Abstract
Administration of a clade C/B′ candidate HIV-1 DNA vaccine, ADVAX, by in vivo electroporation (EP) was safe and more immunogenic than intramuscular administration without EP. The breadth and specificity of T-cell responses to full-length Env were mapped. Responses to multiple Env regions were induced, with most focusing on V3/C4 and V2 regions, including the α4β7 integrin-binding domain. The breadth of responses induced by this DNA vaccine regimen was comparable to that of viral-vectored vaccine regimens.
TEXT
DNA-based vaccines have several theoretical and practical advantages over other vaccine types, including safety, relative ease of construction and manufacture, and an ability to revaccinate not limited by induced antivector immunity. However, DNA-based vaccines have not always elicited robust immune responses in humans. The ADVAX vaccine candidate, which is composed of a DNA construct incorporating a clade B′/C HIV-1 recombinant insert isolated from the predominant circulating recombinant form in Yunnan Province, China (10), administered by intramuscular electroporation (EP) to 24 individuals (0.2, 1, and 4 mg), was safe and well tolerated and considered acceptable for a prophylactic vaccine (4). EP delivery of ADVAX increased the magnitude of HIV-1-specific cell-mediated immunity by up to 70-fold over IM injection, as measured by gamma interferon (IFN-γ) ELISpot assays. The number of antigens to which a response was detected improved with EP and increasing dosage (22).
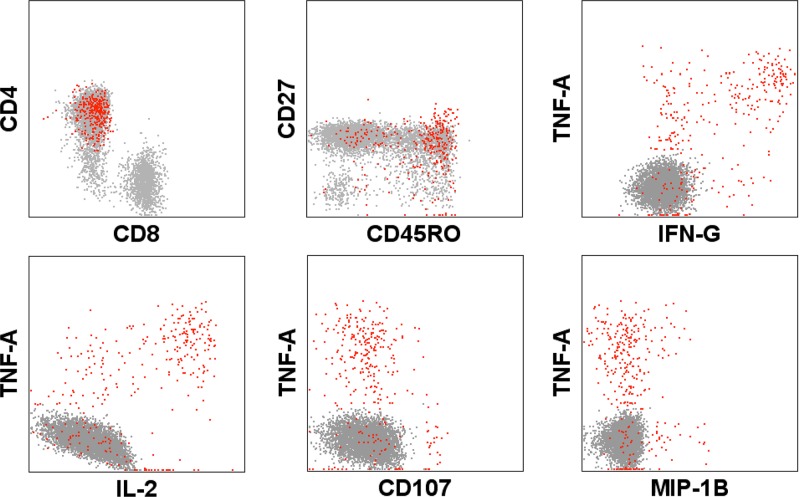
In the first study, we showed that, following stimulation with ADVAX insert-matched peptide pools, seven of eight volunteers in the 4-mg-dose group had demonstrable flow cytometry responses. Four had CD3+ CD4+ responses alone, two had both CD3+ CD4+ and CD3+ CD8+ responses, and one had only a CD3+ CD8+ T cell response. The majority of cellular responses were induced in CD4+ T cells that were of a polyfunctional phenotype and targeted HIV gp120 (22). Figure 1 shows a representative example of a gp120-specific response induced in ADVAX vaccinees; the responses tended to be of a CD27+ CD45RO+ central memory phenotype, producing predominantly interleukin-2 (IL-2), tumor necrosis factor alpha (TNF-α), and IFN-γ, with low levels of MIP1β and little or no degranulation (22).
Fig 1.
Vaccination-induced Env pool-specific responses in CD4+ CD8− CD3+ cells illustrated using a combinatorial Boolean gate of IFN-γ-, TNF-α-, and IL-2-producing populations (red) overlaid onto memory (CD45RO+ CD27+) and functional phenotypes as evaluated by polychromatic flow cytometry (representative example).
