Abstract
A novel, whole-cell enzyme-linked immunosorbent assay (ELISA) based on a non-type-specific anti-human papillomavirus (HPV) E6 antibody was tested on 182 residual cytological specimens. For samples with a designation of more severe than cervical intraepithelial neoplasia grade 3 (CIN3+), 83% tested positive for E6; in a subset with paired testing for E6 ELISA and HPV DNA, 72% tested E6 positive and 92% tested high-risk (HR)-HPV DNA positive (P = 0.2). Among the women with a less than CIN3 diagnosis, 31% and 47% tested positive for E6 and HR-HPV DNA, respectively (P = 0.0006).
INTRODUCTION
Cervical cancer is the second most common cause of cancer deaths in women worldwide. Routine screening and treatment have substantially decreased the cervical cancer mortality rate in the United States. However, according to the World Health Organization, there are approximately 500,000 new cases and 250,000 deaths from cervical cancer every year worldwide. Although Pap tests and colposcopy have contributed tremendously to the decreased mortality of cervical cancer, they are subjective tests, prone to human error, and not always conclusive. Therefore, it is extremely important to develop more objective screening tools that can identify patients who are most at risk for developing cervical cancer (8).
Human papillomavirus (HPV) infection is a major cause of virtually all invasive cervical cancers (2, 4, 26, 28, 33). Of the 40 HPV types that infect the genital tract, only a subset of HPV subtypes are classified as “high-risk” HPV (HR-HPV) types that were found in cancers (33). Most of these HPV infections are transient, are resolved by the body's immune system, and have no major clinical consequences. However, persistent HPV infections are found in 5 to 10% of infected women and represent a high risk factor for progression to cervical cancer (3, 25). Thus, it is important to identify the small percentage of women with HPV infections who are truly at risk for developing cervical cancer. Unfortunately, current screening tests cannot accurately predict the risk of dysplasia or cancer. Therefore, there is a significant need to develop a test that could better predict progression to these outcomes.
The current paradigm for cervical cancer screening is based on the Pap test, which is a cytologically based test using cells scraped from the cervix that are examined microscopically to detect dysplastic lesions (9, 15a, 20, 23). There are approximately 4 million abnormal Pap tests each year in the United States. Under current practice guidelines, most of these patients are referred for colposcopy and cervical biopsy to identify the subset that has clinically significant high-grade precancers, such as cervical intraepithelial neoplasia grade 2/3 (CIN2/3) (15a). However, the Pap test is subjective, with significant interobserver variability, and is limited by low sensitivity. In addition, high false-positive rates, defined as a positive Pap result with no clinically significant disease by subsequent biopsy (i.e., histology results of less than CIN2/3), were observed in two-thirds of patients with abnormal Pap smears (6, 7, 18, 23, 34). As a result, approximately 3 million colposcopic examinations performed each year, at a cost exceeding $2 billion dollars annually, may not be necessary.
In the last few years, HR-HPV DNA testing has been included in routine screening to increase the sensitivity and negative predictive value of the Pap test. While these tests can detect the presence of HPV DNA, they cannot differentiate a true precancerous state from self-limited HPV infection (1, 8, 22, 24), which represents the majority of infections. The low specificity of HPV DNA testing potentially results in overdiagnosis and inefficient disease management (26). In addition to DNA tests, a number of host cellular proteins, including p16INK4a, Ki67, and ProExC, have also been identified as biomarkers for cervical cancer diagnosis. However, they are considered surrogate markers and not specific to HPV.
HPVs are DNA viruses that code for several functional (E1 to E7) genes and two late structural (L1 and L2) genes. When high-risk HPV types integrate into the host genome, loss of negative-feedback control results in increased expression of viral E6 and E7 oncogenes, which in turn inactivate tumor suppressor genes that operate at key cell cycle checkpoints (10–12, 19, 29). Since these two oncogenes are integral to the development of cervical cancer, their gene products could potentially serve as highly specific biomarkers to identify high-grade precancerous lesions that may progress to cervical cancer if left untreated. Indeed, evidence suggests that elevated levels of the E6 and E7 oncoproteins are better indicators of increased risk for cervical cancer than the presence of HPV DNA (6, 15). The recent FDA approval of the Aptima HPV E6/E7 RNA test is a significant milestone for the application of E6/E7 as specific biomarkers for cervical cancer screening. However, RNA is prone to degradation, and its detection requires expensive instrumentation and cumbersome procedures; tests based on the detection of E6/E7 mRNA may have limited clinical application in routine gynecological practice. Diagnostic tests based on the direct detection of the E6/E7 oncoproteins may have advantages over detection of HPV DNA or HPV E6/E7 mRNA. Until recently, most of the antibodies developed against the HPV E6 or E7 protein used either peptides or denatured proteins, as it has been difficult to purify recombinant nondenatured E6 and E7 proteins suitable for antibody production (31). Antibodies produced using these denatured proteins do not have sufficient sensitivity for clinical use (16). We have recently overcome the technical hurdles and have purified recombinant HPV E6 and E7 proteins in their native form to generate monoclonal antibodies that recognize HPV E6 and E7 from many high-risk HPV types.
