarcoidosis, a novel association to this gene is supported by previous expression and disease studies.
We replicated associations for several previously reported sarcoidosis susceptibility risk loci in our AA collection including MHC Class II region genes (HLA-DRA, HLA-DRB5, HLA-DRB1, and HLA-DQA1), BTNL2, RAB23, and ANXA11 [9], [11], [25], [27], [49]. These regions were also replicated in our EA dataset except for RAB23. It is known that the MHC Class II region plays a major role in immune-mediated disorders, including associations to celiac disease, insulin-dependent diabetes mellitus, rheumatoid arthritis, multiple sclerosis, and systemic lupus erythematosus (SLE) [50], [51]. Similarly, BTNL2, RAB23, and ANXA11 have been suggested to play a role in T-cell activation [9], antibacterial defense processes [27], and apoptosis [11]. It is worth noting that we did not replicate the association with C10orf67 [12] as identified in a joint GWAS of German patients with either sarcoidosis or Crohn’s disease.
Additional regions with suggestive evidence of association in both AAs and EAs include TRAK1, SLC44A4, GLI3-C7orf25, ATP8A2, and TGM3. While the biological relevance of most of these genes to sarcoidosis is still unknown, GLI3-C7orf25 and TGM3 may warrant further investigation. Although C7orf25 is a hypothetical gene with unknown function, GLI3 encodes zinc finger protein Gli3 that has a bipotential function as a transcriptional activator or repressor of the sonic hedgehog pathway [52], [53]. This pathway contains RAB23 (discussed above) and has been suggested to play a role in the sarcoidosis pathophysiology [27]. TGM3 (Transglutaminase 3) encodes protein involved in the later stages of cell envelope formation in the epidermis and hair follicle [54] and has been associated with celiac disease [55], [56] and psoriasis [57], [58].
Despite the overlap of compelling signals across populations, we did find evidence of genetic heterogeneity between ethnic groups in this disease (see Tables 2 and 3). The previously reported MHC Class I region [24] including HLA-C and HLA-B (associated with psoriasis [59] and ankylosing spondylitis [60], respectively) was associated only in the EA dataset. Other noteworthy genes with suggestive association specific to EAs included CASP10, RARB, and NCR3. CASP10 (caspase 10) plays a role in apoptosis and has been associated with autoimmune lymphoproliferative syndrome [61] and non-Hodgkin lymphoma [62]. In addition, RARB (retinoic acid receptor beta) and NCR3 (natural cytotoxicity triggering receptor 3) have been associated with pulmonary function based on a recent GWAS of European Caucasians [63]. Suggestive associations specific to AAs include FHIT, FRMD3, DMBT1, and PRDM1. FHIT (fragile histidine triad) is involved in various intracellular functions and a putative tumor suppressor for various cancers including lung cancer [64], [65]. FRMD3 (FERM domain containing 3) is over-expressed in normal human lung tissue compared with tissue from lung tumors of lung carcinoma patients suggesting its important role in the origin and progression of lung cancer [66]. DMBT1 (deleted in malignant brain tumors 1) is overexpressed in epithelial cells [67] and has been found associated with ulcerative colitis [68] and Crohn’s disease [67], [69]. PRDM1 (PR domain containing protein 1) plays a role as a repressor of beta-interferon gene expression [70] and had been associated with rheumatoid arthritis [71], inflammatory bowel disease (IBD) [72], [73], and SLE [74], [75]. We also observed variants with suggestive associations specific to AAs in a region containing ZSCAN2, SCAND2, WDR73, NMB, SEC11A, ZNF592, and ALPK3 as well as a region identified in our linkage studies [10], [28], [30] on 5q11.2 (a region between SNX18 and ESM1). However, the actual biological functions of these genes are largely unknown.
In summary, this is the first report of GWAS in an American sample and the first report of a significant association between sarcoidosis and NOTCH4. We have replicated several previously reported sarcoidosis susceptibility loci in both our EA and AA samples as well as report several biologically plausible effects at loci with suggestive statistical evidence. We report sarcoidosis associations both shared between ethnicities as well as those unique to either our AA or EA dataset, supporting genetic heterogeneity of this disease. The presence of genetic heterogeneity may well serve as a useful tool in the isolation of the causal variants associated with this disease as it has in other complex disorders [76], [77]. Finally, this study demonstrates both the usefulness of and need for genetic studies of sarcoidosis in diverse populations and further elucidates potential pathogenic mechanisms of this disease. Future replication, sequencing and functional studies are required to further elucidate the causal variants that may underlie these associations as well as to discover rare variants that may have yet to be identified.