Certain regions of the HIV-1 proteome remain highly conserved due to their structural and/or functional importance. Vaccine-induced priming of T-cell responses to such functionally important and conserved regions may limit early virus replication either through destruction of infected cells or through decreasing virus fitness as a consequence of mutating these sites to escape recognition (12, 15, 21, 23, 26, 27). It is important to determine the specificity of immune responses induced following vaccination, to assess whether conserved, functionally important regions are targeted and, if so, how many (5).
A total of 101 15-mer peptides, sequentially overlapping by 11 amino acids and corresponding to the envelope sequence contained within ADVAX, were manufactured to >90% purity by AnaSpec (Fremont, CA). Peptide matrix pools were designed using Deconvolute this! software v1.0 (Mario Roederer, Vaccine Research Center [VRC], NIH); each individual peptide was present in 3 unique pools (18).
The gamma interferon (IFN-γ) ELISpot assay was performed as previously described (6). To facilitate mapping, vaccinees were selected on the basis of ADVAX envelope-specific IFN-γ ELISpot responses above a threshold of >100 spot-forming cells/million peripheral blood mononuclear cells (PBMC). Stimulations used preprepared matrix pools at a final peptide concentration of 1.5 μg/ml.
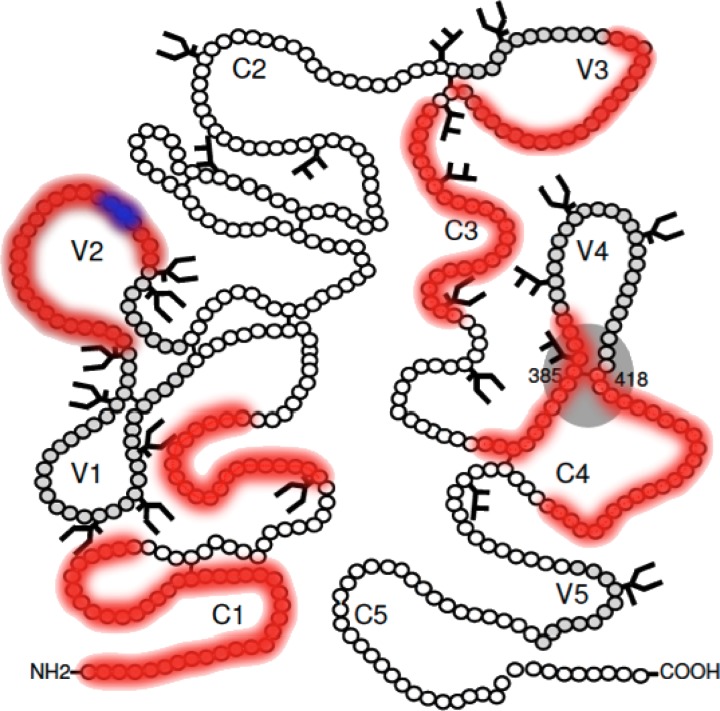
16 vaccinees were found to fulfill the ELISpot threshold criteria outlined above, with 7, 6, and 3 vaccinees from the 4-mg, 1-mg, and 0.2-mg DNA EP groups, respectively; no vaccinees receiving DNA alone without EP met the criteria. Responses were deconvoluted in 11 of 16 vaccinees, resulting in a total of 22 individual peptides being recognized across the ENV pool (Table 1). Vaccinees responded to up to a total of 5 epitopes, with the majority focusing responses on three regions of the vaccine insert: 5 of 11 volunteers mounted responses against the V2 region spanning peptides VYALFYRLDIVPLNK and FYRLDIVPLNKKNSS, 5 of 11 focused responses against peptides VTENFNMWKNDMVNQ and/or FNMWKNDMVNQMHED found within the C1 region, and 4 of 11 individuals targeted the sequence TGDIIGDIRQAHCNI present within the V3/C3 region (Fig. 2).
Table 1.