In this study, we describe a simple whole-cell enzyme-linked immunoabsorbent assay (ELISA) using a pan-HPV anti-E6 monoclonal antibody to detect HPV E6 protein in previously collected and frozen cytology samples. Using cervical biopsy results as the gold standard, ELISA results are compared to HPV DNA test results. The ability to detect the E6 oncoprotein in clinical samples is a critical advance that will facilitate the development of diagnostic testing to distinguish benign HPV infections from precancers, thus preventing unnecessary colposcopies and biopsies.
MATERIALS AND METHODS
Purification of recombinant HPV proteins.
HPV E6 and E7 cDNAs containing the respective coding regions were produced by PCR amplification and cloned into a histidine tag expression vector. The proteins were then expressed in Escherichia coli BL21(DE3) using isopropyl-β-d-thiogalactopyranoside (IPTG)-driven induction. In order to produce HPV E6 and E7 proteins in soluble, nondenatured form, full-length HPV type 18 (HPV18) E6 and E7 were expressed at 25°C and purified at low concentration using affinity chromatography without denaturation and refolding (Amersham and New England BioLabs). Recombinant HPV18 E6 protein, estimated to be >90% pure based on PAGE analysis, was used as an immunogen for generation of polyclonal and monoclonal antibodies.
Mouse monoclonal anti-HPV E6 antibody.
Anti-HPV E6 antibodies were generated using the purified native forms of recombinant E6 protein as immunogens in the BALB/c mouse strain at 1-mg/ml concentration using Freund's adjuvant. Monoclonal antibodies were screened by ELISA using HPV-related or non-HPV-related protein. To obtain non-HPV type-specific monoclonal antibodies, HPV16 and HPV18 E6 proteins were used to screen hybridoma cell lines. Monoclonal antibodies produced from ascites fluid were purified on a protein G column (ThermoScientific, IL).
Western blot analysis of cell lines.
Human cervical epithelial cell lines, HeLa (ATCC CCL-2), SiHa (ATCC HTB-35), and C33A (ATCC HTB-31), were purchased from ATCC and were used within 10 passages of purchase. Proteins from these cell extracts were prepared using 3% NP-40 lysis buffer. The protein concentration was determined by Bradford protein analysis. Proteins were separated by SDS-PAGE and then transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad) previously blocked with 5% (wt/vol) bovine serum albumin (BSA). The primary antibodies, mouse monoclonal anti-HPV E6 (1:1,000 dilution; NeoDiagnostic Laboratories Inc.) and anti-actin (1:5,000 dilution; Chemicon), were incubated overnight at 4°C, followed by secondary antibody (horseradish peroxidase-conjugated anti-mouse from Biobasic Inc., Canada; 1:5,000) and detected with an ECL detection kit (Biobasic Inc.).
HPV E6 whole-cell ELISA.