Materials and Methods
Ethics Statement
The study and sample collection were approved by the Institutional Review Board (IRB) at all participating institutions including A Case Control Etiologic Study of Sarcoidosis (ACCESS) Group, Sarcoidosis Genetic Analysis (SAGA) study, Henry Ford Health System in Detroit, Michigan, and Oklahoma Medical Research Foundation (OMRF), Oklahoma City, Oklahoma, Institutional Review Boards (IRBs). Only individuals who signed informed consent forms were included in this study. No minors or children were involved in our study.
Subjects
Our AA sample collection, which comprises 1487 cases and 1504 controls (Figure1, Table 1), was taken from an extensive cohort of AA sarcoidosis patients, family members and controls assembled from 1) case-control pairs collected as a part of a 10 center collaborative study (ACCESS Group) [78], 2) the SAGA sample ascertained through affected sib pairs [79], 3) a nuclear family-based sample ascertained through single sarcoidosis-affected offspring from the Henry Ford Health System in Detroit, Michigan [80], and 4) healthy controls from the OMRF Lupus Family Registry and Repository (LFRR) [81]. The AA cases and their family members were grouped into a discovery set of 818 cases and 908 related and unrelated controls and the other 455 independent cases and 557 independent controls were selected for a replication set after applying quality control measures as described below (Figure 1, Table 1). In addition, genotype data from 180 HapMap controls from Yoruba in Ibadan, Nigeria (YRI) and of African ancestry in Southwest USA (ASW) were obtained from the Illumina HumanOmni1-Quad iControlDB (http://www.illumina.com/science/icontroldb.ilmn) and included into the control group of the AA discovery set, as is common practice in order to increase statistical power [82], [83], [84]. The EA dataset consisted of 518 independent cases and 379 independent controls from the ACCESS and the Henry Ford Health System studies mentioned above. We also assembled external genotype data on 3208 healthy Caucasian controls from the Illumina iControlDB (175), the dbGaP (Accession: phs000187.v1.p1) GENEVA Melanoma study (1047), and the dbGAP (Accession: phs000196.v2.p1) CIDR: NGRC Parkinson’s Disease Study (1986) (Figure 1, Table 1). Each sample collection site received the IRB approval to recruit samples. All samples were processed and genotyped at the OMRF under the auspice of the OMRF IRB.
Genotyping and Quality Control
Genotyping was performed at the OMRF using the Illumina HumanOmni1-Quad array for ∼1.1M variants across the genome. SNPs had to meet the following quality control criteria for inclusion for each population: well-defined cluster plots by visual inspections, call rate >95%, minor allele frequency >0.01, Hardy-Weinberg proportion tests P>0.0001 in cases and P>0.001 in controls, and case-control differences in missingness P>0.001. Copy number variations, X, Y, XY, and mitochondrial chromosomes were not included in the analysis. A total of 864,829 and 682,921 SNPs passed our quality controls in the AA discovery and replication sets and the EA dataset, respectively. We found 657,350 successfully genotyped SNPs that overlap between the panels. Samples were removed from analysis if they were determined to be a duplicate of another sample, cryptic relatedness in the independent datasets (the proportion of alleles shared identical by descent >0.25), displayed low call rates (<90%), exhibited extreme heterozygosity (>5 standard deviations from the mean), demonstrated either outlying principal component values of population membership calculated by EIGENSOFT 3.0 [85] or global ancestry estimates calculated by ADMIXMAP [86], [87], or revealed discrepancies between reported gender and genetic data (Table S1). For the EA dataset, we assigned to each sarcoidosis case the five best-matched controls as determined by identity-by-state (IBS) allele sharing using PLINK v1.07 [88] resulting in a large drop-out of external controls in the EA dataset.
Imputation Method
Imputation was performed in each population at 5 Mb bins across the genome using the IMPUTE2 program [89], [90]. The 1000 Genomes Project Phase I data release (June 2011), which contains haplotypes derived from 1,094 individuals from Africa, Asia, Europe, and the Americas, was used as the reference [89], [90]. IMPUTE2 estimated the posterior probabilities for the three possible genotypes (i.e. AA, AB, and BB). The posterior probabilities were then converted to the most likely genotypes with a threshold of 0.9. Imputed SNPs with either low imputation accuracy (information measure <0.5 and the average maximum posterior genotype call probability <0.9) and that failed the SNP quality control standards described above were removed in order to minimize false positives. After imputation, 10,948,298 SNPs in the AA discovery set, 11,160,451 SNPs in the AA replication set, and 6,620,482 SNPs in the EA replication set passed quality control measures for analysis.
