Abstract
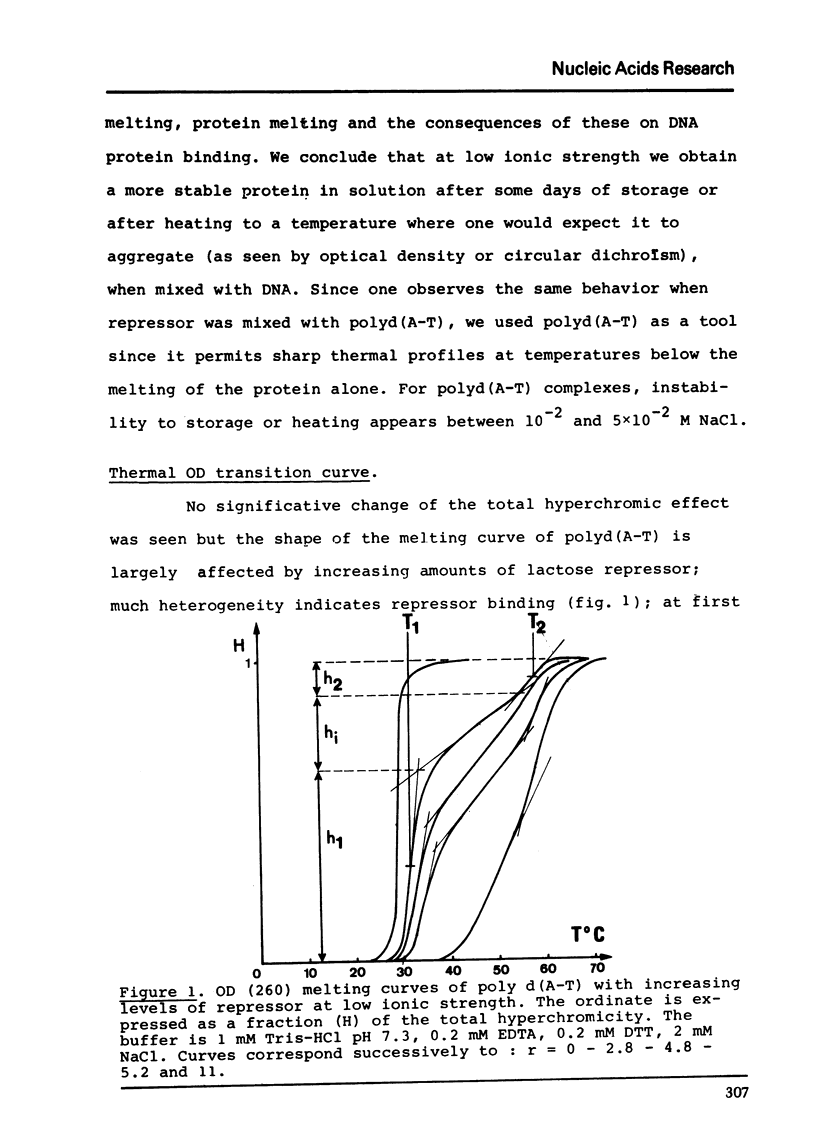
The binding of lactose repressor to poly d(A-T) at low ionic strength has been investigated by heat denaturation. The poly d(A-T) melting is monitored by optical density and the protein melting by circular dichroism. From the modification of the poly d(A-T) melting curve we estimate a maximum binding ratio of about one tetrameric repressor to about 20 bases pairs. The repressor melting can be interpreted as a global shift from α to β structure of about 25 residues per subunit. The melting curves of poly d(A-T) and repressor have not a shape easy to interpret; nevertheless both show a cooperative transition in the same temperature range where we can evaluate that about 3.8 aminoacid residues shift from α to β structure when 1 basespair melt.
