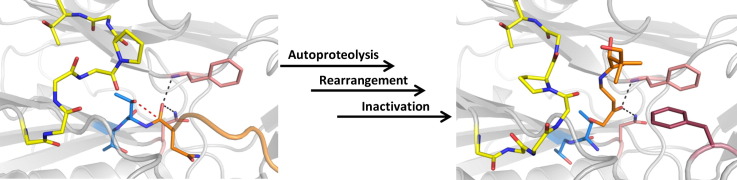
Abstract
cis-Autoproteolysis is a post-translational modification necessary for the function of ThnT, an enzyme involved in the biosynthesis of the β-lactam antibiotic thienamycin. This modification generates an N-terminal threonine nucleophile that is used to hydrolyze the pantetheinyl moiety of its natural substrate. We determined the crystal structure of autoactivated ThnT to 1.8 Å through X-ray crystallography. Comparison to a mutationally inactivated precursor structure revealed several large conformational rearrangements near the active site. To probe the relevance of these transitions, we designed a pantetheine-like chloromethyl ketone inactivator and co-crystallized it with ThnT. Although this class of inhibitor has been in use for several decades, the mode of inactivation had not been determined for an enzyme that uses an N-terminal nucleophile. The co-crystal structure revealed the chloromethyl ketone bound to the N-terminal nucleophile of ThnT through an ether linkage, and analysis suggests inactivation through a direct displacement mechanism. More importantly, this inactivated complex shows that three regions of ThnT that are critical to the formation of the substrate binding pocket undergo rearrangement upon autoproteolysis. Comparison of ThnT with other autoproteolytic enzymes of disparate evolutionary lineage revealed a high degree of similarity within the proenzyme active site, reflecting shared chemical constraints. However, after autoproteolysis, many enzymes, like ThnT, are observed to rearrange in order to accommodate their specific substrate. We propose that this is a general phenomenon, whereby autoprocessing systems with shared chemistry may possess similar structural features that dissipate upon rearrangement into a mature state.
Abbreviations: CMK, chloromethyl ketone; PβS, proteasome β‐subunit; GA, glycosylasparaginase; CA, cephalosporin acylase; HMK, halomethyl ketone; PDB, Protein Data Bank
Keywords: thienamycin biosynthesis, cis‐autoproteolysis, mechanism of chloromethyl ketone inhibition, N‐terminal nucleophile, X‐ray crystallography
Graphical Abstract
Highlights
► A 1.8-Å X-ray structure shows that peptide bond isomerization accompanies autoproteolysis. ► The threonine γ-methyl facilitates formation of a reactive N-terminal nucleophile. ► A halomethyl ketone inhibitor is shown to bind through an ether linkage. ► Computational modeling suggests inactivation through a direct displacement mechanism. ► Comparison with the proteasome β-subunit reveals general features of autoactivation.
Introduction
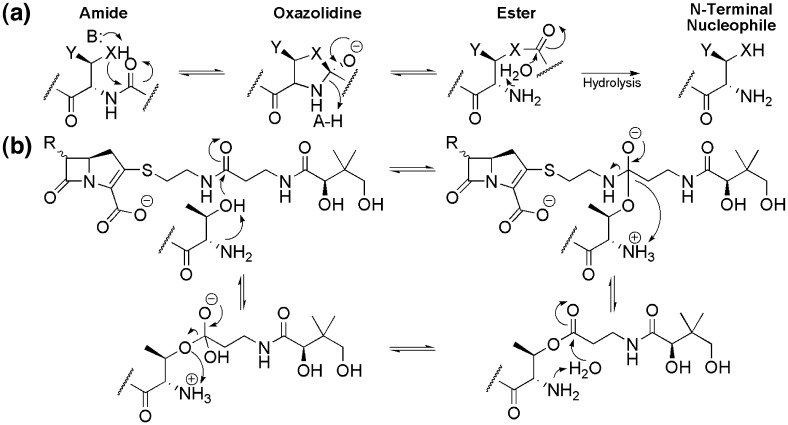
Autoproteolytic proteins pose unique challenges to understanding structure–function relationships because their cleavage is an intrinsic consequence of folding. This post-translational modification is involved in the regulation of protein folding,1 cell signaling,2 nucleoporin biogenesis,3 apoptosis,4 and the function of many enzymes.5., 6., 7. These studies have established that self-cleavage proceeds through an N–O(S) acyl shift mechanism: an active-site nucleophile (Thr, Ser, or Cys) attacks into its N-terminal peptide bond generating a (thia)oxazolidine intermediate (Fig. 1a).8 This transient species collapses to a (thio)ester, which is subsequently hydrolyzed to yield two subunits bearing new N- and C-termini. The N-terminal nucleophile (Ntn) generated by autoproteolysis is used by enzymes, such as the Thr-utilizing proteasome β-subunit (PβS),9 in a catalytic diad (or triad) for catalysis. It was originally believed that all Ntn-utilizing enzymes belonged to the same superfamily (dubbed the Ntn superfamily).5., 10. However, subsequent research showed that a second group of enzymes exists (the D/O superfamily), which is related to the Ntn superfamily through convergent evolution.11 Together, these enzymes perform a diverse array of hydrolysis and acyl transfer reactions.