Association Analyses
Because our discovery set contained related individuals, association analysis to any single marker in this set was performed using the Efficient Mixed-Model Association eXpedited (EMMAX) software [91], [92]. EMMAX was chosen because it implements a variance component approach in the linear mixed-model that simultaneously adjusts for both pairwise genetic relatedness between individuals and corrects for population stratification using an empirical kinship matrix based on the proportion of alleles at all genome-wide SNPs shared identical-by-state between all pairs of individuals in the study [91]. We assumed an additive model [91], [92] and adjusted the statistics for gender. Since EMMAX does not calculate odds ratios (ORs), we estimated these using logistic regression as implemented in PLINK using independent samples (480 cases and 367 controls) ascertained from the AA discovery set. The association analyses of the independent sets of AAs and EAs were calculated using logistic regression in PLINK. We assumed the additive genetic model and adjusted the statistics for gender and the first five principal components of each population (calculated using EIGENSOFT 3.0). Meta-analyses were performed using the weighted Z-score method that accounts for the direction of effects and sample-size as implemented in METAL [93]. Both the Cochran’s Q test statistic and I 2 index were used to test for heterogeneity in the meta-analysis of all samples. The Cochran’s Q test calculates the weighted sum of the squared deviations between each study effects and the overall effect across studies [94], whereas the I 2 index quantifies the percentage of inconsistency across studies due to heterogeneity rather than by chance [95]. The Q test with P<0.05 or I 2>50% indicates the presence of heterogeneity. Stepwise conditional association analysis in AAs was conducted for SNPs with P<5×10−8 using EMMAX adjusting for gender and SNPs of interest, a SNP added at a time. We required a SNP threshold of P<5×10−8 to be considered significantly associated and P<1×10−4 to be considered suggestively associated with sarcoidosis [96], [97], [98].
The power calculations for different minor allele frequencies and odds ratios for each dataset were performed using the Genetic Power Calculator program [99] and have been summarized in Figure S2. The assumptions are a disease prevalence of 0.05%, complete linkage disequilibrium between SNP and predisposing loci, an additive genetic model and a type I error rate α = 5×10−8. To present power curves that are comparable across sets, we used a power calculator that assumes independence, but adjusted the analysis of the AA discovery set (family-based set) assuming a familial correlation of 0.25 since most pairs are siblings (and thus smaller equivalent count or 75% of the total cases and controls in this set).
Supporting Information
The quantile-quantile (Q–Q) plots of the observed and expected distributions of P-values. (A–C) The Q–Q plots for (A) the AA discovery set (genomic control inflation factor [λGC] = 0.980), (B) the AA replication set (λGC = 1.030), and (C) the EA dataset (λGC = 1.027).
(DOC)
Power calculation plots of the GWAS datasets. (A–C) Power calculation plots for the AA discovery set (A), the AA replication set (B), and the EA dataset (C).
(DOC)
Regional association plots of SNP-sarcoidosis association test results within NOTCH4. (A–D) Association results in the AA discovery set (A), AA replication set (B), a meta-analysis between the AA discovery and AA replication sets including the LD (D’) plot (C), and the EA dataset including the LD (D’) plot (D). Each SNP is colored according to its LD (r 2) with the top SNP. The blue solid line denotes the recombination rate.
(DOC)
Summary of dropped samples after QC.
(DOC)
Association results with P<5×10−8 in either dataset.
(XLS)
Stepwise conditional analysis in AA samples for SNPs in the MHC region with P<5×10−8.
(XLS)
Stepwise conditional analysis in EA samples for SNPs in the MHC region with P<5×10−8.
(XLS)
Association results with P<1×10−4 in either dataset.
(XLS)
Shared or Ethnic Specific Suggestive Association Regions supported by the heterogeneity test results and list of inflammatory or lung diseases associated with these regions.
(DOC)
Acknowledgments
We are grateful to all sarcoidosis patients and controls for participation in this study. We would like to express our gratitude to the research assistants, coordinators and physicians that helped in the recruitment of subjects.