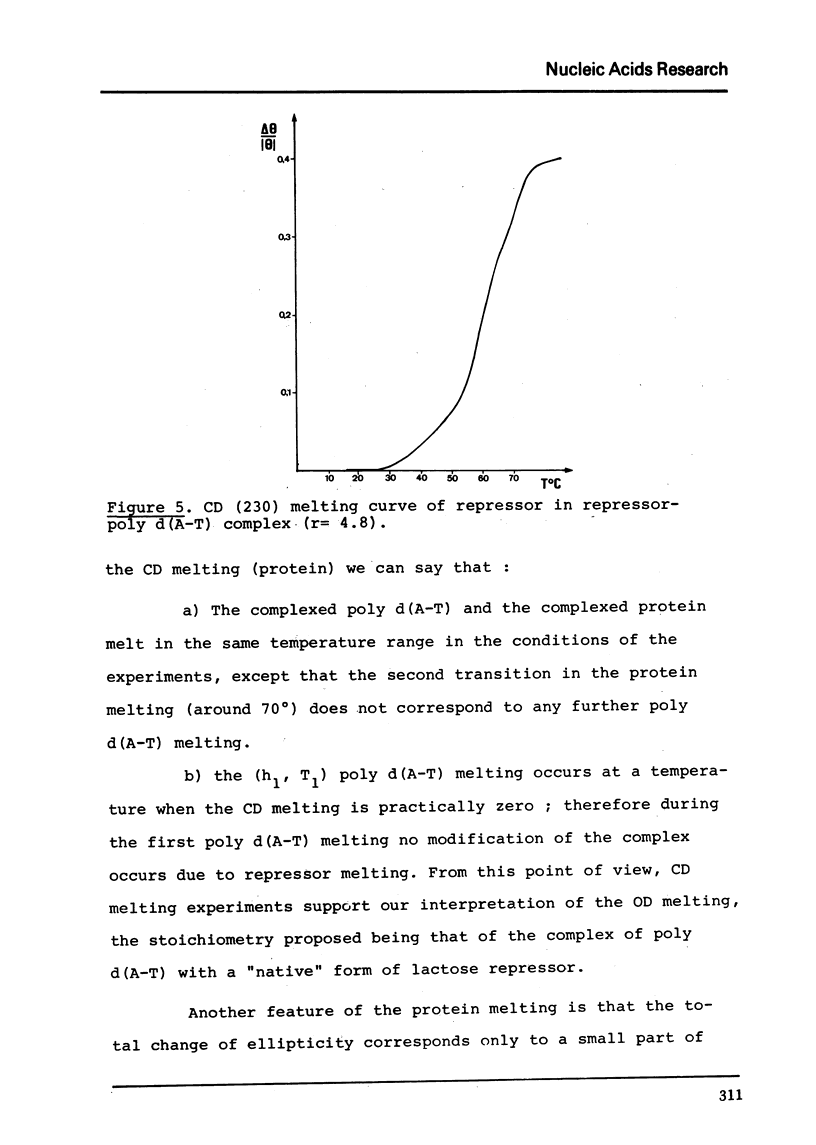
Full text
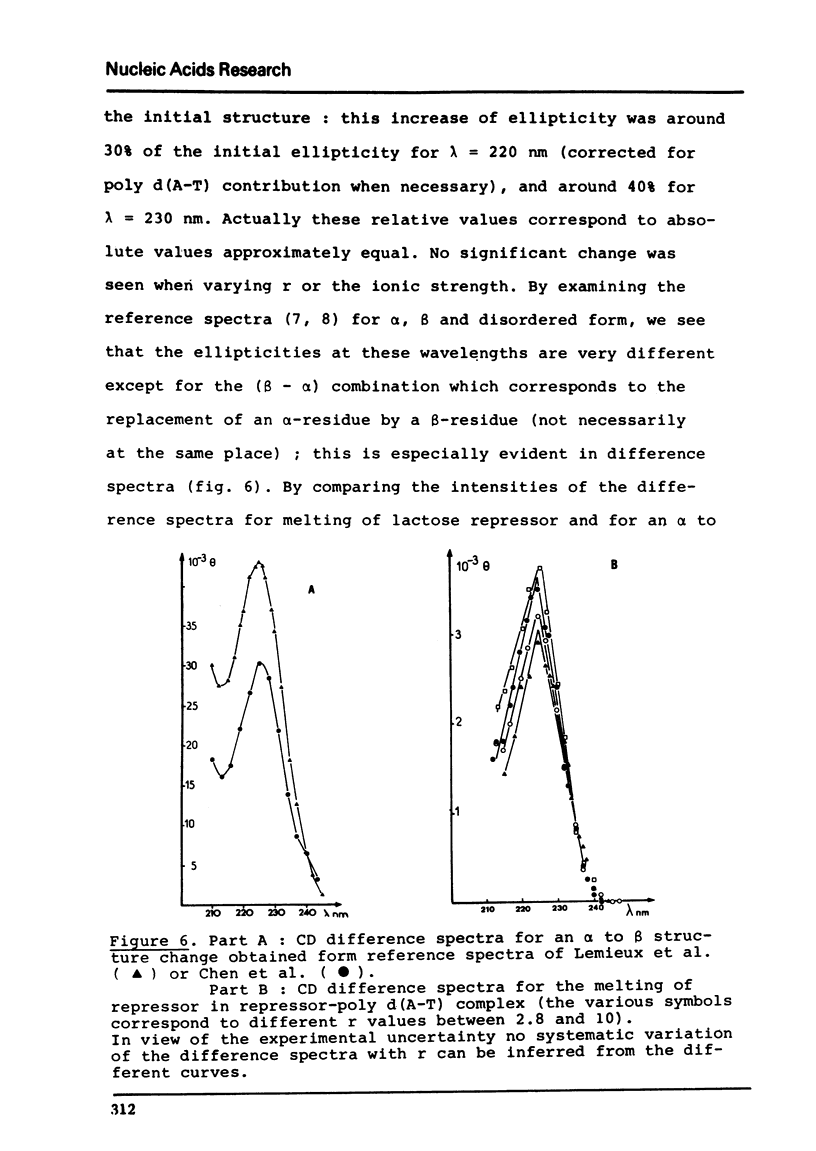
PDF









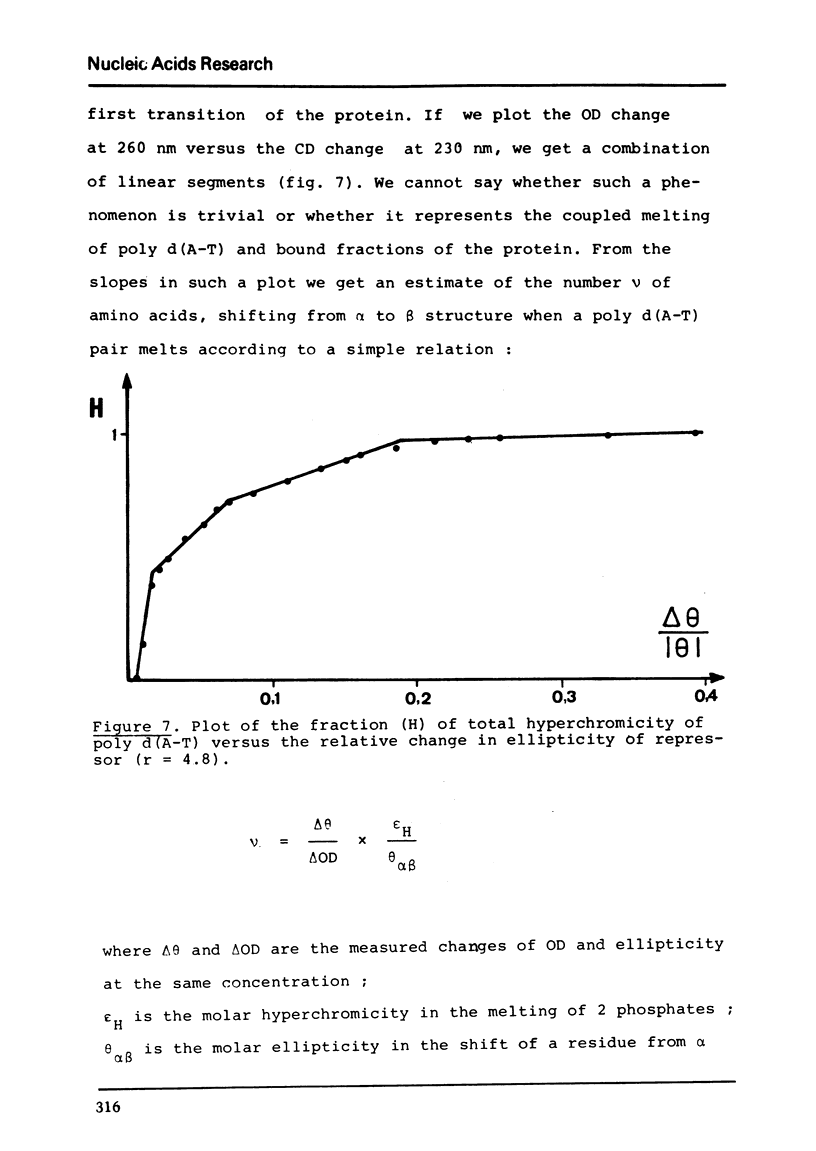


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barth G., Bunnenberg E., Djerassi C. Magnetic circular dichroism studies. XIX. Determination of the tyrosine: tryptophan ratio in proteins. Anal Biochem. 1972 Aug;48(2):471–479. doi: 10.1016/0003-2697(72)90100-5. [DOI] [PubMed] [Google Scholar]
- Beyreuther K., Adler K., Geisler N., Klemm A. The amino-acid sequence of lac repressor. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3576–3580. doi: 10.1073/pnas.70.12.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. H., Yang J. T., Martinez H. M. Determination of the secondary structures of proteins by circular dichroism and optical rotatory dispersion. Biochemistry. 1972 Oct 24;11(22):4120–4131. doi: 10.1021/bi00772a015. [DOI] [PubMed] [Google Scholar]
