Abstract
Routine antigenic characterization of swine influenza virus isolates in a high-throughput serum neutralization (HTSN) assay found that approximately 20% of isolates were not neutralized by a panel of reference antisera. Genetic analysis revealed that nearly all of the neutralization-resistant isolates possessed a seasonal human-lineage hemagglutinin (HA; δ cluster). Subsequent sequencing analysis of full-length HA identified a conserved N144 residue present only in neutralization-resistant strains. N144 lies in a predicted N-linked glycosylation consensus sequence, i.e., N-X-S/T (where X is any amino acid except for proline). Interestingly, neutralization-sensitive viruses all had predicted N-linked glycosylation sites at N137 or N142, with threonine (T) occupying position 144 of HA. Consistent with the HTSN assay, hemagglutination inhibition (HI) and serum neutralization (SN) assays demonstrated that migration of the potential N-linked glycosylation site from N137 or N142 to N144 resulted in a >8-fold decrease in titers. These results were further confirmed in a reverse genetics system where syngeneic viruses varying only by predicted N-glycosylation sites at either N142 or N144 exhibited distinct antigenic characteristics like those observed in field isolates. Molecular modeling of the hemagglutinin protein containing N142 or N144 in complex with a neutralizing antibody suggested that N144-induced potential glycosylation may sterically hinder access of antibodies to the hemagglutinin head domain, allowing viruses to escape neutralization. Since N-linked glycosylation at these sites has been implicated in genetic and antigenic evolution of human influenza A viruses, we conclude that the relocation of the hemagglutinin N-linked glycosylation site from N142 to N144 renders swine influenza virus δ-cluster viruses resistant to antibody-mediated neutralization.
INTRODUCTION
Influenza viruses possess a number of mechanisms to elude host humoral immunity against hemagglutinin (HA), which mediates viral entry. Reassortment of genome segments can lead to antigenic shift, while gradual accumulation of mutations leads to genetic drift (10, 21). In addition to mutations in the antigenic epitopes of HA that can impact the ability of preexisting antibodies to recognize mutant HA, mutations in N-linked glycosylation of HA can promote viral evasion of antibody recognition by altering the oligosaccharide layer surrounding HA (22). Glycan residues can restrict the binding of some antibodies to their epitopes, leading to loss of antibody recognition and immunogenicity in a phenomenon known as glycan shielding (30, 31). Additionally, mutation and genetic drift occur preferentially at positions in the HA globular head that are not protected by glycans (9). Glycan residues also influence receptor binding and consequently can affect viral replication kinetics.
The evolution of influenza viruses frequently includes modification of the number and position of glycosylation sites (18). For H3N2 viruses, the number of glycosylation sites has increased from 2 to 10 over the last 40 years (4). However, increasingly glycosylated HA has been correlated with decreased virulence in mice and reduced viral fitness (8, 27). While increasing glycosylation can shield HA from neutralizing antibodies, the viral affinity for cellular receptors is obligatorily decreased (1, 8). Also, compensatory mutations in either the HA or neuraminidase (NA) are required to balance receptor binding and release activities (29). These compensatory mutations in NA have been associated with the acquisition of natural resistance to NA inhibitors (14).
Numerous lineages of swine influenza virus circulate concurrently in pigs, including β-, γ-, and δ-cluster H1N1 and H1N2 viruses, as well as the H3N2 subtype (19). The δ cluster was originally subdivided into two subclusters (δ-I and δ-II), but more recent work demonstrated five distinct genetic subclusters (δ-A, δ-B, δ-C, δ-D, and δ-E) representing at least three antigenically distinct groups (11, 28). In our laboratory, routine antigenic characterization of swine influenza viruses isolated from pigs displaying influenza-like illness found that approximately 20% of field isolates in 2011 were not neutralized by a panel of antisera (H1 clusters α, β, γ, δ-A, δ-B, δ-C, δ-D, and δ-E, 2009 pandemic H1N1 virus, and H3 clusters I, III, and IV) representing circulating subtypes and clusters of influenza virus in North America. Genetic analysis found that nearly all of the nonneutralized viruses were located in the δ cluster. Interestingly, the HA genes of the nonneutralized viruses were nearly identical to those of others that were neutralized by reference antisera. The aim of this study was to determine the molecular basis for the apparent variability observed in δ-cluster antigenic characterization to allow for more effective vaccine formulations.
MATERIALS AND METHODS
Cells and viruses.
Clinical samples (nasal swabs or lung tissue) were collected from pigs exhibiting influenza-like illness and were submitted to Newport Laboratories for viral isolation and characterization as part of routine diagnostic testing. Samples originated from various commercial swine production sites in North Carolina in 2010 and 2011 (designated by virus identifier prefix; for example, isolate 10-0036-2 was isolated in 2010). Virus isolation was performed on swine testicle (ST) cells grown in Dulbecco's modified Eagle's medium (DMEM) containing 5% fetal bovine serum (FBS) at 37°C with 5% CO2. For viral propagation, fetal bovine serum was omitted from the DMEM. 293T and MDCK cells were propagated in DMEM containing 10% fetal bovine serum.
Genetic analysis.
RNA was harvested from infected cell culture harvest fluids by use of a 5× MagMax-96 viral isolation kit (Life Technologies). Complete viral genomes were amplified using a previously described multisegment reverse transcription-PCR method (33). Viral cDNA libraries were prepared using an NEB Next Fast DNA fragmentation and library prep set 4 kit according to the manufacturer's instructions (New England BioLabs), with the exception that the kit adaptors were replaced with bar-coded adaptors (Ion Xpress Barcode Adaptor 1-16 kit; Life Technologies). DNA sequencing templates were prepared using an Ion Xpress template kit, version 2.0 (Life Technologies), and sequenced using an Ion Torrent personal genome machine (Life Technologies). Contigs were assembled using SeqMan NGen software (DNAStar). Contigs encoding full-length HA were identified by BLAST analysis. Full-length HA DNA sequences were aligned using the ClustalW method. Phylogenetic analyses were performed using MEGA 5.0 via the neighbor-joining method, and tree topology was verified by use of 1,000 bootstrap replicates (26).