Funding Statement
This work was supported by the United States National Institutes of Health (NIH) grants: 1RC2HL101499 to CGM, R56-AI072727 and R01-HL092576 to BAR, R01-HL54306 and U01-HL060263 awarded to MCI, R01-AR043274 to KLM, R37-AI024717, R01-AR042460, and P20-RR020143 to JBH, N01-AR62277 to JBH and KLM, P01-AR049084 to JBH and RPK, P01-AI083194 to JBH, KMK and RPK. Other institution supports include the United States Department of Veterans Affairs to JBH and KMK and the United States Department of Defense PR094002 to JBH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Iannuzzi MC, Rybicki BA, Teirstein AS (2007) Sarcoidosis. N Engl J Med 357: 2153–2165. [DOI] [PubMed] [Google Scholar]
- 2. Iwai K, Tachibana T, Takemura T, Matsui Y, Kitaichi M, et al. (1993) Pathological studies on sarcoidosis autopsy. I. Epidemiological features of 320 cases in Japan. Acta Pathol Jpn 43: 372–376. [DOI] [PubMed] [Google Scholar]
- 3. James DG (1997) Descriptive definition and historic aspects of sarcoidosis. Clin Chest Med 18: 663–679. [DOI] [PubMed] [Google Scholar]
- 4. Newman LS, Rose CS, Bresnitz EA, Rossman MD, Barnard J, et al. (2004) A case control etiologic study of sarcoidosis: environmental and occupational risk factors. Am J Respir Crit Care Med 170: 1324–1330. [DOI] [PubMed] [Google Scholar]
- 5. Kucera GP, Rybicki BA, Kirkey KL, Coon SW, Major ML, et al. (2003) Occupational risk factors for sarcoidosis in African-American siblings. Chest 123: 1527–1535. [DOI] [PubMed] [Google Scholar]
- 6. Rybicki BA, Amend KL, Maliarik MJ, Iannuzzi MC (2004) Photocopier exposure and risk of sarcoidosis in African-American sibs. Sarcoidosis Vasc Diffuse Lung Dis 21: 49–55. [DOI] [PubMed] [Google Scholar]
- 7. Rybicki BA, Iannuzzi MC, Frederick MM, Thompson BW, Rossman MD, et al. (2001) Familial aggregation of sarcoidosis. A case-control etiologic study of sarcoidosis (ACCESS). Am J Respir Crit Care Med 164: 2085–2091. [DOI] [PubMed] [Google Scholar]
- 8. Schurmann M, Reichel P, Muller-Myhsok B, Schlaak M, Muller-Quernheim J, et al. (2001) Results from a genome-wide search for predisposing genes in sarcoidosis. AmJRespirCrit Care Med 164: 840–846. [DOI] [PubMed] [Google Scholar]
- 9. Valentonyte R, Hampe J, Huse K, Rosenstiel P, Albrecht M, et al. (2005) Sarcoidosis is associated with a truncating splice site mutation in BTNL2. Nat Genet 37: 357–364. [DOI] [PubMed] [Google Scholar]
- 10. Iannuzzi MC, Iyengar SK, Gray-McGuire C, Elston RC, Baughman RP, et al. (2005) Genome-wide search for sarcoidosis susceptibility genes in African Americans. Genes Immun 6: 509–518. [DOI] [PubMed] [Google Scholar]
- 11. Hofmann S, Franke A, Fischer A, Jacobs G, Nothnagel M, et al. (2008) Genome-wide association study identifies ANXA11 as a new susceptibility locus for sarcoidosis. Nat Genet 40: 1103–1106. [DOI] [PubMed] [Google Scholar]
- 12. Franke A, Fischer A, Nothnagel M, Becker C, Grabe N, et al. (2008) Genome-wide association analysis in sarcoidosis and Crohn’s disease unravels a common susceptibility locus on 10p12.2. Gastroenterology 135: 1207–1215. [DOI] [PubMed] [Google Scholar]
- 13. Siltzbach LE, James DG, Neville E, Turiaf J, Battesti JP, et al. (1974) Course and prognosis of sarcoidosis around the world. Am J Med 57: 847–852. [DOI] [PubMed] [Google Scholar]
- 14. James DG, Sherlock S (1994) Sarcoidosis of the liver. Sarcoidosis 11: 2–6. [PubMed] [Google Scholar]
- 15. Cozier YC, Berman JS, Palmer JR, Boggs DA, Serlin DM, et al. (2011) Sarcoidosis in black women in the United States: data from the Black Women’s Health Study. Chest 139: 144–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rybicki BA, Major M, Popovich J Jr, Maliarik MJ, Iannuzzi MC (1997) Racial differences in sarcoidosis incidence: a 5-year study in a health maintenance organization. Am J Epidemiol 145: 234–241. [DOI] [PubMed] [Google Scholar]
- 17. Sartwell PE, Edwards LB (1974) Epidemiology of sarcoidosis in the U.S. Navy. Am J Epidemiol 99: 250–257. [DOI] [PubMed] [Google Scholar]
- 18. Cummings MM, Dunner E, Schmidt RH Jr, Barnwell JB (1956) Concepts of epidemiology of sarcoidosis; preliminary report of 1,194 cases reviewed with special reference to geographic ecology. Postgrad Med 19: 437–446. [DOI] [PubMed] [Google Scholar]
- 19. Gundelfinger BF, Britten SA (1961) Sarcoidosis in the United States Navy. Am Rev Respir Dis 84(5)Pt 2: 109–115. [DOI] [PubMed] [Google Scholar]