To test the hypothesis that E6 protein can serve as a valuable biomarker for HPV disease progression, we developed a whole-cell ELISA in which the residual cells from liquid-based cytology samples are directly immobilized onto 96-well microtiter plates. This whole-cell ELISA allows objective measurement of the HPV E6 oncoprotein expression level in cervical cancer cell lines or clinical specimens. Cells from cell lines or from cervical scrapes were immobilized by passive adsorption on 96-well plates for 30 min at room temperature (RT). Each plate was then washed 3 times with phosphate-buffered saline (PBS) for 5 min following each incubation, unless otherwise specified. After the washes, the cells were fixed with 25 μl 100% ethanol and air blow dried at RT, followed by cell permeabilization with chilled (−20°C) 90% methanol. To decrease the background signal and to block endogenous hydrogen peroxidase, the wells were incubated with 3% H2O2 for 20 min at RT, washed, and blocked with 100 μl of 10% normal goat serum for 2 h at RT. A proprietary anti-E6 monoclonal antibody developed by OncoHealth (diluted 1:200 in 10% normal goat serum) was added, and the plate was incubated for 1 h at RT, washed, and then incubated with biotinylated secondary antibody (50 μl/well; 1:500 in 5% normal goat serum; Vector Laboratories, Burlingame, CA) for 30 min at RT. After further washes, the wells were incubated with 50 μl of Streptavidin conjugated with horseradish peroxidase (HRP) (1:600; Vector Laboratories) for 45 min at RT, washed, and incubated with 50 μl of 3,3′,5,5′-tetramethylbenzidine substrate (BD Bioscience, San Jose, CA) for 10 min. The reaction was stopped by addition of 25 μl of acid stop solution, and the signal intensity was measured as the optical density at 450 nm (OD450) using a plate reader.
To determine the detection limits of the whole-cell ELISA and to generate standard curves using recombinant HPV E6 protein, a high-binding 96-well microtiter plate was directly coated with serial titrations of purified HPV E6 protein (0.4 pg to 4 μg/ml of PBS) overnight at 4°C. Blocking of each well was performed on the second day with 100 μl of 10% normal goat serum for 2 h at RT. Assay procedures were performed as described above to generate a standard curve for HPV18 E6.
Clinical samples.
The cervical samples used in this study were obtained from collaborative clinical laboratories from Asia and North America in compliance with institutional review board (IRB)-approved protocols. These clinical laboratories performed colposcopy/biopsy and HR-HPV testing and provided us the cytological diagnosis and HR-HPV DNA results. The histology diagnosis was categorized as benign, CIN grade 1, 2, or 3, or cancer. CIN3 and the more severe CIN3+ (n = 42), the precancerous and cancer stage where treatment takes place in current clinical practice, were analyzed as one group. Clinical samples with diagnoses of less than CIN3 (n = 140) were analyzed as another group. CIN2 was not analyzed separately from CIN1, since there were only five CIN2 cases in the study. The HR-HPV DNA test results (assigned as either positive or negative) were based on the presence of the HR-HPV DNA according to the protocol used and validated at the clinical sites provided by the manufacturer of the test kits. Samples (182) were collected in liquid-based Pap test vials (preserved in ThinPrep from Hologic, Inc., Marlborough, MA, or SurePath from BD Diagnostics, Burlington, NC) and used in the current study. Specimens were normalized to the cellular volume of a 25-μl cell pellet per ml of solution, and all 182 specimens were tested in duplicate by E6 whole-cell ELISA. Of these samples, the clinical laboratories had results on HR-HPV DNA testing (using Hybrid Capture 2 from Qiagen) for 165 specimens. The 17 cases that were not tested for HR-HPV DNA were cancer cases (Tables 1 and 2). The results of our E6 ELISA were compared to those of the previously performed HR-HPV DNA test using histology diagnoses as a gold standard. An exact version of the McNemar chi-square test was used to test for differences between testing positive for CIN3+ and less than CIN3 for paired-test results.
Table 1.
Single-test results of the HPV E6 whole-cell ELISA and HR-HPV DNA
| HC2 DNA result | E6 ELISA result | All |
CIN3+ |
<CIN3 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. tested | No. positive | % Positive | No. tested | No. positive | % Positive | No. tested | No. positive | % Positive | ||
| Positive | Negative | 165 | 89 | 49 | 25 | 23 | 92 | 140 | 66 | 47 |
| Negative | Positive | 182 | 78 | 42 | 42 | 35 | 83 | 140 | 43 | 31 |
Table 2.
Paired-test results of the HPV E6 whole-cell ELISA and HR-HPV DNA
| HC2 DNA result | E6 ELISA result | All |
CIN3+ |
<CIN3 |
|||
|---|---|---|---|---|---|---|---|
| No. positive | % Cola | No. positive | % Col | No. positive | % Col | ||
| Positive | Positive | 49 | 36 | 16 | 38 | 33 | 24 |
| Positive | Negative | 40 | 21 | 7 | 17 | 33 | 24 |
| Negative | Positive | 12 | 7 | 2 | 5 | 10 | 7 |
| Negative | Negative | 64 | 35 | 0 | 0 | 64 | 46 |
| NAc | Positive | 17b | 9 | 17b | 40 | 0 | |
| NA | Negative | 0 | 0 | 0 | |||
% Col, column percentage.