Serological assays.
Antibody cross-reactivity was determined using a panel of reference antisera in hemagglutination inhibition (HI) and high-throughput serum neutralization (HTSN) assays (11). Antiserum against an isolate from each of the five subclusters of the δ cluster was generated in pigs and normalized to a homologous HI titer of 1,280 as previously described (11). Antiserum was also generated against the ST cell line as a control. All sera were heat inactivated at 56°C for 30 min prior to use.
HI assays were performed following standard procedures (7). In brief, sera were treated with a receptor-destroying enzyme for 24 h at 37°C and then adsorbed with a 20% suspension of turkey erythrocytes in phosphate-buffered saline (PBS) for 30 min at room temperature. Virus suspensions containing 4 to 8 HA units of virus were incubated for 1 h with serial 2-fold dilutions of antiserum, and the HI titer was determined as the reciprocal of the highest dilution that showed complete inhibition of hemagglutination using 0.5% washed turkey erythrocytes. Antigenic cartography was performed using the program AntigenMap (http://sysbio.cvm.msstate.edu/AntigenMap), which utilizes matrix completion multidimensional scaling (MC-MDS) to map HI titers in two dimensions (5).
Serum neutralization assays were performed as previously described, with slight modifications (32). Two-fold serial dilutions in PBS, from 1:4 to 1:32,768, were performed on the sera. Next, 100 μl of 40 to 200 median tissue culture infective doses (TCID50)/100 μl of virus was added to each serum dilution and incubated for 1 h at 37°C in 5% CO2. The serum-virus mixture was next transferred to a confluent monolayer of ST cells and incubated at 37°C in 5% CO2 for 4 days. Viral neutralization was assessed by a lack of cytopathic effects in each well by microscopy. Back titrations were performed on ST cells to confirm a virus concentration of 40 to 200 TCID50/100 μl.
The HTSN assay was performed as previously described (11, 12). Briefly, antisera were diluted 1:800 in DMEM, combined with 1,000 median TCID50 of virus, and incubated at 37°C for 1 h. The virus-antiserum mixture was next transferred to a confluent monolayer of ST cells and incubated for 4 days at 37°C. Nonneutralized virus was assayed by incubating cell culture supernatants for 2 h with methylumbelliferyl-N-acetylneuraminic acid and measuring fluorescence with excitation and emission filters of 355 and 460 nm, respectively. The mean of duplicate fluorescence values was used to calculate the serum neutralization ratio, as follows: neutralization ratio = (sample value − virus-only control value)/(no-virus control value − virus-only control value). A serum neutralization ratio of >0.31 was previously determined to indicate positivity for neutralization (12).
Reverse genetics.
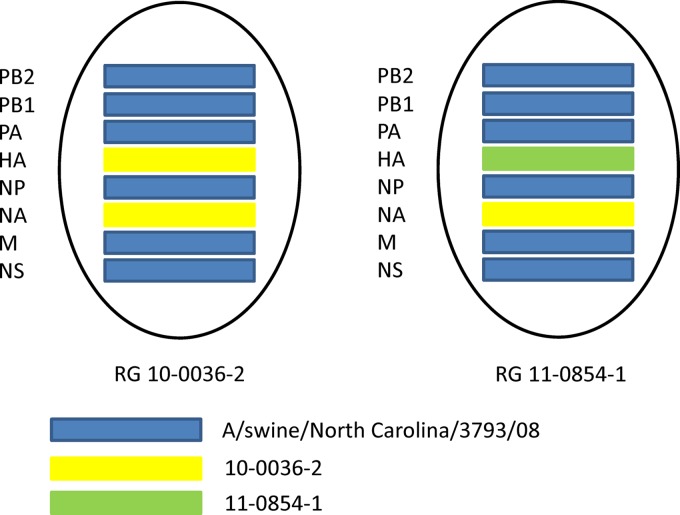
A triple-reassortant internal gene cassette (TRIG) swine influenza virus (A/swine/North Carolina/3793/08 [H1N1]) was used as the template to create a TRIG swine influenza virus reverse genetics system. All eight segments were PCR amplified, digested with BsmBI, and ligated into similarly digested pHW2000 as previously described (15). Plasmids bearing an insert were identified by restriction digestion and sequenced to verify identity with A/swine/North Carolina/3793/08. The HA and NA genes from neutralization-sensitive isolate 10-0036-2 were also cloned into pHW2000 and transfected along with plasmids bearing polymerase basic 2 (PB2), polymerase basic 1 (PB1), polymerase acid (PA), nucleoprotein (NP), matrix (M), and nonstructural (NS) genes derived from A/swine/North Carolina/3793/08 to produce a reverse genetics-derived isolate (RG 10-0036-2) (Fig. 1). Additionally, the HA gene from neutralization-resistant isolate 11-0854-1 was cloned and transfected along with plasmids containing the NA gene from isolate 10-0036-2 and the PB2, PB1, PA, NP, M, and NS genes from A/swine/North Carolina/3793/08 to create RG 11-0854-1. Site-directed mutagenesis was performed on plasmids containing HA genes from isolates 10-0036-2 and 11-0854-1 to create mutants RG 10-0036-2 T144N and RG 11-0854-1 N144T, respectively, using a QuikChange II site-directed mutagenesis kit (Agilent Technologies).
Fig 1.
Genotypes of viruses created using reverse genetics. Blue bars represent genes from A/swine/North Carolina/3793/08, yellow bars represent genes from field isolate 10-0036-2, and the green bar represents a gene from field isolate 11-0854-1.