- 20. Edmondstone WM, Wilson AG (1985) Sarcoidosis in Caucasians, Blacks and Asians in London. Br J Dis Chest 79: 27–36. [DOI] [PubMed] [Google Scholar]
- 21. Brewerton DA, Cockburn C, James DC, James DG, Neville E (1977) HLA antigens in sarcoidosis. Clin Exp Immunol 27: 227–229. [PMC free article] [PubMed] [Google Scholar]
- 22. Berlin M, Fogdell-Hahn A, Olerup O, Eklund A, Grunewald J (1997) HLA-DR predicts the prognosis in Scandinavian patients with pulmonary sarcoidosis. American journal of respiratory and critical care medicine 156: 1601–1605. [DOI] [PubMed] [Google Scholar]
- 23. Rossman MD, Thompson B, Frederick M, Maliarik M, Iannuzzi MC, et al. (2003) HLA-DRB1*1101: a significant risk factor for sarcoidosis in blacks and whites. Am J Hum Genet 73: 720–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grunewald J, Eklund A, Olerup O (2004) Human leukocyte antigen class I alleles and the disease course in sarcoidosis patients. Am J Respir Crit Care Med 169: 696–702. [DOI] [PubMed] [Google Scholar]
- 25. Rybicki BA, Walewski JL, Maliarik MJ, Kian H, Iannuzzi MC (2005) The BTNL2 gene and sarcoidosis susceptibility in African Americans and Whites. Am J Hum Genet 77: 491–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li Y, Wollnik B, Pabst S, Lennarz M, Rohmann E, et al. (2006) BTNL2 gene variant and sarcoidosis. Thorax 61: 273–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofmann S, Fischer A, Till A, Muller-Quernheim J, Hasler R, et al. (2011) A genome-wide association study reveals evidence of association with sarcoidosis at 6p12.1. Eur Respir J. [DOI] [PubMed]
- 28. Rybicki BA, Levin AM, McKeigue P, Datta I, Gray-McGuire C, et al. (2011) A genome-wide admixture scan for ancestry-linked genes predisposing to sarcoidosis in African-Americans. Genes Immun 12: 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miretti MM, Walsh EC, Ke X, Delgado M, Griffiths M, et al. (2005) A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polymorphisms. Am J Hum Genet 76: 634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gray-McGuire C, Sinha R, Iyengar S, Millard C, Rybicki BA, et al. (2006) Genetic characterization and fine mapping of susceptibility loci for sarcoidosis in African Americans on chromosome 5. Hum Genet 120: 420–430. [DOI] [PubMed] [Google Scholar]
- 31. Song W, Nadeau P, Yuan M, Yang X, Shen J, et al. (1999) Proteolytic release and nuclear translocation of Notch-1 are induced by presenilin-1 and impaired by pathogenic presenilin-1 mutations. Proc Natl Acad Sci U S A 96: 6959–6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maillard I, Adler SH, Pear WS (2003) Notch and the immune system. Immunity 19: 781–791. [DOI] [PubMed] [Google Scholar]
- 33. Noseda M, McLean G, Niessen K, Chang L, Pollet I, et al. (2004) Notch activation results in phenotypic and functional changes consistent with endothelial-to-mesenchymal transformation. Circulation research 94: 910–917. [DOI] [PubMed] [Google Scholar]
- 34. Noseda M, Chang L, McLean G, Grim JE, Clurman BE, et al. (2004) Notch activation induces endothelial cell cycle arrest and participates in contact inhibition: role of p21Cip1 repression. Molecular and cellular biology 24: 8813–8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu ZJ, Shirakawa T, Li Y, Soma A, Oka M, et al. (2003) Regulation of Notch1 and Dll4 by vascular endothelial growth factor in arterial endothelial cells: implications for modulating arteriogenesis and angiogenesis. Molecular and cellular biology 23: 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Quillard T, Devalliere J, Coupel S, Charreau B (2010) Inflammation dysregulates Notch signaling in endothelial cells: implication of Notch2 and Notch4 to endothelial dysfunction. Biochemical pharmacology 80: 2032–2041. [DOI] [PubMed] [Google Scholar]
- 37. Uyttendaele H, Marazzi G, Wu G, Yan Q, Sassoon D, et al. (1996) Notch4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development 122: 2251–2259. [DOI] [PubMed] [Google Scholar]
- 38. Collins BJ, Kleeberger W, Ball DW (2004) Notch in lung development and lung cancer. Seminars in cancer biology 14: 357–364. [DOI] [PubMed] [Google Scholar]