Fig. 1.
Mechanism of autoactivation and substrate hydrolysis. (a) Abstraction of a proton by a general base (B:) initiates cis-autoproteolysis with Thr (X = O, Y = CH3), Ser (X = O, Y = H), or Cys (X = S, Y = H) nucleophile. The activated residue attacks into its N-terminal amide bond, resulting in a (thia)oxazolidine ring that collapses to a (thio)ester with concomitant protonation of the amine leaving group by a general acid (AH). The α-amine activates a water molecule for hydrolysis of the (thio)ester, creating a new N-terminal residue. (b) Mechanism of ThnT hydrolysis of a pantetheinylated carbapenem. The N-terminal amine acts as a general base to activate the nucleophile for attack into the carbonyl of the pantetheine side chain. The α-ammonium donates a proton back to the amine leaving group, generating an acyl-enzyme intermediate. In a manner directly comparable to autoactivation, the α-amine again activates a water molecule to hydrolyze the substrate from the enzyme, returning it to its resting state. This hydrolytic chemistry is standard among Ntn-utilizing enzymes.
We recently reported use of the pantetheinyl hydrolase ThnT (Fig. 1b), a D/O enzyme involved in the biosynthesis of the potent β-lactam antibiotic thienamycin,12 as a model system with which to study autoproteolysis.13 Our analysis showed that a local conformational rearrangement is necessary for autoactivation and indicated that the majority of reported uncleaved structures from the Ntn superfamily do not, in fact, represent a cleavage‐competent state. Consequently, a comparison between the functional active site before and after autoactivation is possible for only two systems, glycosylasparaginase14., 15. (GA) and PβS.9., 16. This lack of mechanistically relevant structural information constitutes a fundamental gap in our understanding of how a single protein sequence is optimized to facilitate both autoproteolysis and enzymatic catalysis in its mature form. Because cephalosporin acylase (CA) and penicillin acylase, both members of the Ntn superfamily, are currently the subject of directed evolution experiments to improve the industrial production of β-lactam antibiotics,17., 18. further insights may provide valuable lessons in the development of enzymes with improved activity. Addressing this problem will rely on the ability to compare and contrast structural changes upon autoactivation for both Ntn and D/O superfamilies. To this end, we sought to extend our analysis of ThnT by determining the mature structure of the enzyme and elucidating the features necessary for substrate binding and catalysis.
Results and Discussion
Structural transitions accompanying autoproteolysis
A 6-h room temperature incubation of ThnT purified from heterologous expression afforded autoproteolytically activated enzyme suitable for X-ray crystallography. Wild-type ThnT crystallized under similar conditions and in the same space group as the mutationally inactive precursor ThnT T282C. Diffraction to high resolution and phasing by molecular replacement yielded the 1.8-Å crystal structure of wild-type ThnT. For clarity, all residues of ThnT referenced hereafter will be numbered according to their position in the precursor protein. Autoproteolysis generates two subunits, α and β, composed of residues 1–281 and 282–399, respectively. Like most structurally characterized Ntn enzymes, the subunits of ThnT remain intimately intertwined after activation with global features comparable to their uncleaved forms (Fig. 2a).11., 19. The N-6His tag and the first 23 residues of the α‐subunit were not observed, as was the case for the T282C structure. A short linker sequence leading into the scissile bond of the proenzyme, residues 274–281, showed few tertiary interactions and modeled with very high average B-factors (41.4 Å2).13 These disordered residues comprise the C-terminus of the α‐subunit, and as we anticipated, they are not observed in the structure of mature ThnT. Interestingly, comparable disorder in precursor proteins and nascent C-termini has been found in many autoproteolytic systems.14., 20. This observation is readily explained as a strategy employed to promote dissociation of the C-terminus, which must occur to expose the active site of the mature enzyme.
