Abstract
Scrub typhus, caused by Orientia tsutsugamushi infection, is one of the main causes of acute febrile illness in the Asian-Pacific region. Although early diagnosis and immediate antibiotic treatment are critical for reducing disease severity and mortality, current diagnostic methods using serological and molecular approaches have some limitations in sensitivity and applicability in clinical laboratories. In this study, we identified and characterized O. tsutsugamushi surface cell antigen (sca) family genes encoding autotransporter proteins in order to test them as novel diagnostic targets. We evaluated antibody responses against the Sca proteins in scrub typhus patient sera and examined the genetic diversity of these genes in different strains after PCR amplification. Specific antibody responses against ScaA and ScaC were observed in patients with high indirect immunofluorescence assay titers (≥1:640), whereas specific responses against ScaB and ScaE were relatively low. Genetic analysis using genomic DNAs revealed the sca genes to be quite variable among the different strains. In contrast to scaA, scaC, and scaD, which were detected in all of the tested strains, scaB and scaE were amplified differentially from the different strains, suggesting a differential presence of the genes in the genomes. Among the members of the gene family, the sequence of scaC is the most highly conserved between the different strains, and the size of scaD is the most variable due to the presence of different numbers of internal repeat sequences. These results suggest that the sca genes of O. tsutsugamushi may be valuable targets for use in combination with classical assay methods for scrub typhus diagnosis.
INTRODUCTION
Scrub typhus is an acute febrile illness caused by Orientia tsutsugamushi infection following the bite of an infected larval mite (9). It has been estimated that 1 billion people are at risk, and 1 million new cases arise annually in the Asian-Pacific region (23). This infectious disease has recently become an important public health issue due to regional outbreaks (16, 18) and new emergence (6, 28). Clinical presentations of scrub typhus, typically characterized by eschar, fever, rash, lymphadenopathy, and myalgia, can vary in severity from a mild and self-limiting flu-like syndrome to a life-threatening disease (13, 25). Although early diagnosis and immediate antibiotic treatment are important to prevent severe complications of scrub typhus (27), the clinical discrimination of scrub typhus from other undifferentiated fevers, such as dengue and leptospirosis, is often very difficult because the clinical symptoms of these illnesses are similar (15, 20). In addition, diagnosis of scrub typhus requires laboratory confirmation, usually by serologic detection of antibodies against the bacterial pathogen during the acute and convalescent phases of the disease, and the gold standard is the indirect immunofluorescence assay (IFA), which requires laborious bacterial culture in a biosafety level 3 facility (15). Moreover, despite their widespread use, all of the currently available serologic tests and PCR-based nucleic acid amplification methods have limitations which clinicians need to be aware of (15). For example, the bacterial antigen used for IFA is of high importance due to the risk of significant bias depending on the antigenic and genetic variation (14) of local strains in different regions of endemicity. The sensitivity of PCR-based nucleic acid amplification methods, which mainly target the gene for a major outer membrane protein, TSA56, has been reported to be as low as 50% (15). Although PCR targeting of the tsa56 gene has been shown to be highly specific, sequence variability may affect primer annealing and test sensitivity (15). Therefore, it has been proposed that new diagnostic assays using a panel of both serological and antigen detection systems targeting multiple antigens need to be developed to improve sensitivity (15, 20).
Recently, our group reported that one gene (scaC) among a group of genes encoding autotransporter proteins in the O. tsutsugamushi genome is involved in bacterial adhesion to eukaryotic host cells, potentially through binding to host fibronectin (12). The scaC gene is highly conserved among different strains, and antibodies against the ScaC protein are detected in scrub typhus patients (12). Rickettsia species, comprising a sister group of Orientia, also possess multiple sca genes in their genomes (2). The rickettsial Sca proteins are involved in bacterial adhesion or the invasion process (3, 24) and have been targeted for vaccine development (4). In order to discover whether the sca genes of O. tsutsugamushi can be used as novel diagnostic targets, we analyzed for the first time the antibody responses against diverse Sca proteins in scrub typhus patients and sequence variations of sca genes of different strains of O. tsutsugamushi.
MATERIALS AND METHODS
Ethics statements.
Ethical approval for this work was granted by the institutional review boards of both Seoul National University Hospital (IRB no. 0-1001-039-307) and Chungnam National University Hospital (IRB no. 2008-10-08). All patients and healthy volunteers provided written informed consent prior to sample collection.
Patient samples.
Human sera were collected during the scrub typhus endemic season (from September to January) from healthy volunteers (n = 10) and patients with acute febrile disease (n = 100) at Chungnam National University Hospital in Daejeon, Republic of Korea, after informed consent was obtained. All of the patients resided in the Chungcheong Province, in the middle part of the Republic of Korea, where the Boryong, Gilliam, and Karp strains are the most prevalent (5, 22). Primary diagnosis of scrub typhus was performed by IFA, and the O. tsutsugamushi-specific IFA titer was determined.
Preparation of O. tsutsugamushi and its genomic DNA.