- 39. Miniati D, Jelin EB, Ng J, Wu J, Carlson TR, et al. (2010) Constitutively active endothelial Notch4 causes lung arteriovenous shunts in mice. American journal of physiology Lung cellular and molecular physiology 298: L169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li X, Howard TD, Moore WC, Ampleford EJ, Li H, et al. (2011) Importance of hedgehog interacting protein and other lung function genes in asthma. The Journal of allergy and clinical immunology 127: 1457–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xu K, Moghal N, Egan SE (2012) Notch signaling in lung development and disease. Advances in experimental medicine and biology 727: 89–98. [DOI] [PubMed] [Google Scholar]
- 42. Barcellos LF, May SL, Ramsay PP, Quach HL, Lane JA, et al. (2009) High-density SNP screening of the major histocompatibility complex in systemic lupus erythematosus demonstrates strong evidence for independent susceptibility regions. PLoS Genet 5: e1000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Duvefelt K, Anderson M, Fogdell-Hahn A, Hillert J (2004) A NOTCH4 association with multiple sclerosis is secondary to HLA-DR*1501. Tissue Antigens 63: 13–20. [DOI] [PubMed] [Google Scholar]
- 44. Gorlova O, Martin JE, Rueda B, Koeleman BP, Ying J, et al. (2011) Identification of novel genetic markers associated with clinical phenotypes of systemic sclerosis through a genome-wide association strategy. PLoS genetics 7: e1002178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fellay J, Ge D, Shianna KV, Colombo S, Ledergerber B, et al. (2009) Common genetic variation and the control of HIV-1 in humans. PLoS Genet 5: e1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Grigorian A, Hurford R, Chao Y, Patrick C, Langford TD (2008) Alterations in the Notch4 pathway in cerebral endothelial cells by the HIV aspartyl protease inhibitor, nelfinavir. BMC Neurosci 9: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Luo X, Klempan TA, Lappalainen J, Rosenheck RA, Charney DS, et al. (2004) NOTCH4 gene haplotype is associated with schizophrenia in African Americans. Biol Psychiatry 55: 112–117. [DOI] [PubMed] [Google Scholar]
- 48. Sklar P, Schwab SG, Williams NM, Daly M, Schaffner S, et al. (2001) Association analysis of NOTCH4 loci in schizophrenia using family and population-based controls. Nat Genet 28: 126–128. [DOI] [PubMed] [Google Scholar]
- 49. Dubaniewicz A, Moszkowska G (2007) DQA1*03011 allele: protective or an adverse effect on the development of sarcoidosis; preliminary study. Respiratory medicine 101: 2213–2216. [DOI] [PubMed] [Google Scholar]
- 50. Todd JA, Acha-Orbea H, Bell JI, Chao N, Fronek Z, et al. (1988) A molecular basis for MHC class II–associated autoimmunity. Science 240: 1003–1009. [DOI] [PubMed] [Google Scholar]
- 51. Grusby MJ, Glimcher LH (1995) Immune responses in MHC class II-deficient mice. Annual review of immunology 13: 417–435. [DOI] [PubMed] [Google Scholar]
- 52. Taipale J, Beachy PA (2001) The Hedgehog and Wnt signalling pathways in cancer. Nature 411: 349–354. [DOI] [PubMed] [Google Scholar]
- 53. Jacob J, Briscoe J (2003) Gli proteins and the control of spinal-cord patterning. EMBO Rep 4: 761–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kim IG, Gorman JJ, Park SC, Chung SI, Steinert PM (1993) The deduced sequence of the novel protransglutaminase E (TGase3) of human and mouse. J Biol Chem 268: 12682–12690. [PubMed] [Google Scholar]
- 55. Alaedini A, Green PH (2008) Autoantibodies in celiac disease. Autoimmunity 41: 19–26. [DOI] [PubMed] [Google Scholar]
- 56. Uemura N, Nakanishi Y, Kato H, Saito S, Nagino M, et al. (2009) Transglutaminase 3 as a prognostic biomarker in esophageal cancer revealed by proteomics. Int J Cancer 124: 2106–2115. [DOI] [PubMed] [Google Scholar]
- 57. Mehul B, Bernard D, Brouard M, Delattre C, Schmidt R (2006) Influence of calcium on the proteolytic degradation of the calmodulin-like skin protein (calmodulin-like protein 5) in psoriatic epidermis. Exp Dermatol 15: 469–477. [DOI] [PubMed] [Google Scholar]
- 58. Candi E, Oddi S, Paradisi A, Terrinoni A, Ranalli M, et al. (2002) Expression of transglutaminase 5 in normal and pathologic human epidermis. J Invest Dermatol 119: 670–677. [DOI] [PubMed] [Google Scholar]