Rescue of recombinant viruses was performed as previously described (16). In brief, 293T and MDCK cells were cocultured in Opti-MEM I containing 5% FBS in 6-well plates containing approximately 1 × 106 cells each of 293T and MDCK cells approximately 18 h prior to transfection. One hundred nanograms of each of the eight plasmids was pooled in 100 μl of Opti-MEM I, combined with 100 μl Opti-MEM containing 3 μl Lipofectamine (Invitrogen), and incubated at room temperature for 15 min before being diluted to 1 ml with Opti-MEM I and transferred to a single well of the 6-well plate. Plates were incubated at 37°C with 5% CO2 for 6 h before the transfection mixture was replaced with Opti-MEM I. At 24 h posttransfection, 1.5 ml of each mixture was transferred to a 6-well plate of confluent MDCK cells, and 1.5 ml of DMEM containing 1 μg/ml of tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin was added. Viruses were harvested on day 5 postinfection, and their titers were determined by HA assay. The HA genes of rescued viruses were sequenced to verify the correct sequence.
Hemagglutinin modeling.
Homologs of neutralization-sensitive isolate 10-0036-2 with either T or N at position 144 were searched against the Protein Data Bank (PDB) structure database by using BLASTP with default parameters (2). The structures of HA proteins with T144 or N144 in complex with neutralizing antibody were built using Modeller, with PDB structure 3SM5 as the template (10-0036-2 and 3SM5 sequence identity was 92%) (20). Homologs with glycosylation sites at N142 or N144 were glycosylated artificially with a complex N-glycan (Glycom-DB accession no. 8793) by using Glyprot (3). The preferred orientation of the attached N-glycan relative to the glycosylation site was chosen using default parameters, based on knowledge of the orientation preference of glycans in known glycoproteins (3). We superimposed and replaced the HA in structure 3SM5 with our modeled HA by using Pymol.
Nucleotide sequence accession numbers.
HA gene sequences were deposited in GenBank under accession numbers JQ638655 to JQ638665.
RESULTS
Genetic analysis identified a conserved N144 mutation present only in nonreactive viruses in HTSN and HI assays.
During our regular surveillance of genetic diversity and antigenic variation of swine influenza virus HA from 2010 to 2011, using an HTSN assay incorporating a panel of reference antisera generated against representative viruses of contemporary circulating influenza viruses, we found that neutralization-resistant viruses accounted for approximately 20% of virus isolates studied. In order to understand the molecular basis of antigenic variation, we randomly selected five neutralization-sensitive isolates and five neutralization-resistant isolates for further genetic and antigenic analysis.
Consistent with our previous observation (data not shown), isolates 10-1287-1, 10-3025-1, 10-3215-1, 11-1035-1, and 11-0376-1 were all neutralized by antisera generated against δ-subclusters C, D, and E (Table 1). Similarly, the same isolates had high HI titers with the same antisera (mean titer = 1,255) (Table 2). In contrast, isolates 10-2858-1, 11-0667-1, 11-0669-1, 11-0854-1, and 11-1553-1 were all negative in the HTSN assay and had low HI titers, against δ-subclusters B and E only (mean titers = 13 and 64, respectively).
Table 1.
Summary of HTSN ratios of δ-subcluster E field isolates tested against δ-subcluster antiseraa
| Viral isolate | N siteb | HTSN ratio with indicated antiserum |
|||||
|---|---|---|---|---|---|---|---|
| δ-A | δ-B | δ-C | δ-D | δ-E | ST cell antiserum | ||
| 10-2858-1 | 144 | 0.08 | 0.02 | 0.01 | 0.02 | 0.00 | 0.03 |
| 11-0667-1 | 144 | 0.12 | 0.07 | 0.03 | 0.05 | 0.01 | 0.06 |
| 11-0669-1 | 144 | 0.11 | 0.07 | 0.04 | 0.07 | 0.03 | 0.04 |
| 11-0854-1 | 144 | 0.05 | 0.02 | 0.01 | 0.00 | 0.04 | 0.00 |
| 11-1553-1 | 144 | 0.06 | 0.03 | 0.02 | 0.02 | 0.01 | 0.01 |
| 10-1287-1 | 142 | 0.06 | 0.02 | 0.98 | 0.98 | 0.99 | 0.02 |
| 10-3025-1 | 142 | 0.05 | 0.02 | 1.00 | 1.00 | 1.00 | 0.00 |
| 10-3215-1 | 142 | 0.05 | 0.01 | 0.98 | 0.98 | 0.99 | 0.00 |
| 11-0376-1 | 137 | 0.09 | 0.00 | 0.96 | 0.96 | 0.97 | 0.01 |
| 11-1035-1 | 142 | 0.13 | 0.04 | 0.97 | 0.97 | 0.98 | 0.02 |
Positive ratios are shown in bold. Results are means for duplicate analyses.
Amino acid position of N-linked glycosylation.
Table 2.
HI antibody titers of δ-subcluster E isolates against δ-subcluster antiseraa
| Viral isolate | N siteb | HI antibody titer against indicated antiserum |
|||||
|---|---|---|---|---|---|---|---|
| δ-A | δ-B | δ-C | δ-D | δ-E | ST cell antiserum | ||
| 10-2858-1 | 144 | 0 | 17 | 0 | 0 | 80 | 0 |
| 11-0667-1 | 144 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11-0669-1 | 144 | 0 | 10 | 0 | 0 | 80 | 0 |
| 11-0854-1 | 144 | 0 | 20 | 0 | 0 | 80 | 0 |
| 11-1553-1 | 144 | 0 | 20 | 0 | 0 | 80 | 0 |
| 10-1287-1 | 142 | 0 | 0 | 2,560 | 640 | 853 | 0 |
| 10-3025-1 | 142 | 0 | 0 | 2,560 | 640 | 1,067 | 0 |
| 10-3215-1 | 142 | 0 | 10 | 1,707 | 640 | 1,067 | 0 |
| 11-0376-1 | 137 | 0 | 0 | 2,133 | 160 | 320 | 0 |
| 11-1035-1 | 142 | 0 | 0 | 2,560 | 640 | 1,280 | 0 |
Means for triplicate analyses are shown.