All cervical cancers.
NA, not applicable.
Data analysis.
Purified recombinant HPV18 E6 protein was used as a positive control. Pap-negative PreservCyt liquid-based cytology (LBC) samples were pooled to be used as a negative control. Blank, empty wells were used as assay background controls. The OD of the blank well was subtracted from the OD450 raw data collected from the plate reader (BioTek ELX800) to obtain an average signal intensity as a readout for data analysis. Absorbance of individual samples obtained from whole-cell ELISA using anti-E6 antibody, as well as the average absorbance from each group, was determined. E6 protein levels were compared to the severity of the histologic grade using a Kruskal-Wallis test. Receiver operating characteristic (ROC) analysis (ROC curve) (data not shown) with a preliminary cutoff threshold of 0.33 was used to obtain the optimal sensitivity and specificity of the ELISA. The sensitivity was calculated as follows: number of true positives/(number of true positives + number of false negatives). The specificity was calculated as follows: number of true negatives/(number of true negatives + number of false positives).
RESULTS
Pan-HPV E6 antibody recognizes the E6 protein in cell lines infected with different high-risk HPV types and in clinical samples.
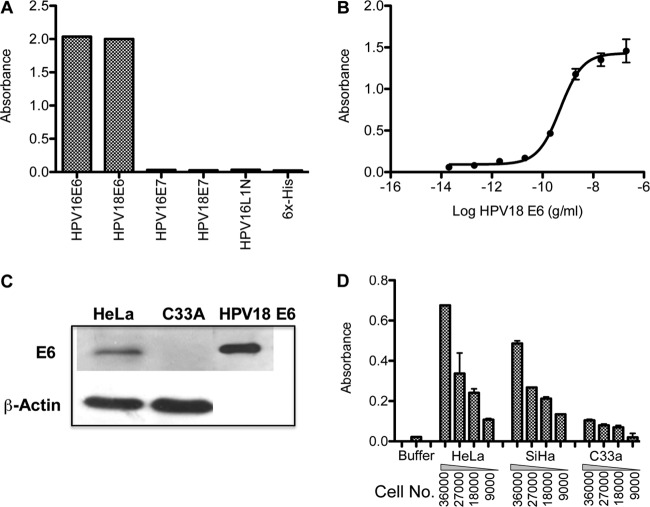
Currently available anti-HPV antibodies were produced against either small synthetic peptides or denatured recombinant proteins. These antigens often do not react with naturally occurring HPV antibodies (13, 14, 27, 30, 32). In order to obtain antibodies that recognize HPV E6 proteins in clinical samples, antibodies were generated using the purified native form of recombinant E6 protein as the immunogen. The mouse anti-E6 monoclonal antibody recognized recombinant HPV E6 proteins from HPV16 and -18, but not recombinant E7 proteins from HPV16 or -18 or the N-terminus of the HPV16 recombinant major capsid protein, L1 protein (Fig. 1A). The limit of detection for this anti-E6 antibody on recombinant HPV18 E6 protein ranged from 10 to 100 pg/ml (0.5 to 5 pg) (Fig. 1B) in an ELISA. The antibody also recognized E6 protein from HPV18-positive cervical-cancer-derived HeLa cells, but not that from HPV-negative cervical-cancer-derived C33A cells, in immunoblot analysis (Fig. 1C). Using whole-cell ELISA, the anti-E6 antibody detected E6 protein from SiHa and HeLa cells expressing HPV16 and HPV18, respectively, with signal strengths dependent on cell density (9,000 to 36,000 cells/well) (Fig. 1D). The E6 protein was minimally detected in HPV-negative C33A cells (Fig. 1D). The anti-E6 antibody examined in the current study possesses a unique binding site recognizing a common epitope of the E6 proteins that are accessible in clinical samples (S. Cheng, U.S. patent application US 2010/033944). In addition, the antibody is not HPV type specific and detects E6 proteins from the high-risk types HPV16, -18, -31, -33, -45, -51, -52, -58, and -59 (S. Cheng, U.S. patent application 2010/0003704 A1), which were available in the cases tested by immunohistochemistry and genotyping on formalin-fixed, paraffin-embedded (FFPE) cervical tissues (S. Cheng, U.S. patent application 2010/0003704 A1).