O. tsutsugamushi strains Boryong, Gilliam, Karp, and Kato were propagated in L929 cells (NTCT929; ATCC) and used for indirect immunofluorescence assays (see below) and the preparation of genomic DNA. O. tsutsugamushi was purified using a modification of a Percoll gradient purification method (17). At 3 to 4 days postinfection, infectivity was determined by IFA. When an infection rate of >90% was achieved, cells were harvested by centrifugation at 6,000 × g for 20 min. The cell pellet was resuspended with 6.5 ml of Tris-sucrose (TS) buffer (33 mM Tris-Cl [pH 7.4] and 0.25 M sucrose). Resuspended cells were homogenized with a Polytron homogenizer (Wheaton Inc., Millville, NJ) for 100 strokes and centrifuged at 200 × g for 5 min. The supernatant was mixed with 40% Percoll (Pharmacia Fine Chemicals, Uppsala, Sweden) in TS buffer and centrifuged at 25,000 × g for 60 min. The bacterial band was collected and centrifuged at 77,000 × g for 30 min. O. tsutsugamushi was collected and used for genomic DNA extraction by use of an RBC genomic DNA extraction kit (RBC Bioscience Co., Taipei, Taiwan) according to the manufacturer's instructions.
Cloning, expression, and purification of proteins.
The tsa56, p47, and sca genes were PCR amplified from O. tsutsugamushi genomic DNA by use of the specific primer pairs presented in Table 1. The amplified region of each gene is summarized in Table 1 and Fig. 1A. The amplified DNAs were then cloned into a pET28a vector for sequence analysis and protein expression. All of the genes used for protein purification were derived from the O. tsutsugamushi Boryong strain, and the sca genes used for sequence analysis were amplified from the genomic DNAs of the Boryong, Gilliam, Karp, and Kato strains. Escherichia coli BL21(DE3) (Novagen, Darmstadt, Germany) was transformed with the recombinant plasmids for protein expression and purification. Protein expression was induced by adding 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG; Duchefa, Zwijndrecht, Netherlands) for 4 h at 37°C. Bacteria were harvested by centrifugation at 1,000 × g for 10 min, resuspended in binding buffer (300 mM NaCl, 50 mM sodium phosphate buffer, 10 mM imidazole) containing 1 mg/ml of lysozyme, incubated at 4°C for 30 min, and disrupted by sonication on ice for 5 min. The sonicated lysates were centrifuged at 1,600 × g for 20 min at 4°C, and the supernatants were applied to a Ni-nitrilotriacetic acid (Ni-NTA) His-binding resin (Novagen) preequilibrated with binding buffer. His-tagged proteins bound to the Ni-NTA resin were eluted with elution buffer (300 mM NaCl, 50 mM sodium phosphate buffer, 250 mM imidazole) and serially dialyzed against elution buffer to remove any free imidazole. Finally, the identity and purity of the proteins were assessed by Western blotting and Coomassie blue staining, respectively.
Table 1.
Primer sequences used in this study
| Gene | Primer direction | Primer sequencea | Product size (bp) (amplified region [nt positions])b |
|---|---|---|---|
| tsa56 | Forward | GGCGGATCCCCAGGATTTAGAGCAGAG | 1,344 (253–1596) |
| Reverse | CGGTCGACGAAGTTATAGCGCACA | ||
| p47 | Forward | CGGGATCCATTAGTGTAAATAGTTTATCC | 1,278 (121–1398) |
| Reverse | CGGTCGACCTTATTAATATTAGGTAAAGC | ||
| scaA | Forward | CGGGATCCTCATCTGCTAGAGGTGAGATG | 1,169 (2371–3540) |
| Reverse | CGGTCGACATCGCCTTTTAAACCCGGGTTACT | ||
| Forward | GGCGGATCCTTACTTGGAGATTATACTG | 1,121 (2414–3534) | |
| Reverse | CGCTCGAGTTGTTTATTGATATTAGATCC | ||
| scaB | Forward | GGCGGATCCAGTACAACTCAAAGGATATTAGG | 1,047 (70–1116) |
| Reverse | CGGTCGACACTACTACAAATGTTTGATCC | ||
| scaC | Forward | GGCGGATCCAAAAGTATAACTCCAGAAAAGTG | 600 (97–696) |
| Reverse | CGGTCGACGTTTAATTTAGCACGATTTAT | ||
| Forward | GGCGGATCCATGTACCAATCTAAT | 1,578 (1–1578) | |
| Reverse | CGGTCGACAAAATTAGTTCCTATATG | ||
| scaD | Forward | AAGGATCCCAATTAAGTGAGCGACTA | 1,926 (136–2061) |
| Reverse | CGGAATTCTACCGTAGCAACAGTAACAGC | ||
| scaE | Forward | GGCGGATCCAATGTAAATGCACAGCCCAATAG | 1,332 (91–1422) |
| Reverse | CGGTCGACCTGTACTGTTGCTATTTTAGA | ||
| Forward | GGCGGATCCAATGTAAATGCACAGCCCAA | 1,291 (111–1401) | |
| Reverse | CGGTCGACCTGTACTGTTGCTATTTTAGA |
Restriction enzyme sites are underlined.
For Boryong strain genes.
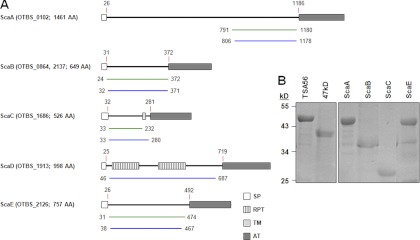
Fig 1.
Schematic representation of the five Sca proteins whose genes were identified in the O. tsutsugamushi genome and demonstration of purified bacterial antigens. (A) Domain structures were predicted using SMART (simple modular architecture research tool [http://smart.embl-heidelberg.de/]). The amino acid positions (indicated by numbers) are based on the sequences from the Boryong strain. The green lines indicate the cloned region for protein expression, and the blue lines show the passenger domains used for sequence analysis after PCR amplification. SP, signal peptide; RPT, internal repeat sequence; TM, transmembrane domain; AT, autotransporter domain; AA, amino acids. (B) The bacterial antigens were cloned into the pET28a expression vector by use of the specific primers summarized in Table 1 and were expressed in E. coli. The His-tagged proteins were purified using Ni-NTA His-binding resin and then visualized by SDS-PAGE and Coomassie brilliant blue staining.