Fig. 2.
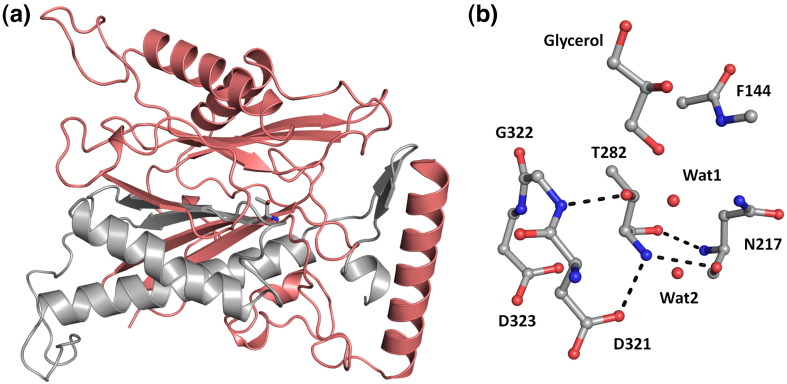
Active site of autoproteolytically activated ThnT. (a) Heterodimeric ThnT has extensive quaternary interactions between the α‐subunit (red) and the β‐subunit (silver). The immediate environment of the catalytic, N-terminal threonine residue (shown) is composed of elements from each subunit. For clarity, only a single heterodimer of the asymmetric unit is shown. (b) Residues Phe144 and Asn217 from the α‐subunit form the oxyanion hole of ThnT and the three residues D321, G322, and D323 are positioned to activate the γOH of Thr282, allowing for its role as the catalytic nucleophile. The position of a bound glycerol molecule suggests an orientation for the pantetheine moiety of the substrate. Two waters observed in the active site may be displaced upon substrate binding or repositioned to hydrolyze the acyl-enzyme intermediate generated during catalysis.
The N-terminal residue of the β‐subunit is the characteristic N-terminal threonine residue central to autoprocessing and substrate catalysis (Fig. 2b). Surprisingly, the peptide ω angle of Thr282 consistently refined to a strained 167°. Comparable strain was observed at this position in the proenzyme inactive state, and we hypothesized that this destabilization was used to help drive formation of the reactive conformation.13 Hence, it appears that retention of this unfavorable geometry is a consequence of the selection for an active site that can promote two different chemical reactions. The energetic penalty of straining this bond may be offset by the hydrogen bond between the carbonyl of Thr282 and the amide nitrogen of Asn217, which would be disrupted upon relaxation of the ω angle. The amide proton of Phe144 and δNH2 of Asn217 comprise the oxyanion hole of ThnT for autoactivation.13 In the major conformer of the precursor structure, a single water molecule occupied the oxyanion hole, taking the place of the reactive carbonyl. A similar placeholder is not observed for mature ThnT; instead, a glycerol molecule and additional water molecules are bound in the active site (Fig. 2b).