Fig 1.
Anti-E6 antibody binds specifically to HPV E6 protein. (A) ELISA results showing that anti-E6 antibody recognizes recombinant E6 proteins from HPV16 and -18 but not recombinant E7 proteins from HPV16 or -18, recombinant HPV16 L1 protein, or the 6×His tag. (B) Titration curves of recombinant HPV18 E6 protein detected by anti-E6 antibody in ELISA showing that the limits of detection range from 10 to 100 pg/ml. The data are presented as means and standard errors. (C) Immunoblot using cervical cancer cell line lysates. The top blot shows that anti-E6 antibody recognizes recombinant HPV18 E6 protein (right lane) and E6 protein from cell lysates from HeLa (HPV-positive) but not C33A (HPV-negative) cells. At the bottom is an immunoblot using anti-β-actin for the corresponding cell lysates. (D) Whole-cell ELISA using anti-E6 antibody detected E6 proteins from HeLa and SiHa cells expressing HPV18 and HPV16, respectively, with signal strengths dependent on the cell density. E6 protein was minimally detected in HPV-negative C33A cells.
HPV E6 detection in whole-cell ELISA correlates with the disease grade.
To determine whether this whole-cell ELISA could detect HPV E6 protein in clinical samples and whether the level of HPV E6 detected by the assay correlated with the disease grade, we tested 182 previously collected liquid-based cytology specimens (Fig. 2). We compared the results of the E6 whole-cell ELISA to the following categories defined by histology and HR-HPV results: (i) histology and HPV DNA negative (n = 62), (ii) histology negative and HPV DNA positive (n = 27), (iii) CIN1/2 (n = 51), and (iv) CIN3+ (n = 42; 15 CIN3 and 27 cancers). The absorbance of individual samples and the average absorbance obtained from each group are shown in Fig. 2A and B, respectively. E6 whole-cell ELISA detected significantly more E6 protein in cases of CIN3+ than the low absorbance levels observed from clinical samples that were less than CIN3 (P < 0.0001) (Fig. 2B). We set a positive threshold for absorbance of 0.33 for subsequent analyses.
Fig 2.
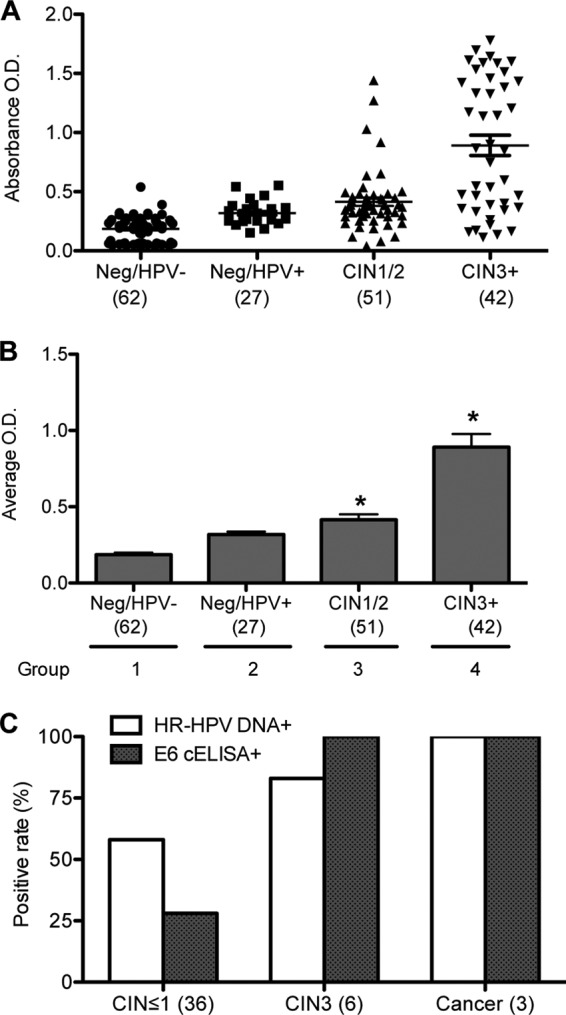
Whole-cell ELISA using anti-E6 antibody. (A) Scatter dot plot of the individual absorbance signals of clinical samples in a whole-cell ELISA using anti-E6 antibody. E6 whole-cell ELISA was performed on cells from 182 cervical scrapes that were categorized into the following 4 groups: group 1, histology negative and HPV DNA negative (Neg/HPV−) (n = 62); group 2, histology negative and HPV DNA positive (Neg/HPV+) (n = 27); group 3, CIN1/2 (n = 51); and group 4, CIN3+ (n = 42). Lines show means and standard errors of the means (SEM). (B) Average absorbances obtained from the ELISA in panel A graphed as means and SEM. (C) Results from E6 whole-cell ELISA using 45 SurePath fresh samples obtained within 1 to 2 weeks of collection. The data are presented as percent positive rates for histological designations of CIN ≤ 1 (36 samples), CIN3 (6 samples), and cervical cancer (3 samples) and compared to the positive rates for HPV DNA.