Env sequences targeted following ADVAX administration
| Peptide sequencea | Region(s) | Location (amino acid positions)b | No. of responders (n = 11) |
|---|---|---|---|
| SAAENLWVTVYYGVP | C1 | 29–43 | 1 |
| GVPVWKEAKTTLFCA | C1 | 41–55 | 1 |
| KTTLFCASDAKAYEK | C1 | 49–63 | 3 |
| FCASDAKAYEKEVHN | C1 | 53–67 | 1 |
| YEKEVHNVWATHACV | C1 | 61–75 | 1 |
| VLENVTENFNMWKND | C1 | 85–98 | 1 |
| VTENFNMWKNDMVNQ | C1 | 89–103 | |
| FNMWKNDMVNQMHED | C1 | 93–107 | 5c |
| FNATTVLRDRKQTVYA | V2 | 159–173 | |
| VLRDRKQTVYALFYR | V2 | 163–177 | 3c |
| RKQTVYALFYRLDIV | V2 | 167–181 | 1 |
| VYALFYRLDIVPLNK | V2 | 171–185 | |
| FYRLDIVPLNKKNSS | V2 | 175–189 | 5c |
| GPGQTFYATGDIIGD | V3 | 310–324 | 1 |
| TGDIIGDIRQAHCNI | V3/C3 | 319–333 | 4 |
| WNETLQRVGKKLAEHF | C3 | 338–352 | 2 |
| LQRVGKKLAEHFPNKT | C3 | 342–356 | 1 |
| FNCRGEFFYCNTSSL | C3 | 376–390 | 1 |
| CRIKQIINMWQEVGR | C4 | 424–438 | 1 |
| KQIINMWQEVGRAMYA | C4 | 428–442 | 1 |
| MWQEVGRAMYAPPIE | C4 | 432–446 | 1 |
The α4b7 binding motif is indicated in bold.
Location data are based on the HIV-Env gp120 HXB2 sequence.
Data include responses to both of two contiguous 15-mers sharing an 11-aa overlap.
Fig 2.
Locations of targeted regions (red) superimposed on a schematic gp120 backbone. The α4β7 integrin binding motif is highlighted in blue. (Adapted from Retrovirology [19] with permission of the publisher.)
Delivery of DNA vaccines by EP has been shown to increase their immunogenicity (4, 20, 22, 25). In this report, administration of the DNA-based ADVAX vaccine candidate through EP is shown to induce a breadth of ENV-specific T-cell responses (mean, 2.8 regions/patient) comparable to that seen with vaccination using DNA prime and adenovirus vector boost (11) and NYVAC (8) (mean responses, 3.3 and 4.2, respectively). Furthermore, the preferential targeting of certain sequences, in particular, V2 (FYRLDIVPLNK), C1 (FNMWKNDMVNQ), and V3/C4 (TGDIIGDIRQAHCN) sequences, to one or more of which 10 of 11 individuals mounted responses, supported by data from Harari et al. (8) where equivalent CD4 epitopes were targeted following vaccination with NYVAC containing a clade C insert, suggests the potential for vaccine-induced immune focusing.
A phase III clinical trial involving >16,000 adult volunteers conducted in Thailand (RV144) demonstrated that a prime-boost regimen of administration of the canarypox-based vaccine ALVAC plus gp120 protein was 31.2% efficacious in preventing HIV infection, as judged at 3.5 years of follow-up (17). Initial results of the correlate discovery team revealed that levels of antibodies to the first and second variable loops (V1/V2) of gp120 correlated inversely with subsequent risk of HIV-1 acquisition in RV144, while levels of plasma IgA binding to a panel of HIV-1 envelope proteins showed a direct correlation with risk of infection (9). While these preliminary results require further verification and analysis, it is encouraging that ADVAX delivered by EP can also elicit immune responses to the V2 loop, a functionally important region of the HIV-1 envelope involved in the formation of the glycoprotein trimer, masking of neutralizing epitopes, and interaction with cellular coreceptors (28).