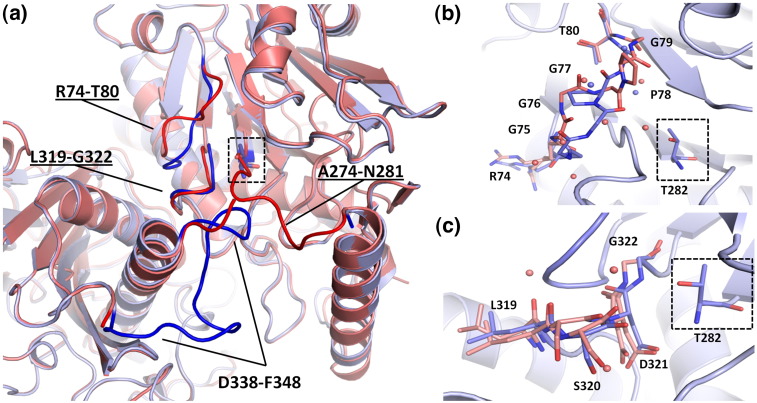
In addition to dissociation of the nascent C-terminus, three structural rearrangements in the vicinity of the active site were identified (Fig. 3a). Notably, the solvent-filled groove that may represent the substrate binding site is not present prior to autoactivation and is only revealed upon a conformational change in residues Arg74–Thr80, which includes cis–trans isomerization of the Gly77–Pro78 peptide bond (Fig. 3b). This dramatic structural rearrangement is promoted electrostatically by reorienting the carbonyl of Arg74 away from Asp321 and positioning it 1.4 Å closer to the side chain of Lys300. The new conformation allows the amide nitrogens of Gly76 and Gly77 to hydrogen bond with the backbone of Leu318 and Asp321, respectively. Additionally, there is a net release of four water molecules, which is entropically favorable.21 These transitions stabilize the loop relative to the uncleaved form, which is corroborated by a decrease in the average B-factor of the loop from 18.5 Å2 prior to cleavage to 11.6 Å2 in the mature enzyme. Together, these data demonstrate that specific structural features of ThnT promote the isomerization of the Arg74–Thr80 loop.
Fig. 3.
Structural changes accompanying autoactivation. (a) Ribbon diagram showing the residues that undergo a major structural change upon autoproteolysis. Residues A274–N281 are disordered after autoproteolysis and were not modeled for wt-ThnT. The uncleaved structure (from PDB ID: 3S3U) is shown in red, and the mature enzyme is shown in blue, with the N-terminal Thr nucleophile boxed. (b) Isomerization of the G76–P77 peptide bond from cis (proenzyme state) to trans (mature state) is accompanied by reorganization of the entire R74–T80 loop. Waters that form hydrogen bonds with this loop in each state are displayed as spheres. For clarity, only the ribbon diagram of the mature enzyme is displayed. (c) The loop harboring S320–D321 was observed in two conformations in one subunit of the uncleaved protein. This converts to a single observable conformation after autoproteolysis. One pre-cleavage conformation would have a steric clash with the trans conformation of the G76–P77 loop, suggesting that the dynamics of these loops may be coupled to one another.
Two other motifs undergo a structural transition upon autoproteolysis. Residues Leu318–Gly322 were observed in two conformations of one subunit in the T282C structure but collapse to a single observable state in both subunits of the wild-type enzyme (Fig. 3c). The role of this apparent conformational flexibility is unclear. However, proximity to the active site and hydrogen bonding interactions with the Arg74–Thr80 loop (described above) suggest its relevance. Residues Asp338–Phe348 were not observed in the structure of the precursor protein but were modeled in the structure of mature ThnT. A subset of these, Pro337–Asp341, refined with a high average B-factor of 43.9 Å2, suggesting that they retain considerable flexibility after autoactivation. Conversely, Ala342–Phe348 were well resolved, with the side chain of Phe348 extending across the dimer interface and dividing the active site of the adjacent heterodimeric unit into two distinct sections. Because this conformation is not sterically accessible prior to dissociation of the newly formed C-terminus, formation of the observed state must occur relatively late in the maturation of the active site.
Active‐site features of ThnT
The structure of autoproteolytically activated ThnT shows that Thr282 has adopted the g− rotamer, which positions the γOH such that it forms a hydrogen bond with the α-amino group. The Ntn was originally proposed to constitute a “single-amino-acid catalytic center”;22 however, subsequent rigorous analysis showed that auxiliary residues are critical to catalysis.23 The α-amine of ThnT is adjacent to two hydrogen bond acceptors: the carboxylate of Asp321 and the carbonyl of Asn217. These residues correctly orient the nitrogen lone pair to accept a proton from the nucleophile (Fig. 2b). Interestingly, Gly322 is part of the network of residues that undergo structural rearrangement upon autoactivation, and the amide nitrogen shifts 0.8 Å to form a tight 2.6‐Å hydrogen bond with the nucleophile. The oxyanion hole residues Phe144 and Asn217 maintain their orientation to stabilize the tetrahedral oxyanion intermediates generated during substrate catalysis. Hence, the hydrogen bond interactions between Asn217 and Thr282 define the proper active‐site geometry for activation of both the nucleophile and the electrophile. Furthermore, this network leaves the γOH of Thr282 with an unsatisfied lone pair directed at the oxyanion hole.