The E6 whole-cell ELISA and HPV DNA test results were compared (Tables 1 and 2). Thirty-five of the 42 (83%) CIN3+ cases tested E6 positive. Among the 25 cases of CIN3+ with paired testing by HR-HPV DNA and E6 whole-cell ELISA, 23 (92%) tested HR-HPV DNA positive and 18 (72%) tested E6 positive (P = 0.2). Among the 140 women without CIN3+ (less than CIN3), 31% tested positive for E6 and 48% tested positive for HR-HPV DNA (P = 0.0006).
Since degradation of E6 protein in archived samples may be responsible for the lower positive rate of E6 whole-cell ELISA than HPV DNA testing (Tables 1 and 2), we conducted a post hoc analysis on a subset of 45 samples that were tested within 1 to 2 weeks of collection using the same positive cutoff (OD = 0.33) (Fig. 2C). Of the 9 women diagnosed with CIN3+, 100% tested positive for HPV E6 whole-cell ELISA and 89% tested positive for HR-HPV DNA. For the 36 women with less than CIN3, 28% tested positive for HPV E6 whole-cell ELISA and 58% tested positive for HR-HPV DNA.
DISCUSSION
We generated a pan-HPV E6 monoclonal antibody that is capable of binding a common epitope among different high-risk HPV types. Based on that monoclonal antibody, we developed a whole-cell ELISA that was more specific but nonsignificantly less sensitive for CIN3+ than HR-HPV DNA detection. This is the first report demonstrating that a pan-HPV anti-E6 antibody used in a whole-cell ELISA can detect cervical precancers and cancer sensitively and specifically. The E6 whole-cell ELISA presented here incorporates a simple detection method that can potentially be used in diagnostic laboratories throughout the world.
In this study, the sensitivity and specificity of our E6 whole-cell ELISA were compared with the results of HPV DNA tests previously performed by the clinical laboratories that provided us with the samples for whole-cell ELISA testing. Both E6 whole-cell ELISA and HPV DNA testing were sensitive in identifying clinical cytologic samples with biopsy-proven CIN3 or cancer (Tables 1 and 2). Post hoc analysis of samples tested within 1 to 2 weeks of collection suggests that testing of more recent samples improves sensitivity (Fig. 2C). For clinical samples with histology results graded less than CIN3, E6 whole-cell ELISA detected a lower percentage of samples than HPV DNA testing (Tables 1 and 2). This result is consistent with published reports showing that HPV DNA testing has poor specificity (or high false-positive rates) despite the fact that it is highly sensitive (5, 21). Thus, E6 whole-cell ELISA is significantly more specific than HPV DNA testing while retaining similar sensitivity for detecting clinically significant disease. This improved specificity is important, since false-positive rates often result in excessive referrals and diagnostic procedures.
One of the limitations of this study is the fact that clinical samples were collected retrospectively. Our E6 whole-cell ELISA detects HPV E6 protein in CIN3 and cancer samples collected in ThinPrep or SurePath. In this 182-case study, it is noted that the E6 whole-cell ELISA is less sensitive for specimens from ThinPrep than for those from SurePath (data not shown). This could be due to the different preservative agents and/or the sample age. A prospective study with larger sample size using both ThinPrep and SurePath for collection from each patient is under way to determine the effect of the medium and sample age on the detection of HPV E6 protein by whole-cell ELISA.