Amino acid position of N-linked glycosylation site.
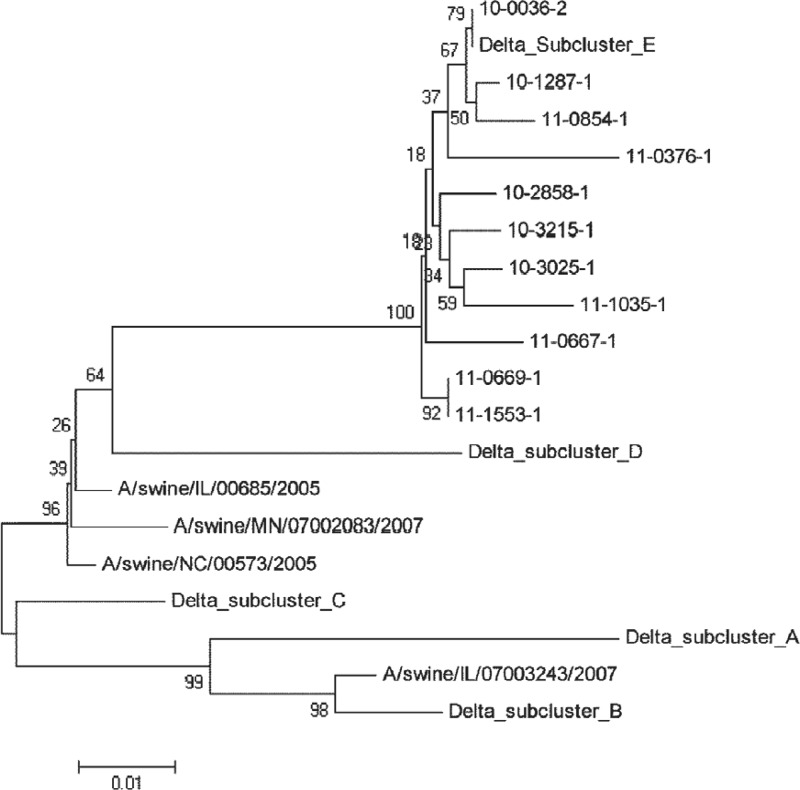
Next, we sequenced the full-length HA genes of all 10 viruses. The results showed that these viruses share >97% amino acid identity and all belong to δ-subcluster E (Fig. 2). Subsequent analysis of predicted N-linked glycosylation sites (N-X-S/T) found conserved sites at N28, N40, N104, N176, N303, N497, and N556 among these viruses (data not shown). Interestingly, we noted that five neutralization-resistant viruses (10-2858-1, 11-0667-1, 11-0669-1, 11-0854-1, and 11-1553-1) all possessed an eighth additional glycosylation site, at N144 (Fig. 3). In contrast, four viruses (10-1287-1, 10-3025-1, 10-3215-1, and 11-1035-1) had the eighth predicted glycosylation site at N142, and one virus (11-0376-1) had the remaining glycosylation site at N137. The last five viruses were neutralized by reference antisera in the HTSN assay. Variation in potential N-linked glycosylation at N142 and N144 has been observed previously in human influenza virus HA proteins (24, 31). Thus, our results appear to suggest a potential link between glycosylation pattern alteration centering at N142/N137 or N144 and susceptibility of viruses to neutralization.
Fig 2.
Phylogenetic analysis of the hemagglutinin proteins of the swine influenza virus δ-cluster isolates used in this study.
Fig 3.
Hemagglutinin amino acid sequences for positions 122 to 162. Potential N-linked glycosylation sites are shaded in gray. The amino acids located in the Sa epitope are P141 and N142. Viruses with the prefix “RG” were engineered using reverse genetics.
Migration of N-linked glycosylation site from N142 to N144 is responsible for discriminating viruses into neutralization-sensitive and neutralization-resistant groups.
To elucidate the genetic basis for the observed antigenic variation, two approaches were pursued in this study. One approach was to develop a reverse genetics system that allowed for rescue of representative viruses, while the other approach was to employ site-specific mutagenesis to study how the presence or absence of N144 impacts viral susceptibility to neutralization.
A TRIG swine influenza virus reverse genetics system was created using H1N1 A/swine/North Carolina/3793/08. We selected neutralization-sensitive virus 10-0036-2, containing N142 and T144, and neutralization-resistant virus 11-0854-1, containing N142 and N144, as representative strains in our study. HA and NA segments derived from isolate 10-0036-2 were combined with a TRIG swine influenza virus backbone consisting of the PB2, PB1, PA, NP, M, and NS genes to rescue the reverse genetics-derived virus RG 10-0036-2 (Fig. 1). RG 11-0854-1 virus was recovered from 293T and MDCK cells transfected with the HA segment derived from isolate 11-0854-1 together with the NA segment of isolate 10-0036-2 in combination with the TRIG swine influenza virus backbone.
A replacement of threonine (T) at HA position 144 with asparagine (N) was made in the context of the neutralization-sensitive isolate RG 10-0036-2, and the resultant virus was designated RG 10-0036-2 T144N. Asparagine substitution at position 144 abrogates N142-associated N-linked glycosylation but provides a new glycosylation site, resembling the case in the neutralization-resistant strains. Conversely, the asparagine at position 144 was replaced by threonine in neutralization-resistant isolate RG 11-0854-1 without altering any other amino acids, including N142. The derived virus, designated RG 11-0854-1 N144T, mimics neutralization-sensitive viruses. We did not perform site-directed mutagenesis on the N-linked glycosylation site at N137, as observed for 11-0376-1, because a search of our influenza virus HA sequence database identified only three instances (<1%) of N137.