Immunofluorescence assay.
Immunofluorescence assay was used to measure antibody titers against O. tsutsugamushi (12, 14). L929 cells infected with each strain of O. tsutsugamushi (Boryong, Karp, or Gilliam) were harvested, mixed in equal amounts, and used as antigens for immunofluorescence assay. Infected L929 cells were harvested, washed with phosphate-buffered saline (PBS, pH 7.2), spotted onto Teflon-coated spot slides, and fixed with cold acetone for 10 min. The slides were stored at −70°C until used. Twofold serially diluted (1:40 to 1:1,280 in PBS) patient sera were added to the antigen-coated spot on the slide and incubated for 30 min in a moist chamber at room temperature. Alexa Fluor 488-conjugated goat anti-human IgG (Molecular Probes), diluted 1:1,000 in PBS, was used as the secondary antibody. The stained slides were examined under an Olympus FV1000 laser scanning confocal microscope (Olympus, Tokyo, Japan).
ELISA.
All of the purified protein antigens used for enzyme-linked immunosorbent assay (ELISA) were derived from the Boryong strain, since it was reported that this strain covers approximately 80% of O. tsutsugamushi infections in the region of endemicity (5). Immunoassay plates (96-well plates; Nunc, Rochester, NY) were coated overnight with 100 μl of purified His-tagged antigens at a concentration of 1 μg/ml in 0.05 M bicarbonate buffer (pH 9.5) and blocked with 3% bovine serum albumin (BSA) at room temperature for 1 h. Human sera from the patients and healthy volunteers were diluted in PBS containing 3% BSA and 0.1% Tween 20, added to the immunoassay wells, and incubated at 37°C for 1 h. After washing six times with PBS containing 0.05% Tween 20, an anti-human IgG–horseradish peroxidase (HRP) conjugate (Promega), diluted 1:10,000 in PBS containing 3% BSA and 0.1% Tween 20, was added and incubated for 1 h. TMB microwell peroxidase substrate solution (KPL, Gaithersburg, MD) was then added to develop color for 7 min, and the reaction was stopped by the addition of 1 M H3PO4 solution. Absorbance was measured at 450 nm using an ELISA plate reader (Beckman).
Sequence analysis and alignment.
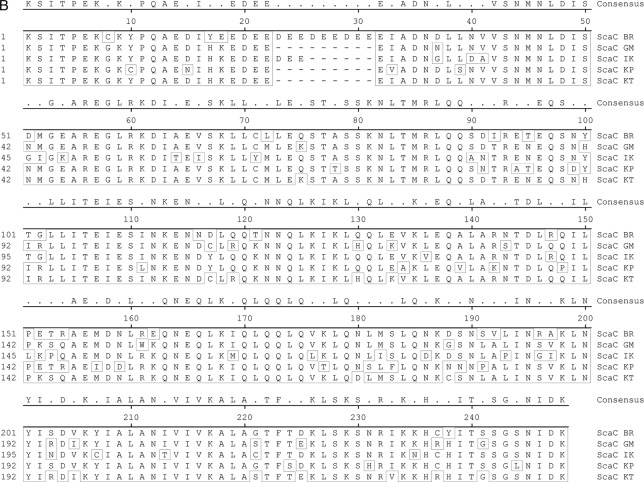
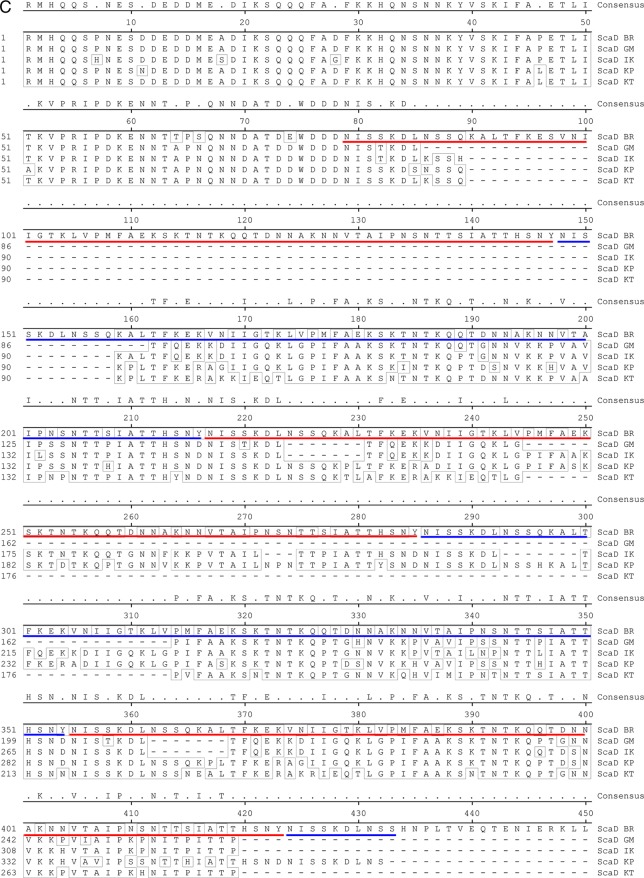
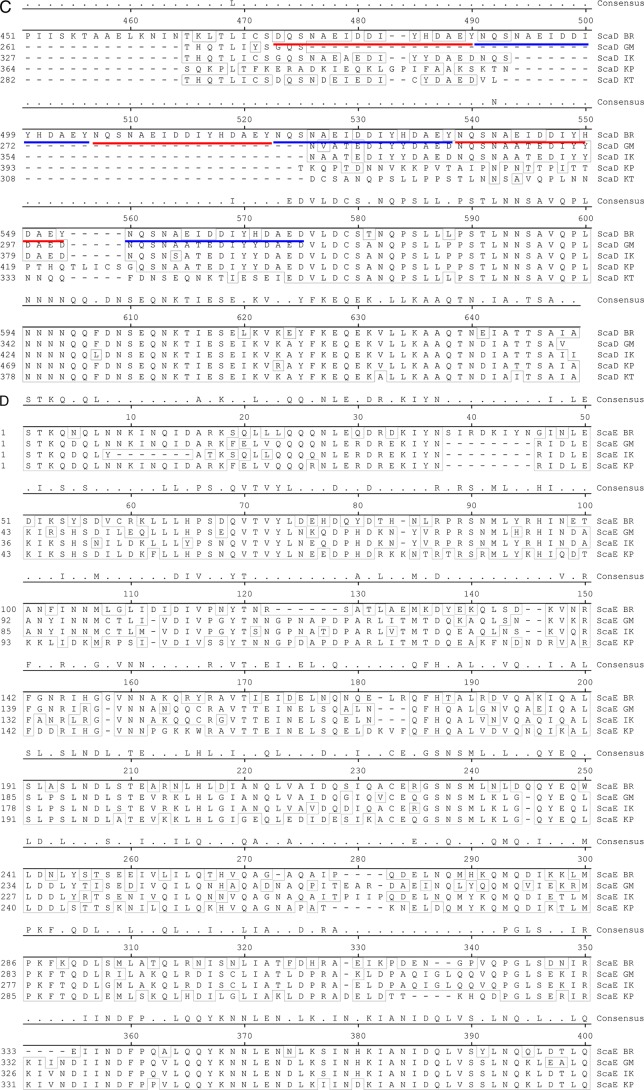
The passenger domains of O. tsutsugamushi sca genes amplified from genomic DNAs of different strains, including Boryong, Gilliam, Karp, and Kato, were sequenced (Macrogen Co., Seoul, Republic of Korea). We also used DNA sequence information for sca genes found in the genomes of O. tsutsugamushi strains Boryong (GenBank accession no. AM494475.1) (9) and Ikeda (GenBank accession no. AP008981.1) (12, 19). The sca gene sequences were aligned for phylogenetic analysis by use of ClustalW embedded within MegAlign software (DNAstar Inc., Madison, WI). The degrees of identity among the Sca protein sequences were determined using calculated alignment. The sequence alignment results for the translated passenger domains from the five different strains are presented in Fig. 5. Phylogenetic analyses of tsa56 genes were performed using sequences deposited in GenBank (accession no. CAM79668 for strain Boryong, ABF50384 for strain Gilliam, YP_001937637 for strain Ikeda, AAA26391 for strain Karp, and AAA26397 for strain Kato).