Currently, HPV E6 and E7 expression can be measured only at the RNA level, which requires a more expensive instrument for analysis and sample processing. Direct measurement of HPV E6 protein expression may avoid the technical challenges associated with RNA testing. The E6 whole-cell ELISA presented in this study uses a simple detection method routinely used in diagnostic laboratories and thus may have advantages over RNA detection. Clinical samples are applied directly to 96-well plates without cell lysis or protein extraction. HPV E6 protein is detected using a monoclonal antibody that recognizes an epitope common to the E6 proteins from most high-risk HPV types. Thus, a single assay using anti-E6 antibody can detect the presence of HPV E6 protein from liquid-based cytology samples.
An essay that detects both HPV E6 and E7 proteins may further improve assay specificity and sensitivity. Prospective studies are under way to validate and determine the clinical utility of the assay using a larger set of recently collected clinical samples. The results will be compared side by side with HR-HPV16/18 DNA testing. Such a whole-cell ELISA (using anti-E6, anti-E7, or a combination of the two antibodies) may be a useful tool to screen for cervical precancer and cancer, either as a primary screening tool with or without Pap testing or as a triage test following an abnormal Pap and/or an HPV-positive result. It may offer an effective tool for physicians to differentiate precancerous lesions from benign HPV infections, thus reducing unnecessary repeat testing and invasive diagnostic procedures.
Footnotes
Published ahead of print 18 July 2012
REFERENCES
- 1. Arbyn M, et al. 2009. Triage of women with equivocal or low-grade cervical cytology results: a meta-analysis of the HPV test positivity rate. J. Cell. Mol. Med. 13:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arbyn M, et al. 2006. Clinical applications of HPV testing: a summary of meta-analyses. Vaccine 24(Suppl. 3):78–89 [DOI] [PubMed] [Google Scholar]
- 3. Bodily J, Laimins LA. 2011. Persistence of human papillomavirus infection: keys to malignant progression. Trends Microbiol. 19:33–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bosch FX, Lorincz A, Munoz N, Meijer CJLM, Shah KV. 2002. The causal relation between human papillomavirus and cervical cancer. J. Clin. Pathol. 55:244–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Castle PE. 2010. Screening: HPV testing for cervical cancer: the good, the bad, and the ugly. Nat. Rev. Clin. Oncol. 7:364–365 [DOI] [PubMed] [Google Scholar]
- 6. Castle PE, Sideri M, Jeronimo J, Solomon D, Schiffman M. 2007. Risk assessment to guide the prevention of cervical cancer. Am. J. Obstet. Gynecol. 197:356.e1–356.e6 doi:10.1016/j.ajog.2007.07.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dehn D, Torkko KC, Shroyer KR. 2007. Human papillomavirus testing and molecular markers of cervical dysplasia and carcinoma. Cancer Cytopathol. 111:1–14 [DOI] [PubMed] [Google Scholar]
- 8. Gravitt PE, et al. 2008. New technologies in cervical cancer screening. Vaccine 265:K42–K52 [DOI] [PubMed] [Google Scholar]
- 9. Kulasingam SL, et al. 2006. Cost-effectiveness of extending cervical cancer screening intervals among women with prior normal pap tests. Obstet. Gynecol. 107:321–328 [DOI] [PubMed] [Google Scholar]
- 10. Lechner MS, Laimins LA. 1994. Inhibition of p53 DNA binding by human papillomavirus E6 proteins. J. Virol. 68:4262–4273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li T-T, Zhao L-N, Liu Z-G, Fan D-M. 2005. Regulation of apoptosis by the papillomavirus E6 oncogene. World J. Gastroenterol. 11:931–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Longworth MS, Laimins LA. 2004. Pathogenesis of human papillomaviruses in differentiaing epithelia. Microbiol. Mol. Biol. Rev. 68:362–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matlashewski G, et al. 1986. The expression of human papillomavirus type 18 E6 protein in bacteria and the production of anti-E6 antibodies. J. Gen. Virol. 67:1909–1916 [DOI] [PubMed] [Google Scholar]
- 14. Meschede W, et al. 1998. Antibodies against early protein of human papillomaviruses as diagnostic markers for invasive cervical cancer. J. Clin. Microbiol. 36:475–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Molden T, et al. 2005. Predicting CIN2+ when detecting HPV mRNA and DNA by PreTect HPV-proofer and consensus PCR: a 2-year follow-up of women with ASCUS or LSIL Pap smear. Int. J. Cancer 114:973–976 [DOI] [PubMed] [Google Scholar]
- 15a. National Comprehensive Cancer Network 2007. Cervical cancer screening. NCCN Clin. Pract. Guidel. Oncol. 2:1–28. [Google Scholar]
- 16. Nindl I, et al. 1994. Antibodies against linear and conformational epitopes of the human papillomavirus (HPV) type 16 E6 and E7 oncoproteins in sera of cervical cancer patients. Arch. Virol. 137:341–353 [DOI] [PubMed] [Google Scholar]
- 17. Reference deleted.