- Gilbert W., Maxam A. The nucleotide sequence of the lac operator. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3581–3584. doi: 10.1073/pnas.70.12.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiken S. L., Gross C. A., Von Hippel P. H. Equilibrium and kinetic studies of Escherichia coli lac repressor-inducer interactions. J Mol Biol. 1972 Apr 28;66(1):143–155. doi: 10.1016/s0022-2836(72)80012-3. [DOI] [PubMed] [Google Scholar]
- Lemieux G., Lefevre J. F., Daune M. Effect of reconstitution conditions on the structure of Escherichia coli 30-S ribosomol-subunit components. Eur J Biochem. 1974 Nov 1;49(1):185–194. doi: 10.1111/j.1432-1033.1974.tb03824.x. [DOI] [PubMed] [Google Scholar]
- Matsuura M., Oshima Y., Horiuchi T. Secondary structure of the lac repressor. Biochem Biophys Res Commun. 1972 Jun 28;47(6):1438–1443. doi: 10.1016/0006-291x(72)90233-1. [DOI] [PubMed] [Google Scholar]
- Steitz T. A., Richmond T. J., Wise D., Engelman D. The lac repressor protein: molecular shape, subunit structure, and proposed model for operator interaction based on structural studies of microcrystals. Proc Natl Acad Sci U S A. 1974 Mar;71(3):593–597. doi: 10.1073/pnas.71.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]