Fig 5.
Alignment of amino acid sequences of Sca proteins detected in the genomes of O. tsutsugamushi strains. The passenger domains of the genes identified in the indicated strains were compared. (A) ScaA; (B) ScaC; (C) ScaD; (D) ScaE. The conserved sequences are boxed, and the internal repeat sequences detected in ScaD are underlined with red and blue lines.
Statistical analysis.
Statistical analyses were performed using the nonlinear regression module embedded in SigmaPlot (Jandel, San Rafael, CA).
Nucleotide sequence accession numbers.
The passenger domains of the O. tsutsugamushi sca genes amplified from genomic DNAs of different strains were deposited in GenBank under accession no. JQ996623 to JQ996633.
RESULTS
Detection of antibodies against Sca proteins in scrub typhus patients.
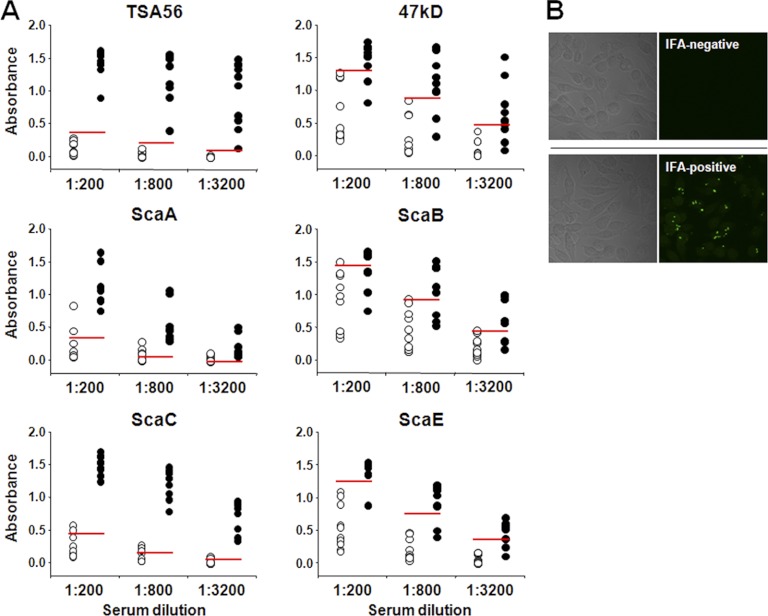
In order to examine anti-Sca protein antibody responses in scrub typhus patients, we screened 100 sera from potential scrub typhus patients during the endemic season in the Republic of Korea. Based on an IFA using L929 cells infected with O. tsutsugamushi, 70 sera with various IFA titers (from 0 to 1,280) were randomly selected and tested in ELISAs using purified Sca proteins (Fig. 1B). IFA-negative sera from 10 healthy volunteers were used to determine the cutoff values. The cutoff value was defined as the mean optical density at 490 nm (OD490) plus 2 standard deviations for each experimental condition (11). To determine the optimal dilution of serum under our ELISA conditions, sera from 10 healthy volunteers, 10 IFA-negative patients, and 10 patients with scrub typhus (IFA titers of ≥1,280) were serially diluted (1:200 to ∼1:3,200) and examined by ELISAs using the different antigens. As shown in Fig. 2, ELISA signals were generally reduced as the sera were diluted, indicating that the antigen-antibody reactions were not saturated within the dilution range. The cutoff values, which indicate the presence of nonspecific antibodies in the sera, were relatively higher for ELISAs using the 47-kD protein, ScaB, or ScaE as the antigen. The sensitivity and specificity of ELISAs using each antigen were determined and are summarized in Table 2. TSA56, ScaA, and ScaC ELISAs showed 100% sensitivity, whereas the ScaB ELISA showed the lowest sensitivity (63%). In terms of specificity, the TSA56 and ScaE ELISAs showed the highest (100%) specificity, in contrast to the ScaC ELISA, which showed 67% specificity.