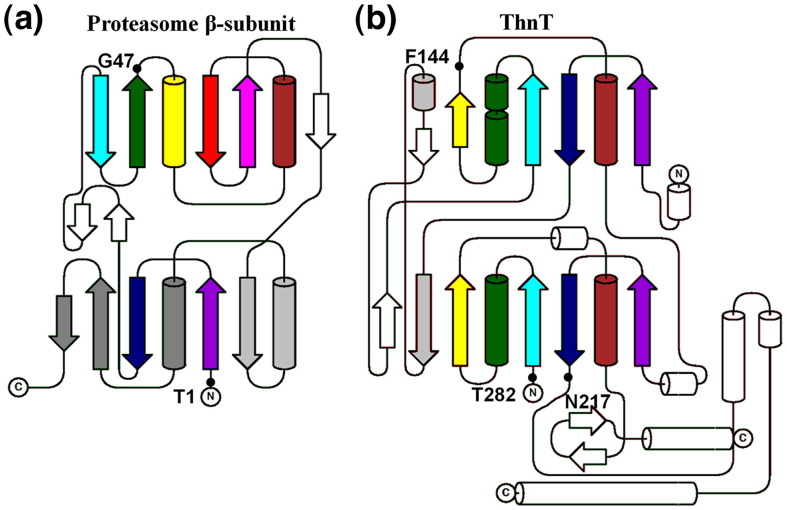
ThnT and PβS have distinct evolutionary lineage but share autoactivation through si-face attack into the scissile bond and amidohydrolase chemistry in their mature form. Hence, it is now possible to distinguish between system-specific features and general features of their active sites. The catalytic activity of PβS relies on a hydrogen bond network very similar to that shown in Fig. 2b. However, the active site of each protein is formed from different parts of their topologically distinct folds (Fig. 4 ), reinforcing their relationship through convergent evolution. Consequently, it appears that this hydrogen bond network represents an optimal balance between facile autoactivation and substrate catalysis. We recently reported that the Thr γ-methyl may accelerate the intramolecular cyclization that occurs during the N–O acyl shift in autoproteolysis.13 This phenomenon is known as the RRE (Reactive Rotamer Effect),25 and a similar effect may apply to any reaction for which the rotamer distribution may be shifted to favor the reactive state by the addition of steric bulk. This is precisely the case for ThnT and PβS, where the inactive g+ rotamer is structurally viable for both Thr and Ser but is destabilized by Thr because the γ-methyl shields a hydrogen bond with Gly322 (in ThnT) or Lys33 (in PβS).9 Similarly, the g+ rotamer cannot be formed for the ornithine acyltransferases (members of the D/O superfamily) because it would cause a strong 1.7-Å steric clash with the Thr111 in the oxyanion hole.26 Hence, comparison of ThnT with PβS and other Ntn enzymes reveals unique features of each active site that promote formation of a reactive state. Combined with the role of the RRE in autoproteolysis, we believe that these effects strongly and specifically contribute to the selection of Thr nucleophiles in autoproteolytic systems.
Fig. 4.