- 59. Nair RP, Stuart PE, Nistor I, Hiremagalore R, Chia NV, et al. (2006) Sequence and haplotype analysis supports HLA-C as the psoriasis susceptibility 1 gene. American journal of human genetics 78: 827–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rubin LA, Amos CI, Wade JA, Martin JR, Bale SJ, et al. (1994) Investigating the genetic basis for ankylosing spondylitis. Linkage studies with the major histocompatibility complex region. Arthritis and rheumatism 37: 1212–1220. [DOI] [PubMed] [Google Scholar]
- 61. Wang J, Zheng L, Lobito A, Chan FK, Dale J, et al. (1999) Inherited human Caspase 10 mutations underlie defective lymphocyte and dendritic cell apoptosis in autoimmune lymphoproliferative syndrome type II. Cell 98: 47–58. [DOI] [PubMed] [Google Scholar]
- 62. Shin MS, Kim HS, Kang CS, Park WS, Kim SY, et al. (2002) Inactivating mutations of CASP10 gene in non-Hodgkin lymphomas. Blood 99: 4094–4099. [DOI] [PubMed] [Google Scholar]
- 63. Soler Artigas M, Loth DW, Wain LV, Gharib SA, Obeidat M, et al. (2011) Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nature genetics 43: 1082–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cecener G, Tunca B, Egeli U, Karadag M, Vatan O, et al. (2008) Mutation analysis of the FHIT gene in bronchoscopic specimens from patients with suspected lung cancer. Tumori 94: 845–848. [DOI] [PubMed] [Google Scholar]
- 65. Demopoulos K, Arvanitis DA, Vassilakis DA, Siafakas NM, Spandidos DA (2002) MYCL1, FHIT, SPARC, p16(INK4) and TP53 genes associated to lung cancer in idiopathic pulmonary fibrosis. J Cell Mol Med 6: 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Haase D, Meister M, Muley T, Hess J, Teurich S, et al. (2007) FRMD3, a novel putative tumour suppressor in NSCLC. Oncogene 26: 4464–4468. [DOI] [PubMed] [Google Scholar]
- 67. Rosenstiel P, Sina C, End C, Renner M, Lyer S, et al. (2007) Regulation of DMBT1 via NOD2 and TLR4 in intestinal epithelial cells modulates bacterial recognition and invasion. J Immunol 178: 8203–8211. [DOI] [PubMed] [Google Scholar]
- 68. Fukui H, Sekikawa A, Tanaka H, Fujimori Y, Katake Y, et al. (2011) DMBT1 is a novel gene induced by IL-22 in ulcerative colitis. Inflamm Bowel Dis 17: 1177–1188. [DOI] [PubMed] [Google Scholar]
- 69. Renner M, Bergmann G, Krebs I, End C, Lyer S, et al. (2007) DMBT1 confers mucosal protection in vivo and a deletion variant is associated with Crohn’s disease. Gastroenterology 133: 1499–1509. [DOI] [PubMed] [Google Scholar]
- 70. Keller AD, Maniatis T (1991) Identification and characterization of a novel repressor of beta-interferon gene expression. Genes Dev 5: 868–879. [DOI] [PubMed] [Google Scholar]
- 71. Raychaudhuri S, Thomson BP, Remmers EF, Eyre S, Hinks A, et al. (2009) Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat Genet 41: 1313–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, et al. (2008) Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet 40: 955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Anderson CA, Boucher G, Lees CW, Franke A, D’Amato M, et al. (2011) Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet 43: 246–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, et al. (2009) Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet 41: 1234–1237. [DOI] [PubMed] [Google Scholar]
- 75. Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, et al. (2009) A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet 41: 1228–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Nath SK, Han S, Kim-Howard X, Kelly JA, Viswanathan P, et al. (2008) A nonsynonymous functional variant in integrin-alpha(M) (encoded by ITGAM) is associated with systemic lupus erythematosus. Nat Genet 40: 152–154. [DOI] [PubMed] [Google Scholar]
- 77. Adrianto I, Wen F, Templeton A, Wiley G, King JB, et al. (2011) Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus. Nat Genet 43: 253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. ACCESS-Group (1999) Design of a case control etiologic study of sarcoidosis (ACCESS). J Clin Epidemiol 52: 1173–1186. [DOI] [PubMed] [Google Scholar]
- 79. Rybicki BA, Hirst K, Iyengar SK, Barnard JG, Judson MA, et al. (2005) A sarcoidosis genetic linkage consortium: the sarcoidosis genetic analysis (SAGA) study. Sarcoidosis Vasc Diffuse Lung Dis 22: 115–122. [PubMed] [Google Scholar]
- 80. Iannuzzi MC, Maliarik MJ, Poisson LM, Rybicki BA (2003) Sarcoidosis susceptibility and resistance HLA-DQB1 alleles in African Americans. Am J Respir Crit Care Med 167: 1225–1231. [DOI] [PubMed] [Google Scholar]