Fig 2.
Antibody responses against the indicated O. tsutsugamushi antigens were analyzed by IgG ELISA. (A) Sera from 10 scrub typhus patients (filled circles; IFA titers of ≥1:1,280) and IFA-negative sera from 10 patients with acute febrile illness (open circles) were diluted as indicated and used for ELISAs. IFA-negative sera from 10 healthy volunteers were used to determine the cutoff values (red lines; mean plus 2 standard deviations). (B) Representative IFA results for a negative serum from a patient with acute febrile illness (top) and a positive serum from a scrub typhus patient (bottom). Differential interference contrast images of O. tsutsugamushi-infected cells are shown in the left panels.
Table 2.
Sensitivity and specificity of ELISAs using different O. tsutsugamushi antigens
| Antigen | Sensitivity (%) | Specificity (%) |
|---|---|---|
| TSA56 | 100 | 100 |
| 47-kDa protein | 77 | 93 |
| ScaA | 100 | 73 |
| ScaB | 63 | 87 |
| ScaC | 100 | 67 |
| ScaE | 80 | 100 |
Correlation of IFA titers with ELISA results obtained using different Sca antigens.
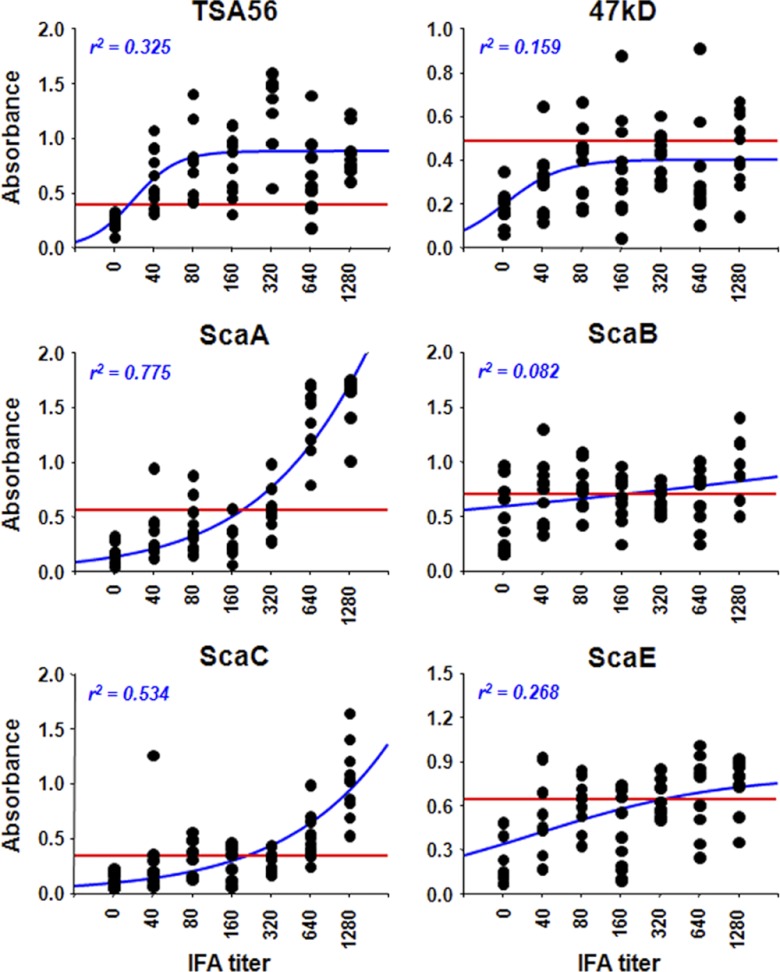
Next, nonlinear regression analysis was performed to compare IFA titers of patient sera after 1:1,000 dilution in PBS with the OD490 values from ELISAs using different O. tsutsugamushi antigens (Fig. 3). The best correlation of ELISA data with IFA titers was observed when ScaA (r2 = 0.775) was used as the antigen, whereas the ScaB test showed the lowest correlation (r2 = 0.082). It is notable that the OD490 values from the ELISA using TSA56 as antigen were rapidly saturated for patient sera starting at a 1:40 IFA titer, although there was a relatively low correlation (r2 = 0.325) compared to those of ELISAs using ScaA or ScaC. These results suggest that specific antibody responses against TSA56 are induced most robustly in scrub typhus patients and that anti-ScaA and anti-ScaC antibody responses are significant only in patients with high IFA titers (≥1:640). In addition, specific antibody responses against the 47-kDa protein, ScaB, and ScaE are relatively weaker in scrub typhus patients.