Topology diagrams of PβS and ThnT. (a) Coloring of PβS from dark (N-terminus) to light (C-terminus) shows that the αββα architecture has unique connectivity at each half of the fold, which is composed of α-helices and a pair of antiparallel β-sheets. The nucleophile is labeled T1. (b) The global fold of ThnT arose from a gene duplication event, as evidenced by the identical connectivity of the central αββα unit (colored) characteristic of the D/O superfamily of autoproteolytic enzymes.11 The nucleophile, T282, is adjacent to a parallel β-strand, and the oxyanion hole element F144 is on a geometrically identical region of the protein as G47 of PβS, whereas N217 is located closer to the nucleophile in the primary sequence. Non-conserved structural motifs are colored white. Image was generated with TopDraw.24
Formation of a potential substrate binding pocket
The position and orientation of the bound glycerol molecule define a groove that could represent part of the substrate binding pocket for the pantetheine moiety of the substrate. Interestingly, residues from all three mobile loops participate in the formation of this pocket. The peptide bond of Gly77, which undergoes cis–trans isomerization, is 5.1 Å away from the catalytic nucleophile, further implicating the rearrangement in enzyme function. Because no other uncleaved D/O structures have been published, we cannot compare the transitions described here with homologues. The well‐characterized enzymes GA and PβS from the Ntn superfamily show minor structural differences after autoproteolysis, other than dissociation of the nascent C-terminus.14., 16. However, these changes are not on the same scale as those observed for ThnT. Interestingly, prolonged incubation of ThnT resulted in further proteolysis of the α-subunit (Fig. S1). This raised the possibility that a secondary cleavage, similar to that observed for CA,27 might also occur for ThnT. The slow timescale of the apparent hydrolysis, however, suggested that the phenomenon could be due to a contaminating protease. Although inclusion of protease inhibitors failed to prevent this cleavage, N-terminal sequencing showed that hydrolysis occurred two residues prior to the first observed residue in the crystal structure. Because this is a disordered region far from the active site, we do not believe that this secondary cleavage is relevant to the function of ThnT in vivo.
The scope of the structural changes revealed by our analysis of ThnT reinforces the general model that autoproteolytic maturation is more sophisticated than the obvious chemistry of bond making and bond breaking. Rather, considerable protein motion must occur as the protein shifts into a new folded state ensemble. Considering the broad implications of this hypothesis, we sought additional evidence to validate the relevance of the groups undergoing significant rearrangement by experimentally demonstrating their importance in substrate binding. To define the binding pocket and interactions with the substrate, we desired to co-crystallize the enzyme in a Michaelis complex. This has been accomplished for GA utilizing a Thr → Cys variant of the nucleophile that was capable of slow autoproteolysis and substrate hydrolysis.15 However, a similar scheme for ThnT was not possible because the comparable Thr → Cys variant does not mature on an accessible timescale.13 Instead, we pursued development of a covalent inhibitor harboring the pantetheine moiety that could be used with the wild-type enzyme.
Design of a covalent chloromethyl ketone inactivator of ThnT
The halomethyl ketones (HMKs) are a potent class of inactivators that have been used to covalently inhibit both serine and cysteine proteases. In the case of Ser proteases, both the catalytic nucleophile and histidine of the catalytic triad can be alkylated by HMKs.28 Alternatively, inhibition of Cys proteases results in alkylation of only the thiol, implying inhibition through a different mechanism.29 Although HMKs have been reported to inactivate the PβS,30 the mode and mechanism of inhibition have not been reported for a Thr protease. Hence, development of an HMK inhibitor of ThnT could simultaneously answer the long-standing question concerning the mode of attachment of HMKs to this class of enzyme while providing detailed information about the substrate binding interactions of ThnT. To this end, we used d-pantothenic acid as the starting material in a four-step synthesis of the chloromethyl ketone (CMK) derivative N-(4-chloro-3-oxobutyl)pantoamide (Fig. 5a) through a diazomethyl ketone intermediate (Scheme S1, see Supplemental Information for details of the synthesis). Incubation of the CMK pantoamide inhibitor with ThnT caused complete loss of pantethine hydrolysis activity, as measured by a qualitative TLC (thin‐layer chromatography) assay. We also tested the effect of the CMK pantoamide on the secondary cleavage events of ThnT and observed no changes (Fig. S1), excluding the possibility that they are caused by ThnT performing low‐efficiency intermolecular proteolysis.
Fig. 5.