Next, we characterized these mutant viruses in parallel with their parental strains in serological assays for susceptibility to neutralization or inhibition by the reference antisera. Reverse genetics-derived viruses were serologically similar to their corresponding parent viruses (Tables 3, 4, and 5). RG 10-0036-2 was neutralized by δ-subcluster C, D, and E antisera in the HTSN assay (Table 3). Similarly, HI titers for these antisera were 1,280, 267, and 640, respectively, against RG 10-0036-2 (Table 4). RG 10-0036-2 has a predicted glycosylation site at N142, and 10-0036-2 is the isolate used to generate δ-subcluster E antisera. In contrast, RG 10-0036-2 T144N, with the disappearance of N142 glycosylation but the appearance of potential N144 glycosylation, was not neutralized by any δ-subcluster antisera in the HTSN assay and had HI titers of 13 and 40 for antisera generated against subclusters B and E, respectively. This result was further supported in the gain-of-function experiment described below. RG 11-0854-1, which is similar to RG 10-0036-2 T144N in that it has a predicted glycosylation site at N144 but not at N142, was also not neutralized by any δ-subcluster antiserum and had similar HI titers (δ-B antibody titer of 20 and δ-E antibody titer of 80). Significantly, however, the N144T mutation and predicted glycosylation site at N142 rendered RG 11-0854-1 susceptible to antibody-mediated neutralization in HTSN assays and to inhibition in HI assays. Additionally, genetically engineered viruses and their parents were assayed by traditional serum neutralization assays. Regression analysis of HI and SN titers found a strong correlation (R2 = 0.86). Antisera generated from δ-C, δ-D, and δ-E viruses had SN titers in excess of 10,000 for viruses with predicted glycosylation at N142 (10-0036-2, RG 10-0036-2, and RG 11-0854-1 N144T) (Table 5). In contrast, the same antisera had a >8-fold drop in titer for viruses with predicted glycosylation at N144 (RG 10-0036-2 T144N, 11-0854-1, and RG 11-0854-1). While neutralizing antibodies were measurable for all reactions, the trends in serum neutralizing titers closely matched the titers measured by the HI assay, i.e., high-titer antibodies present in δ-C, δ-D, and δ-E antisera recognized viruses with predicted glycosylation at N142, and migration of this glycosylation site to N144 resulted in a substantial loss in antibody binding. Thus, the results of our experiments demonstrate that a shift of the predicted N-linked glycosylation site at N142 to N144 is responsible for some swine influenza viruses escaping neutralization by antibody.
Table 3.
HTSN ratios of parent and reverse genetics-derived viruses and mutantsa
| Virus | N siteb | HTSN ratio with indicated antiserum |
ST cell antiserum | ||||
|---|---|---|---|---|---|---|---|
| δ-A | δ-B | δ-C | δ-D | δ-E | |||
| 10-0036-2 | 142 | 0.07 | 0.03 | 0.99 | 0.99 | 1.00 | 0.01 |
| RG 10-0036-2 | 142 | 0.03 | 0.01 | 1.00 | 1.00 | 1.00 | 0.00 |
| RG 10-0036-2 T144N | 144 | 0.04 | 0.02 | 0.01 | 0.01 | 0.01 | 0.02 |
| 11-0854-1 | 144 | 0.05 | 0.02 | 0.01 | 0.00 | 0.04 | 0.00 |
| RG 11-0854-1 | 144 | 0.04 | 0.00 | 0.01 | 0.00 | 0.11 | 0.00 |
| RG 11-0854-1 N144T | 142 | 0.07 | 0.01 | 0.98 | 0.98 | 0.98 | 0.00 |
Positive ratios are shown in bold. Means for duplicate analyses are shown. “RG” denotes strains derived by reverse genetics.
Amino acid position of N-linked glycosylation site.
Table 4.
HI antibody titers of parent and reverse genetics-derived viruses and mutantsa
| Virus | N siteb | HI antibody titer against indicated antiserum |
|||||
|---|---|---|---|---|---|---|---|
| δ-A | δ-B | δ-C | δ-D | δ-E | ST cell antiserum | ||
| 10-0036-2 | 142 | 0 | 0 | 1,280 | 267 | 640 | 0 |
| RG 10-0036-2 | 142 | 0 | 0 | 2,560 | 1,707 | 1,067 | 0 |
| RG 10-0036-2 T144N | 144 | 0 | 13 | 0 | 0 | 40 | 0 |
| 11-0854-1 | 144 | 0 | 20 | 0 | 0 | 80 | 0 |
| RG 11-0854-1 | 144 | 0 | 17 | 0 | 0 | 80 | 0 |
| RG 11-0854-1 N144T | 142 | 10 | 10 | 2,560 | 320 | 1,280 | 0 |
Means for triplicate analyses are shown. “RG” denotes strains derived by reverse genetics.
Amino acid position of N-linked glycosylation site.
Table 5.
SN antibody titers of parent and reverse genetics-derived viruses and mutantsa
| Virus | N siteb | SN antibody titer against antiserum |
|||||
|---|---|---|---|---|---|---|---|
| δ-A | δ-B | δ-C | δ-D | δ-E | ST cell serum | ||
| 10-0036-2 | 142 | 362 | 362 | 23,170 | 11,585 | 23,170 | 11 |
| RG 10-0036-2 | 142 | 362 | 362 | 16,384 | 11,585 | 23,170 | 3 |
| RG 10-0036-2 T144N | 144 | 91 | 181 | 91 | 45 | 724 | 23 |
| 11-0854-1 | 144 | 23 | 362 | 181 | 23 | 2,896 | 11 |
| RG 11-0854-1 | 144 | 45 | 181 | 91 | 23 | 2,896 | 11 |
| RG 11-0854-1 N144T | 142 | 181 | 362 | 23,170 | 23,170 | 23,170 | 0 |
Means for duplicate analyses are shown. “RG” denotes strains derived by reverse genetics.