- 81. Rasmussen A, Sevier S, Kelly JA, Glenn SB, Aberle T, et al. (2011) The lupus family registry and repository. Rheumatology (Oxford) 50: 47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, et al. (2008) Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med 358: 900–909. [DOI] [PubMed] [Google Scholar]
- 83. Genovese G, Tonna SJ, Knob AU, Appel GB, Katz A, et al. (2010) A risk allele for focal segmental glomerulosclerosis in African Americans is located within a region containing APOL1 and MYH9. Kidney Int 78: 698–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Xu Z, Bensen JT, Smith GJ, Mohler JL, Taylor JA (2011) GWAS SNP Replication among African American and European American men in the North Carolina-Louisiana prostate cancer project (PCaP). Prostate 71: 881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, et al. (2006) Principal components analysis corrects for stratification in genome-wide association studies. NatGenet 38: 904–909. [DOI] [PubMed] [Google Scholar]
- 86. Hoggart CJ, Parra EJ, Shriver MD, Bonilla C, Kittles RA, et al. (2003) Control of confounding of genetic associations in stratified populations. Am J Hum Genet 72: 1492–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hoggart CJ, Shriver MD, Kittles RA, Clayton DG, McKeigue PM (2004) Design and analysis of admixture mapping studies. Am J Hum Genet 74: 965–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Howie BN, Donnelly P, Marchini J (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5: e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, et al. (2010) A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kang HM, Sul JH (2010) Service SK, Zaitlen NA, Kong SY, et al (2010) Variance component model to account for sample structure in genome-wide association studies. Nat Genet 42: 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, et al. (2008) Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Willer CJ, Li Y, Abecasis GR (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Cochran WG (1954) The Combination of Estimates from Different Experiments. Biometrics 10: 101–129. [Google Scholar]
- 95. Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273: 1516–1517. [DOI] [PubMed] [Google Scholar]
- 97. McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, et al. (2008) Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 9: 356–369. [DOI] [PubMed] [Google Scholar]
- 98. Dubois PC, Trynka G, Franke L, Hunt KA, Romanos J, et al. (2010) Multiple common variants for celiac disease influencing immune gene expression. Nat Genet 42: 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Purcell S, Cherny SS, Sham PC (2003) Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19: 149–150. [DOI] [PubMed] [Google Scholar]
- 100. Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, et al. (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26: 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The quantile-quantile (Q–Q) plots of the observed and expected distributions of P-values. (A–C) The Q–Q plots for (A) the AA discovery set (genomic control inflation factor [λGC] = 0.980), (B) the AA replication set (λGC = 1.030), and (C) the EA dataset (λGC = 1.027).
(DOC)
Power calculation plots of the GWAS datasets. (A–C) Power calculation plots for the AA discovery set (A), the AA replication set (B), and the EA dataset (C).
(DOC)
Regional association plots of SNP-sarcoidosis association test results within NOTCH4. (A–D) Association results in the AA discovery set (A), AA replication set (B), a meta-analysis between the AA discovery and AA replication sets including the LD (D’) plot (C), and the EA dataset including the LD (D’) plot (D). Each SNP is colored according to its LD (r 2) with the top SNP. The blue solid line denotes the recombination rate.
(DOC)
Summary of dropped samples after QC.
(DOC)
Association results with P<5×10−8 in either dataset.
(XLS)
Stepwise conditional analysis in AA samples for SNPs in the MHC region with P<5×10−8.
(XLS)
Stepwise conditional analysis in EA samples for SNPs in the MHC region with P<5×10−8.
(XLS)
Association results with P<1×10−4 in either dataset.
(XLS)
Shared or Ethnic Specific Suggestive Association Regions supported by the heterogeneity test results and list of inflammatory or lung diseases associated with these regions.
(DOC)