One of the areas targeted within the V2 loop included the α4β7 integrin-binding domain of gp120 (1). HIV-1 gp120 stably binds α4β7 present on CD4 cells, homing to sites of inflammation through an LDI/LDV motif on the V2 loop present in peptide FYRLDIVPLNK (1, 2, 16); recognition of similar peptides by CD4 cells has been previously described with NYVAC (8) and, more recently, in the RV144 trial (3). The ligation event causes the formation of a synapse facilitating events required for HIV-1 entry and resulting in LFA-1 activation, which induces lymphocyte activation and proliferation, further potentiating the formation of a focus of infection (1). Primary HIV-1 infection results in massive CD4 depletion in the gastrointestinal lymphoid tissues (14); one potential reason for this may be HIV-1 infection and destruction of CD4 T cells promoted via the gp120/α4β7 interaction. Targeting this region during early infection may restrict HIV-1 activity. In conclusion, administration of the DNA ADVAX vaccine through EP elicits polyfunctional responses, primarily in CD4+ T-cell populations, to multiple regions of the HIV-1 envelope glycoprotein (22). Here we show that, following epitope mapping, DNA vaccine-induced ENV-specific CD4 responses are of a breadth comparable to the breadth induced by responses elicited by NYVAC (8) and adenovirus-based vectors (11, 13). The immunogenicity of DNA-based vectors may be further improved through incorporation of IL-12 adjuvants and/or boosting with viral vectors or proteins (7, 24, 25).
ACKNOWLEDGMENTS
We thank Mark De Souza (AFRIMS, Thailand) for discussions on the role of V2 in the RV144 study and Patricia Fast (International AIDS Vaccine Initiative [IAVI]) for thorough review of the manuscript and support of the study. We also thank members of the Human Immunology Laboratory in London for technical and administrative assistance.
This study was funded by the International AIDS Vaccine Initiative and its donors, including the generous support of the American people through the United States Agency for International Development (USAID; USAID Cooperative Agreement GPO-A-00-06-00006-00).
The contents of the manuscript are the responsibility of IAVI and do not necessarily reflect the views of USAID or the U.S. government.
The following organizations and institutions played a direct role in study design, data collection and analysis, decision to publish, and preparation of the manuscript: International AIDS Vaccine Initiative (IAVI), New York, the IAVI Human Immunology Laboratory, Imperial College, London, United Kingdom, and the Aaron Diamond AIDS Research Center, New York.
Footnotes
Published ahead of print 25 July 2012
REFERENCES
- 1. Arthos J, et al. 2008. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat. Immunol. 9:301–309 [DOI] [PubMed] [Google Scholar]
- 2. Cicala C, et al. 2009. The integrin {alpha}4{beta}7 forms a complex with cell-surface CD4 and defines a T-cell subset that is highly susceptible to infection by HIV-1. Proc. Natl. Acad. Sci. U. S. A. 106:20877–20882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Souza MS, et al. 2012. The Thai phase III trial (RV144) vaccine regimen induces T cell responses that preferentially target epitopes within the V2 region of HIV-1 envelope. J. Immunol. 188:5166–5176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dolter KE, et al. 2011. Immunogenicity, safety, biodistribution and persistence of ADVAX, a prophylactic DNA vaccine for HIV-1, delivered by in vivo electroporation. Vaccine 29:795–803 [DOI] [PubMed] [Google Scholar]
- 5. Fischer W, et al. 2007. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat. Med. 13:100–106 [DOI] [PubMed] [Google Scholar]
- 6. Gill DK, et al. 2010. Equivalence of ELISpot assays demonstrated between major HIV network laboratories. PLoS One 5:e14330 doi:10.1371/journal.pone.0014330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goonetilleke N, et al. 2006. Induction of multifunctional human immunodeficiency virus type 1 (HIV-1)-specific T cells capable of proliferation in healthy subjects by using a prime-boost regimen of DNA- and modified vaccinia virus Ankara-vectored vaccines expressing HIV-1 Gag coupled to CD8+ T-cell epitopes. J. Virol. 80:4717–4728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Harari A, et al. 2008. An HIV-1 clade C DNA prime, NYVAC boost vaccine regimen induces reliable, polyfunctional, and long-lasting T cell responses. J. Exp. Med. 205:63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Haynes BF, et al. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 366:1275–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang Y, et al. 2008. Design, construction, and characterization of a dual-promoter multigenic DNA vaccine directed against an HIV-1 subtype C/B' recombinant. J. Acquir. Immune Defic. Syndr. 47:403–411 [DOI] [PubMed] [Google Scholar]