Amino acid position of N-linked glycosylation site.
Antigenic cartography was performed to visualize the HI data. Neutralization-resistant viruses clustered within 1.5 log2 of each other (Fig. 4, red dots). A similar small distribution was found for neutralization-sensitive viruses (green dots). The neutralization-resistant and -sensitive clusters were separated by 2 to 3 log2. Reverse genetics-created viruses clustered within 1 log2 of their wild-type parent strains, and mutation of the potential glycosylation site from N142 to N144, or vice versa, led to a 2- to 3-log2 change in cartographic position.
Fig 4.
Antigenic cartograph of influenza viruses, constructed by AntigenMap (http://sysbio.cvm.msstate.edu/AntigenMap) using hemagglutination inhibition titers. Pink, δ-cluster reference antisera; green, neutralization-sensitive influenza virus isolates; red, neutralization-resistant influenza virus isolates; blue, reverse genetics-derived isolate 11-0854 (RG 11-0854-1) and N144T mutant; yellow, reverse genetics-derived isolate 10-0036-2 (RG 10-0036-2) and T144N mutant.
Structural modeling of neutralizing antibody in complex with hemagglutinin further suggests a role for N144 in mediating viral evasion of antibody recognition.
To provide a structural basis for N144-directed viral evasion of neutralization and to explore a new avenue for designing a broad-spectrum antibody response, we conducted structural modeling of neutralizing antibody in complex with swine influenza virus HA antigen containing a predicted glycosylation site at N142 or N144. The structure of the H1 ectodomain and associated neutralizing antibody (PDB accession no. 3SM5) was used as a template to perform homology modeling, while a complex glycan was artificially superimposed on either the N142 or N144 site. As demonstrated in Fig. 5, the glycan at N142 extends away from the contact between the HA head domain and the antibody, thereby exhibiting no interference with the interaction between antibody and viruses observed in the neutralization of viruses such as 10-0036-2 or RG 11-0854-1 N144T. In marked contrast, the complex glycan at N144 forms strong steric clashes with residues of the antibody heavy chain (Fig. 6), which may prevent the binding of the antibody heavy chain to the HA head domain for viruses that result in nonneutralizing phenotypes, such as 11-0854-1 or RG 10-0036-2 T144N. Our model also shows that the distance between residue E102 of the antibody heavy chain and the first and second glycans of N144 (Fig. 6) is shorter than the van der Waals radius of two atoms (∼2.8 Å). Such a short distance might lead to steric clash, and as a result, addition of one or two glycans to HA N144 will affect HA-antibody interaction. Together with the above serology data, this strongly suggests that the presence of an asparagine at HA position 144 and the associated potential N-linked glycosylation may abolish antibody binding in a subset of swine influenza viruses, probably by inducing a steric hindrance effect.
Fig 5.
Structural modeling of neutralizing antibody in complex with swine influenza virus HA antigen containing a predicted glycosylation site at N142. The structure of the H1 ectodomain and associated neutralizing antibody (PDB accession no. 3SM5) was used as a template to perform homology modeling, while a complex glycan was artificially superimposed on the N142 site. Orange, HA; green, antibody heavy chain; cyan, antibody light chain; magenta, glycans.
Fig 6.
Structural modeling of neutralizing antibody in complex with swine influenza virus HA antigen containing a predicted glycosylation site at N144. Panels B and C are magnified images of the antigen-antibody contact surface shown in panel A, with different magnifications focusing on different regions. The antibody-binding pocket on HA is shown using a surface diagram, while antibody and other regions of HA are shown as ribbon diagrams. Orange, HA; green, antibody heavy chain; cyan, antibody light chain. N137, N142, and N144 are colored marine (B), while glycans on N144 are colored magenta (C). In panel C, a residue on the antibody heavy chain that is close to N144 of HA is colored yellow, and the distances from the closest atoms to this residue of the heavy chain and residue N144 are shown (the atoms are antibody heavy chain E102 OE1 and HA N144 ND2). All oxygen atoms and nitrogen atoms shown are colored red and blue, respectively.
DISCUSSION
Killed viral vaccines are commonly used in the United States to control swine influenza virus. Most vaccines are multivalent, containing genetically and antigenically distinct viruses to elicit broadly cross-reactive antibodies. Sequencing of the hemagglutinin gene is a standard practice for assessing genetic relationships and selection of vaccine strains. Variation in hemagglutinin N-linked glycosylation is a known mechanism employed by influenza virus to evade host immune responses; however, analysis of glycosylation variability is not routinely performed for swine influenza viruses in the swine industry. Additionally, the effects of glycosylation pattern changes (in both the number of sites and location) on antigenicity are not well understood for swine influenza virus.
Routine antigenic analysis in our laboratory, using swine influenza virus isolates from clinical submissions, found evidence for considerable antigenic drift, with 20% of isolates not being neutralized by a broad panel of antisera generated against all genetic clusters of influenza virus circulating in the United States. Genetic analysis of the HA genes of these isolates found that nearly all are within the δ cluster, a seasonal human influenza virus lineage (28). Hemagglutinin sequence analysis found a conserved T144N mutation (at the predicted glycosylation site at N144) present in all isolates that were not neutralized in the HTSN assay. Analysis of the glycosylation patterns of seasonal human influenza viruses found that a majority of viruses from 1940 to 1985 were glycosylated at N144 (24). Conversely, nearly all seasonal human influenza viruses from 1986 to the present were glycosylated at N142. Swine influenza viruses containing human seasonal influenza virus HA genes were first identified in 2003, and accordingly, N142 is most common in swine influenza virus isolates with human lineage HA (17).