Fig 3.
Comparison of ELISA data obtained using the indicated antigens and IFA titers. The reactivities of the sera (10 samples per IFA titer) were assessed by ELISA and plotted against IFA titers. The cutoff values (red lines) were determined using IFA-negative sera from 10 healthy volunteers (mean plus 2 standard deviations). Correlations between ELISA results and IFA titers were assessed by nonlinear regression analysis (blue lines), and the r2 values are shown.
Genetic variation of sca genes of different strains of O. tsutsugamushi.
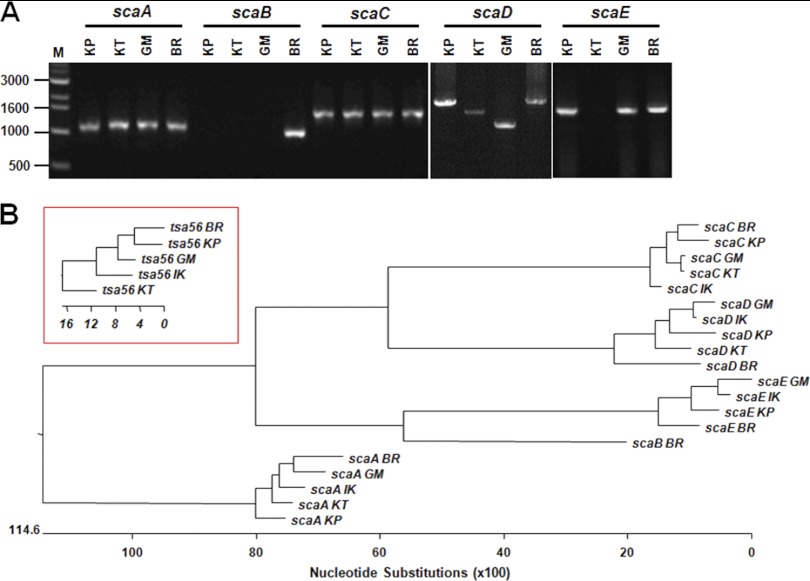
The differential correlation of IFA titers with ELISA results obtained using different O. tsutsugamushi antigens prompted us to examine the presence and variation in the passenger domains of the sca genes of the different strains of O. tsutsugamushi. By using specific primer sets for each gene (Table 1), we were able to amplify the scaA, scaC, and scaD genes from the genomes of three prototype strains (Karp, Kato, and Gilliam strains) as well as from the Boryong strain (Fig. 4A). Interestingly, the sizes of amplified scaD genes were different for all four strains. In the case of scaB, the gene was detected only in the Boryong strain. On the other hand, we could not amplify scaE from the Kato strain. When we compared the sequences of the amplified sca genes, each gene group formed specific clusters in a phylogenetic tree (Fig. 4B). However, nucleotide sequence distances among the strains did not show any consistency among the different Sca groups, i.e., the sequence variation among the strains showed a different pattern within each sca group. The identities of translated amino acid sequences (Fig. 5) among the sca genes of different strains are also quite variable: 65.8 to 81.8% for ScaA, 77.4 to 97.5% for ScaC, 69.7 to 93.8% for ScaD, and 62.8 to 85.3% for ScaE. ScaC proteins are the most conserved, and ScaE proteins show the most variation among the different strains. The size variation observed among scaD genes of different strains is due mainly to the presence of different numbers of internal repeat sequences (Fig. 5). There are two types of amino acid repeat sequences. One is comprised of around 70 amino acids that repeat five times in the ScaD protein of the Boryong strain and three times in that of the Gilliam strain. The other consists of 16 amino acids that were detected five times in the gene of the Boryong strain and once in that of the Gilliam strain. It is also interesting that the phylogenetic distribution of scaC genes of different strains was most similar to that of tsa56 genes (Fig. 4B), which have 81.5 to 90.9% amino acid sequence identity.
Fig 4.
Genetic analysis of the sca genes of different strains of O. tsutsugamushi. (A) Purified O. tsutsugamushi genomes were used as templates, and sca genes were amplified by PCR using specific sets of primers (Table 1). KP, Karp; KT, Kato; GM, Gilliam; BR, Boryong. (B) Phylogenetic tree of the sca genes identified in the genomes of the indicated strains. The red inset box shows a phylogenetic tree for the tsa56 genes of the indicated strains.
DISCUSSION
Diagnosis of scrub typhus is an important issue that determines the administration of rapid and proper antibiotic therapy in local clinics in regions of endemicity (15). However, all currently available serological tests and PCR-based detection methods have limitations in terms of sensitivity, costs, and convenience. Moreover, evaluation of diagnostic methods has been hampered because the current gold standard, IFA, is imperfect (15). Therefore, there is an urgent need for alternative diagnostic methods that use rapid and accurate point-of-care technologies that are readily available, especially for patients in the acute phase of scrub typhus. Recently, combinatorial detection methods consisting of rapid immunochromatographic techniques (ICT) and bacterial DNA amplification have been proposed (1, 20). Combinatorial diagnosis can achieve 50 to 67% sensitivity when applied to a robust reference comparator set, i.e., the scrub typhus infection criteria (STIC), comprised of four parameters: positive cell culture isolation, an admission IgM titer of ≥1:12,800 using the gold standard IFA, a 4-fold increase of IFA IgM titer, and/or a positive result in at least two of three PCR assays. Compared to single conventional detection techniques and methods, the combination of DNA- and antibody-based detection methods has increased sensitivity, with a minimal reduction of specificity, and has expanded the time frame of adequate diagnostic coverage to extend throughout the acute phase of scrub typhus (20).