- 11. Koup RA, et al. 2010. Priming immunization with DNA augments immunogenicity of recombinant adenoviral vectors for both HIV-1 specific antibody and T-cell responses. PLoS One 5:e9015 doi:10.1371/journal.pone.0009015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Letourneau S, et al. 2007. Design and pre-clinical evaluation of a universal HIV-1 vaccine. PLoS One 2:e984 doi:10.1371/journal.pone.0000984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li F, et al. 2011. Mapping HIV-1 vaccine induced T-cell responses: bias towards less-conserved regions and potential impact on vaccine efficacy in the step study. PLoS One 6:e20479 doi:10.1371/journal.pone.0020479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lim SG, et al. 1993. Loss of mucosal CD4 lymphocytes is an early feature of HIV infection. Clin. Exp. Immunol. 92:448–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu Y, et al. 2006. Selection on the human immunodeficiency virus type 1 proteome following primary infection. J. Virol. 80:9519–9529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. O'Rourke SM, et al. 2010. Mutation at a single position in the V2 domain of the HIV-1 envelope protein confers neutralization sensitivity to a highly neutralization-resistant virus. J. Virol. 84:11200–11209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rerks-Ngarm S, et al. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361:2209–2220 [DOI] [PubMed] [Google Scholar]
- 18. Roederer M, Koup RA. 2003. Optimized determination of T cell epitope responses. J. Immunol. Methods 274:221–228 [DOI] [PubMed] [Google Scholar]
- 19. Sanders RW, et al. 2008. The carbohydrate at asparagine 386 on HIV-1 gp120 is not essential for protein folding and function but is involved in immune evasion. Retrovirology 5:10 doi:10.1186/1742-4690-5-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sardesai NY, Weiner DB. 2011. Electroporation delivery of DNA vaccines: prospects for success. Curr. Opin. Immunol. 23:421–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Troyer RM, et al. 2009. Variable fitness impact of HIV-1 escape mutations to cytotoxic T lymphocyte (CTL) response. PLoS Pathog. 5:e1000365 doi:10.1371/journal.ppat.1000365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vasan S, et al. 2011. Electroporation enhances the immunogenicity of an HIV-1 DNA vaccine candidate in healthy volunteers. PLoS One 6:e19252 doi:10.1371/journal.pone.0019252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wagner R, et al. 1999. Molecular and functional analysis of a conserved CTL epitope in HIV-1 p24 recognized from a long-term nonprogressor: constraints on immune escape associated with targeting a sequence essential for viral replication. J. Immunol. 162:3727–3734 [PubMed] [Google Scholar]
- 24. Wang S, et al. 2008. Cross-subtype antibody and cellular immune responses induced by a polyvalent DNA prime-protein boost HIV-1 vaccine in healthy human volunteers. Vaccine 26:1098–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Winstone N, et al. 2011. Enhanced control of pathogenic SIVmac239 replication in macaques immunized with a plasmid IL12 and a DNA prime, viral vector boost vaccine regimen. J. Virol. 85:9578–9587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang OO. 2009. Candidate vaccine sequences to represent intra- and inter-clade HIV-1 variation. PLoS One 4:e7388 doi:10.1371/journal.pone.0007388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang OO, Daar ES, Ng HL, Shih R, Jamieson BD. 2011. Increasing CTL targeting of conserved sequences during early HIV-1 infection is correlated to decreasing viremia. AIDS Res. Hum. Retroviruses 27:391–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zolla-Pazner S, Cardozo T. 2010. Structure-function relationships of HIV-1 envelope sequence-variable regions refocus vaccine design. Nat. Rev. Immunol. 10:527–535 [DOI] [PMC free article] [PubMed] [Google Scholar]