- 18. O'Sullivan JP, et al. 1998. A case-control study of true-positive versus false-negative cervical smears in women with cervical intraepithelial neoplasia (CIN) III. Cytopathology 9:155–161 [DOI] [PubMed] [Google Scholar]
- 19. Park T-W, Fujiwara H, Wright TC. 1995. Molecular biology of cervical cancer and its precursors. Cancer 76:1902–1913 [DOI] [PubMed] [Google Scholar]
- 20. Partridge EE, et al. 2008. Cervical cancer screening. J. Natl. Compr. Canc. Netw. 6:58–82 [DOI] [PubMed] [Google Scholar]
- 21. Ronco G, et al. 2010. Efficacy of human papillomavirus testing for the detection of invasive cervical cancers and cervical intraepithelial neoplasia: a randomised controlled trial. Lancet Oncol. 11:249–257 [DOI] [PubMed] [Google Scholar]
- 22. Ronco G, Rossi PG. 2008. New paradigms in cervical cancer prevention: opportunities and risks. BMC Women's Health 8:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Saint M, Gildengorin G, Sawaya GF. 2005. Current cervical neoplasia screening practices of obstetriciaqn/gynecologists in the US. Am. J. Obstet. Gynecol. 192:414–421 [DOI] [PubMed] [Google Scholar]
- 24. Sawaya GF. 2008. Adding human papillomavirus testing to cytology for primary cervical cancer screening: shooting first and asking questions later. Ann. Intern. Med. 148:557–559 [DOI] [PubMed] [Google Scholar]
- 25. Schiffman M, Castle PE. 2003. Human papillomavirus: epidemiology and public health. Arch. Pathol. Lab. Med. 127:930–934 [DOI] [PubMed] [Google Scholar]
- 26. Schiffman M, et al. 2011. A long-term prospective study of type-specific human papillomavirus infection and risk of cervical neoplasia among 20,000 women in the Portland Kaiser Cohort Study. Cancer Epidemiol. Biomarkers Prev. 20:1398–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sehr P, Zymbach K, Pawlita M. 2001. A generic capture ELISA for recombinant proteins fused to glutathione S-transferase: validation for HPV serology. J. Immunol. Methods 253:153–162 [DOI] [PubMed] [Google Scholar]
- 28. Snijders PJF, Steenbergen RDM, Heideman DAM, Meijer CJLM. 2006. HPV-mediated cervical carcinogenesis: concepts and clinical implications. J. Pathol. 208:152–164 [DOI] [PubMed] [Google Scholar]
- 29. Stoppler MC, et al. 1996. Natural variants of the human papillomavirus type 16 E6 protein differ in their abilities to alter keratinocyte differentiation and to induce p53 degradation. J. Virol. 70:6987–6993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sun Y, et al. 1994. Comparison of peptide enzyme-linked immunisorbent assay and radioimmunoprecipitation assay with in vitro-translated proteins for detection of serum antibodies to human papillomavirus type 16 E6 and E7 proteins. J. Clin. Microbiol. 32:2216–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tungteakkhun SS, Duerksen-Hughes PJ. 2008. Cellular binding partners of the human papillomavirus E6 protein. Arch. Virol. 153:397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Veress G, Konya J, Csiky-Meszaros T, Czegledy J, Gergely L. 1994. Human papillomavirus DNA and anti-HPV secretory IgA antibodies in cytologically normal cervical specimens. J. Med. Virol. 43:201–207 [DOI] [PubMed] [Google Scholar]
- 33. Woodman CBJ, Collins SI, Young LS. 2007. The natural history of cervical HPV infection: unresolved issues. Nat. Rev. Cancer 7:11–22 [DOI] [PubMed] [Google Scholar]
- 34. Yim E-K, Park J-S. 2007. Biomarkers in cervical cancer. Biomark. Insights 1:215–225 [PMC free article] [PubMed] [Google Scholar]