Although the combinatorial detection method can improve the sensitivity of diagnosis during the acute phase, several limitations still remain. First, sensitivity is still less than 70% for applying the STIC to define scrub typhus with a high level of confidence. Second, PCR detection and rapid ICT are largely dependent on a single gene, tsa56, encoding a 56-kDa major outer membrane protein, and on antibody responses against TSA56. Since tsa56 is highly variable, the variation may affect PCR sensitivity (26), and more critically, multiple antigenic variants should be used to cover the diverse serotypes for serological detection of specific antibodies (10). Third, the method cannot differentiate between genetic variations of O. tsutsugamushi strains if restriction fragment length polymorphism (RFLP) mapping or sequencing is not performed. To date, more than 20 antigenically distinct strains have been reported (14). In addition to antigenic variation, multiple studies have shown great interstrain variability in virulence in humans and rodents (14). Therefore, differentiation of genetic variation is an important issue in scrub typhus diagnosis and epidemiology.
Here we report that Sca proteins of O. tsutsugamushi can induce antibody responses in scrub typhus patients. Specific antibody responses against ScaA and ScaC were observed mainly in scrub typhus patients with high IFA titers, so a serological test using the Sca antigens may not be efficient if used solely, especially during the early phase of infection. However, it is notable that ELISAs using ScaA or ScaC showed 100% sensitivity (Fig. 2 and Table 2), and the use of these antigens with TSA56 may synergistically enhance the sensitivity of serological diagnostic tests such as ICT. Previously, it was suggested that the 47-kDa protein, an HtrA homolog of O. tsutsugamushi, might be a useful antigen if combined with TSA56 for serological diagnosis (8, 15). However, specific antibody titers against the 47-kDa antigen in human patients are relatively low during the acute phase of infection (8), which might be due to antibody cross-reaction with human serine protease or HtrAs of other bacteria (7). We also consistently observed relatively low levels of specific antibody responses against the 47-kDa antigen in our human patients (Fig. 3). Given that the amino acid sequences of Sca passenger domains are highly specific to Orientia and show low levels of sequence identity (<30%) with other rickettsial Sca proteins (12), these bacterial antigens may be effective targets for scrub typhus diagnosis. Although we may need to further confirm the antigenic variation of the diverse strains and the antibody responses against Sca antigens of different strains in human scrub typhus patients, the sequence conservation observed in this study supports the potential benefits of using Sca antigens as novel diagnostic targets in combination with TSA56.
In addition to their use in serological methods, the sca genes could also be useful targets for molecular diagnostic methods such as PCR-based detection. Currently, PCR-based methods targeting tsa56, the 47-kDa protein gene, and groEL have been used for species-specific amplification (15, 21). The sensitivity of these DNA detection methods maximizes at around 40% (15, 20). Although targeting the 16S rRNA gene yields a higher sensitivity (37.5 to 52.3%) under real-world conditions (15), all of these DNA-based techniques require sequencing after PCR amplification in order to identify the specific strain that has infected the patient. In this report, we found that two of the sca genes, scaB and scaE, were amplified differentially in different strains, suggesting a differential presence of the genes in the genomes of different strains. In fact, the scaB gene was present in the Boryong strain but absent in the Ikeda strain when their complete genome sequences were examined (12). Therefore, the molecular detection of sca genes could provide direct clues about O. tsutsugamushi strains as well as the species, without sequencing. Furthermore, the size of the passenger domain of the scaD gene is highly variable among the different strains due to the different numbers of internal repeat sequences (Fig. 4A and 5). This characteristic of the gene can provide valuable information on the genotypes of O. tsutsugamushi without any sequencing. Currently, we have examined three prototype strains, the Boryong strain, and the Ikeda strain for sequence comparison. Considering the geographic distribution of various genotypic variants, further studies on the genetic variation of sca genes in a wide geographic area may facilitate the usage of this gene family as molecular targets for scrub typhus diagnosis.
Finally, we propose for the first time that a panel of serological and molecular detection methods using the sca gene family as a novel target, combined with classical assay methods, can provide rapid and cost-effective technologies for scrub typhus diagnosis as well as for epidemiologic studies.
ACKNOWLEDGMENT
This study was supported by a grant (A111503) from the Korea Healthcare Technology R&D Project, Ministry for Health, Welfare & Family Affairs, Republic of Korea.