In this study, we confirmed that viruses containing N144, which is located in a consensus glycosylation sequence motif, are not neutralized by antisera generated against genetically similar viruses exhibiting a predicted glycosylation site at N142. Four field isolates with N142 and one with N137 were all neutralized by well-matched antisera in HTSN assays and effectively inhibited in HI and SN assays. Conversely, viruses with the T144N change were not neutralized in the HTSN assay and exhibited much less inhibition in HI and SN assays. N142 is located in the well-characterized Sa epitope, while N144 is immediately adjacent. The Sa epitope is located on the top part of the HA globular head, and mutations in this region have previously been shown to have a pronounced effect on viral antigenicity (6, 23, 31). To eliminate the possibility of other genetic differences, viruses possessing glycosylation consensus sequences at N142 or N144 were created by reverse genetics and subsequently mutated to migrate the glycosylation consensus sequence to N144 or N142, respectively. Cross-reaction of the engineered viruses was consistent with observations from field isolates with respect to the glycosylation sequence located at either N142 or N144. Antigenic cartography also illustrated that the neutralization-sensitive and -resistant phenotypes were determined by differential glycosylation at N142 and N144.
In this study, we also attempted to address whether the N142 and N144 mutations altered the glycosylation patterns of the HA proteins of both viruses. Examination of purified virions from both viruses by SDS-PAGE and Western blotting revealed a shift in mobility of HA proteins when peptide N-glycosidase F-treated and nontreated samples were compared directly, indicating that N142- and N144-carrying HA proteins are modified by N-linked glycans (data not shown). A previous study convincingly demonstrated N142-linked glycosylation (8). No visible differences in HA mobility were observed for viruses bearing N-linked glycosylation sites at N142 or N144. This result was expected, as the numbers of potential glycosylation sites in HA proteins containing N142 or N144 are equivalent. Alternatively, it could be suggested that the low-resolution SDS-PAGE/Western blotting assay using polyclonal antisera may not detect any quantitative difference in glycosylation patterns that may occur in HA proteins with an N142 or N144 mutation. In this regard, a high-resolution mass spectrometry approach will be pursued in our future work.
To elucidate the structural basis for the lack of cross-reactivity of antibodies generated against viruses with N142 to those with N144, models of hemagglutinin in complex with neutralizing antibody were developed. The complex glycan attached to N144 is projected from the hemagglutinin head domain, which may sterically hinder binding of the antibody heavy chain. Conversely, N142-carrying viruses had complex glycans extended away from the antigen-antibody contact interface, which may not interfere with antibody binding to HA protein. Additionally, a recent genetic analysis of influenza virus glycosylation site migrations suggested that glycosylation at N142 better protects the antigenic Sa site, while glycosylation at N144 is more effective at shielding the antigenic Sb site (25). Thus, the modeling data appear to explain the observed results of our HTSN, HI, and SN assays, showing that antibody is sterically hindered from binding to the head domain of HA for viruses with N144-associated glycans, as well as supporting other predictions based on genetic and structural studies.
Genetic analysis of δ-cluster hemagglutinin sequences in the Newport Laboratories swine influenza virus database found a low prevalence of viruses with N144 prior to 2009 (3.6%). From 2009 to 2011, viruses with N144 represented 18.5% of δ-cluster isolates, and viruses with N144 were identified in each of the five δ subclusters. It can be postulated that these viruses are neutralization resistant, though further experiments are needed to prove this hypothesis. Phylogenetic analysis shows a close relationship between neutralization-resistant and neutralization-sensitive viruses, suggesting that the neutralization-resistant viruses evolved from the neutralization-sensitive δ-cluster viruses circulating in swine since 2003, possibly due to immune selection pressure. Given that swine influenza viruses within the δ cluster contain an HA gene derived from human seasonal influenza virus and that viruses with N-linked glycosylation at position 144 have not circulated in humans since 1985, individuals born thereafter may be more vulnerable to zoonotic transmission of these viruses, as previous work has shown no cross-protection between H1 vaccine strains produced prior to 1986 and vaccines produced in subsequent years (13). Importantly, vaccines for swine influenza virus may require multiple strains of δ-cluster isolates representing glycosylation variants. This work also highlights the limitations of genetic analysis alone, illustrating the need for coupled antigenic surveillance of influenza virus.
Finally, our observation that relocation of potential N-linked glycosylation sites protects swine influenza virus from rapid neutralization by antibodies is interesting and novel. It suggests a role for immune selection in influenza virus in swine. While this study clearly demonstrates the effect of glycosylation on in vitro antigenicity, little is known about the role of glycosylation on in vivo immunogenicity, which will be a focus of future investigations.
ACKNOWLEDGMENTS
We thank Emily Collin, Jen Iverson, and Amy Shirbroun for technical assistance.