Footnotes
Published ahead of print 11 July 2012
REFERENCES
- 1. Blacksell SD, et al. 2012. Prospective evaluation of commercial antibody-based rapid tests in combination with a loop-mediated isothermal amplification PCR assay for detection of Orientia tsutsugamushi during the acute phase of scrub typhus infection. Clin. Vaccine Immunol. 19:391–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blanc G, et al. 2005. Molecular evolution of rickettsia surface antigens: evidence of positive selection. Mol. Biol. Evol. 22:2073–2083 [DOI] [PubMed] [Google Scholar]
- 3. Chan YG, Cardwell MM, Hermanas TM, Uchiyama T, Martinez JJ. 2009. Rickettsial outer-membrane protein B (rOmpB) mediates bacterial invasion through Ku70 in an actin, c-Cbl, clathrin and caveolin 2-dependent manner. Cell. Microbiol. 11:629–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chan YG, Riley SP, Chen E, Martinez JJ. 2011. Molecular basis of immunity to rickettsial infection conferred through outer membrane protein B. Infect. Immun. 79:2303–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chang WH, Kang JS, Lee WK, Choi MS, Lee JH. 1990. Serological classification by monoclonal antibodies of Rickettsia tsutsugamushi isolated in Korea. J. Clin. Microbiol. 28:685–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chattopadhyay S, Richards AL. 2007. Scrub typhus vaccines: past history and recent developments. Hum. Vaccin. 3:73–80 [DOI] [PubMed] [Google Scholar]
- 7. Chen HW, et al. 2009. Identification of cross-reactive epitopes on the conserved 47-kilodalton antigen of Orientia tsutsugamushi and human serine protease. Infect. Immun. 77:2311–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen HW, et al. 2011. Kinetics and magnitude of antibody responses against the conserved 47-kilodalton antigen and the variable 56-kilodalton antigen in scrub typhus patients. Clin. Vaccine Immunol. 18:1021–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cho NH, et al. 2007. The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host-cell interaction genes. Proc. Natl. Acad. Sci. U. S. A. 104:7981–7986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Choi MS, et al. 1999. Homotypic and heterotypic antibody responses to a 56-kilodalton protein of Orientia tsutsugamushi. Infect. Immun. 67:6194–6197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Greiner M, Sohr D, Gobel P. 1995. A modified ROC analysis for the selection of cut-off values and the definition of intermediate results of serodiagnostic tests. J. Immunol. Methods 185:123–132 [DOI] [PubMed] [Google Scholar]
- 12. Ha NY, Cho NH, Kim YS, Choi MS, Kim IS. 2011. An autotransporter protein from Orientia tsutsugamushi mediates adherence to nonphagocytic host cells. Infect. Immun. 79:1718–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jeong YJ, et al. 2007. Scrub typhus: clinical, pathologic, and imaging findings. Radiographics 27:161–172 [DOI] [PubMed] [Google Scholar]
- 14. Kelly DJ, Fuerst PA, Ching WM, Richards AL. 2009. Scrub typhus: the geographic distribution of phenotypic and genotypic variants of Orientia tsutsugamushi. Clin. Infect. Dis. 48(Suppl 3): S203–S230 [DOI] [PubMed] [Google Scholar]
- 15. Koh GC, Maude RJ, Paris DH, Newton PN, Blacksell SD. 2010. Diagnosis of scrub typhus. Am. J. Trop. Med. Hyg. 82:368–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kweon SS, et al. 2009. Rapid increase of scrub typhus, South Korea, 2001–2006. Emerg. Infect. Dis. 15:1127–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee JH, et al. 2008. Fibronectin facilitates the invasion of Orientia tsutsugamushi into host cells through interaction with a 56-kDa type-specific antigen. J. Infect. Dis. 198:250–257 [DOI] [PubMed] [Google Scholar]
- 18. Mathai E, et al. 2003. Outbreak of scrub typhus in southern India during the cooler months. Ann. N. Y. Acad. Sci. 990:359–364 [DOI] [PubMed] [Google Scholar]
- 19. Nakayama K, et al. 2008. The whole-genome sequencing of the obligate intracellular bacterium Orientia tsutsugamushi revealed massive gene amplification during reductive genome evolution. DNA Res. 15:185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Paris DH, et al. 2011. Diagnostic accuracy of a loop-mediated isothermal PCR assay for detection of Orientia tsutsugamushi during acute scrub typhus infection. PLoS Negl. Trop. Dis. 5:e1307 doi:10.1371/journal.pntd.0001307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Park HS, et al. 2008. Rapid identification of rickettsiae using real-time PCR. J. Bacteriol. Virol. 38:221–226 [Google Scholar]
- 22. Park SW, et al. 2010. Antigenic drift of Orientia tsutsugamushi in South Korea as identified by the sequence analysis of a 56-kDa protein-encoding gene. Am. J. Trop. Med. Hyg. 83:930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Richards AL. 2004. Rickettsial vaccines: the old and the new. Expert Rev. Vaccines 3:541–555 [DOI] [PubMed] [Google Scholar]
- 24. Riley SP, et al. 2010. The Rickettsia conorii autotransporter protein Sca1 promotes adherence to nonphagocytic mammalian cells. Infect. Immun. 78:1895–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Seong SY, Choi MS, Kim IS. 2001. Orientia tsutsugamushi infection: overview and immune responses. Microbes Infect. 3:11–21 [DOI] [PubMed] [Google Scholar]
- 26. Sonthayanon P, et al. 2006. Rapid diagnosis of scrub typhus in rural Thailand using polymerase chain reaction. Am. J. Trop. Med. Hyg. 75:1099–1102 [PubMed] [Google Scholar]
- 27. Yasunaga H, Horiguchi H, Kuwabara K, Hashimoto H, Matsuda S. 2011. Delay in tetracycline treatment increases the risk of complications in tsutsugamushi disease: data from the Japanese Diagnosis Procedure Combination database. Intern. Med. 50:37–42 [DOI] [PubMed] [Google Scholar]
- 28. Zhang S, et al. 2010. Scrub typhus in previously unrecognized areas of endemicity in China. J. Clin. Microbiol. 48:1241–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]