Footnotes
Published ahead of print 18 July 2012
REFERENCES
- 1. Abe Y, et al. 2004. Effect of the addition of oligosaccharides on the biological activities and antigenicity of influenza A/H3N2 virus hemagglutinin. J. Virol. 78:9605–9611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altschul SF, Madden TL. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bohne-Lang A, von der Lieth CW. 2005. GlyProt: in silico glycosylation of proteins. Nucleic Acids Res. 33:W214–W219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bragstad K, Nielsen LP, Fomsgaard A. 2008. The evolution of human influenza A viruses from 1999 to 2006: a complete genome study. Virol. J. 5:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cai Z, Zhang T, Wan X-F. 2010. A computational framework for influenza antigenic cartography. PLoS Comput. Biol. 6:e1000949 doi: 10.1371/journal.pcbi.1000949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. 1982. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31:417–427 [DOI] [PubMed] [Google Scholar]
- 7. Cottey R, Rowe CA, Bender BS. 2001. Influenza virus. Curr. Protoc. Immunol. Chapter 19:Unit 19.11 doi:10.1002/0471142735.im1911s42 [DOI] [PubMed] [Google Scholar]
- 8. Das SR, et al. 2011. Fitness costs limit influenza A virus hemagglutinin glycosylation as an immune evasion strategy. Proc. Natl. Acad. Sci. U. S. A. 108:E1417–E1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Das SR, et al. 2010. Glycosylation focuses sequence variation in the influenza A virus H1 hemagglutinin globular domain. PLoS Pathog. 6:e1001211 doi: 10.1371/journal.ppat.1001211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Desselberger U, et al. 1978. Biochemical evidence that “new” influenza virus strains in nature may arise by recombination (reassortment). Proc. Natl. Acad. Sci. U. S. A. 75:3341–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hause BM, Oleson TA, Stine DL, Bey RF, Simonson RR. 2011. Genetic and antigenic characterization of recent human-like H1 (δ-cluster) swine influenza virus isolates. J. Swine Health Prod. 19:268–276 [Google Scholar]
- 12. Hause BM, Oleson TA, Bey RF, Stine DL, Simonson RR. 2010. Antigenic categorization of contemporary H3N2 swine influenza virus isolates using a high-throughput serum neutralization assay. J. Vet. Diagn. Invest. 22:352–359 [DOI] [PubMed] [Google Scholar]
- 13. Hay AJ, Gregory V, Douglas AR, Lin YP. 2001. The evolution of human influenza viruses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:1861–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hensley SE, et al. 2011. Influenza A virus hemagglutinin antibody escape promotes neuraminidase antigenic variation and drug resistance. PLoS One 6:e15190 doi:10.1371/journal.pone.0015190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. 2001. Universal primer set for the full length amplification of all influenza viruses. Arch. Virol. 146:2275–2289 [DOI] [PubMed] [Google Scholar]
- 16. Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. U. S. A. 97:6108–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Karasin AI, Carman S, Olsen CW. 2006. Identification of human H1N2 and human-swine reassortant H1N2 and H1N1 influenza A viruses among pigs in Ontario, Canada (2003–2005). J. Clin. Microbiol. 44:1123–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Long J, et al. 2011. Evolution of H3N2 influenza virus in a guinea pig model. PLoS One 6:e20130 doi:10.1371/journal.pone.0020130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lorusso A, Vincent AL, Gramer MR, Lager KM, Ciacci-Zanella JR. 22 January 2012, posting date. Contemporary epidemiology of North American lineage triple reassortant influenza A viruses in pigs. Curr. Top. Microbiol. Immunol. [Epub ahead of print.] doi:10.1007/82_2011_196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sali A, Potterton L. 1995. Evaluation of comparative protein modeling by MODELLER. Proteins 23:318–326 [DOI] [PubMed] [Google Scholar]
- 21. Schild G, et al. 1974. Antigenic variation in current influenza A viruses: evidence for a high frequency of antigenic ‘drift’ for the Hong Kong virus. Bull. World Health Organ. 51:1–11 [PMC free article] [PubMed] [Google Scholar]
- 22. Schulze IT. 1997. Effects of glycosylation on the properties and functions of influenza virus hemagglutinin. J. Infect. Dis. 176:S24–S28 [DOI] [PubMed] [Google Scholar]
- 23. Strengell M, Ikonen N, Ziegler T, Julkunen I. 2011. Minor changes in the hemagglutinin of influenza A(H1N1)2009 virus alter its antigenic properties. PLoS One 6:e25848 doi:10.1371/journal.pone.0025848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sun S, Wang Q, Zhao F, Chen W, Li Z. 2011. Glycosylation site alteration in the evolution of influenza A (H1N1) viruses. PLoS One 6:e22844 doi:10.1371/journal.pone.0022844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sun S, Wang Q, Zhao F, Chen W, Li Z. 2012. Prediction of biological functions on glycosylation site migrations in human influenza H1N1 viruses. PLoS One 7:e32119 doi:10.1371/journal.pone.0032119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vigerust DJ, et al. 2007. N-linked glycosylation attenuates H3N2 influenza viruses. J. Virol. 81:8593–8600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vincent AL, et al. 2009. Characterization of a newly emerged genetic cluster of H1N1 and H1N2 swine influenza virus in the United States. Virus Genes 39:176–185 [DOI] [PubMed] [Google Scholar]
- 29. Wagner R, Wolff T, Herwig A, Pleschka S, Klenk H-D. 2000. Interdependence of hemagglutinin glycosylation and neuraminidase as regulators of influenza virus growth: a study by reverse genetics. J. Virol. 74:6316–6323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wanzeck K, Boyd KL, McCullers JA. 2011. Glycan shielding of the influenza virus hemagglutinin contributes to immunopathology in mice. Am. J. Respir. Crit. Care Med. 183:767–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wei C-J, et al. 2010. Cross-neutralization of 1918 and 2009 influenza viruses: role of glycans in viral evolution and vaccine design. Sci. Transl. Med. 2:24ra21 doi:10.1126/scitranslmed.3000799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.WHO 2002. WHO manual on animal influenza diagnosis and surveillance. WHO, Geneva, Switzerland: http://whqlibdoc.who.int/hq/2002/WHO_CDS_CSR_NCS_2002.5.pdf [Google Scholar]
- 33. Zhou B, et al. 2009. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and swine origin human influenza A viruses. J. Virol. 83:10309–10313 [DOI] [PMC free article] [PubMed] [Google Scholar]