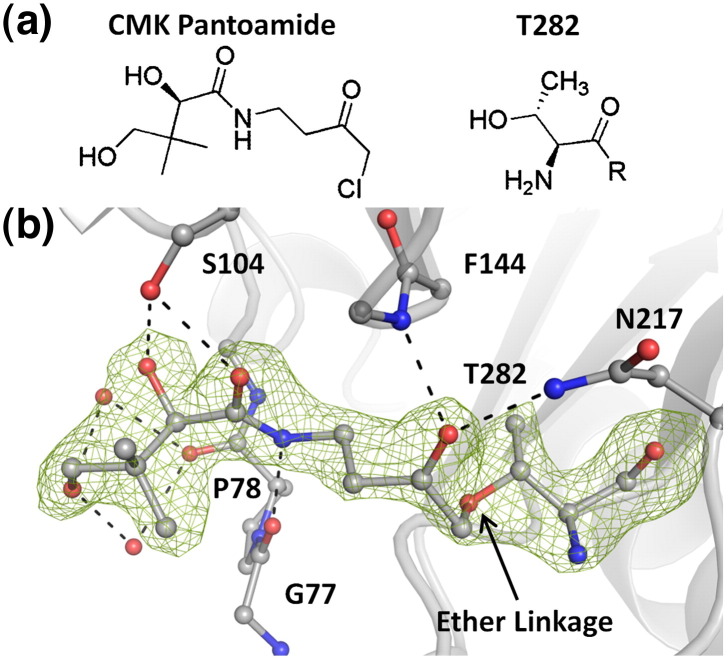
Attachment of a covalently bound inhibitor. (a) Structure of the CMK pantoamide and the N-terminal Thr nucleophile of ThnT. (b) Active site of inactivated ThnT with the (Fo − Fc) difference electron density map contoured at 3.0 σ. The location of the C′ carbon and a methyl group of the inactivator is below the contour level but confirmed in subsequent refinement. Contiguous density extends from T282 onto the inactivator and is consistent with formation of an ether linkage between γO of T282 and C′ of the inhibitor. Hydrogen bonding interactions (black) define the specific mode of binding reported here.
Active‐site features defined by the inhibitor-bound structure
A 10-fold molar excess of the CMK pantoamide was incubated with ThnT for 1 h, which was immediately used for crystallization. Crystals grew under the same conditions as native ThnT and yielded a 2.0-Å X-ray structure. Subsequent model building and refinement clearly showed loss of electron density corresponding to the bound glycerol in the wild-type crystal (Fig. 2b). Instead, we observed continuous electron density extending onto Thr282 corresponding to a nonprotein molecule covalently linked to the γO of the active‐site nucleophile (Fig. 5b). Given the high‐resolution data and the structure of the CMK pantoamide, there was a single possible mode of binding that satisfied the electron density: loss of the chloride and formation of an ether linkage between Thr282 and the inhibitor. Because ThnT and PβS have similar active‐site geometry, we hypothesize that this mode of inactivation is common between them.
The bridging methylene unit itself (C′) is the only component that distinguishes the bound species and the acyl-enzyme intermediate formed during substrate catalysis. Consequently, there is very little measurable distortion in the active site. All three bond angles that include the γO are compressed to ~ 102°. Comparison with the wild-type structure shows that the ψ angle of Thr282 has rotated 10° such that the derivatized side chain is deflected away from the oxyanion hole. Additional rotation of the ψ angle would provide space to allow the γO-bearing bond angles to relax. However, the system appears to have settled in an energetic minimum that maintains the hydrogen bond between Gly322 and Thr282 γOH. The lack of grossly unfavorable structural deviations introduced upon chemical derivatization indicates that inhibitor binding did not disrupt biologically relevant interactions.
The ketone of the inhibitor was designed to mimic the electrophilic amide of the substrate. As was predicted, the inactivated complex shows that this group has bound in the oxyanion hole of the protein, with hydrogen bonds to the backbone amide of Phe144 and the side chain of Asn217. The remainder of the inhibitor extends out toward the protein surface, with its amide nitrogen forming a hydrogen bond to Gly77. Ser104 acts as a hydrogen bond donor to the carbonyl of the amide and a hydrogen bond acceptor to the secondary hydroxyl. Simultaneous satisfaction of these hydrogen bonds restricts the inhibitor to the observed conformation. Lastly, the terminal hydroxyl forms two water-mediated hydrogen bonds with the carbonyl of Pro78 (Fig. 5b).
Evidence supporting inactivation through direct displacement
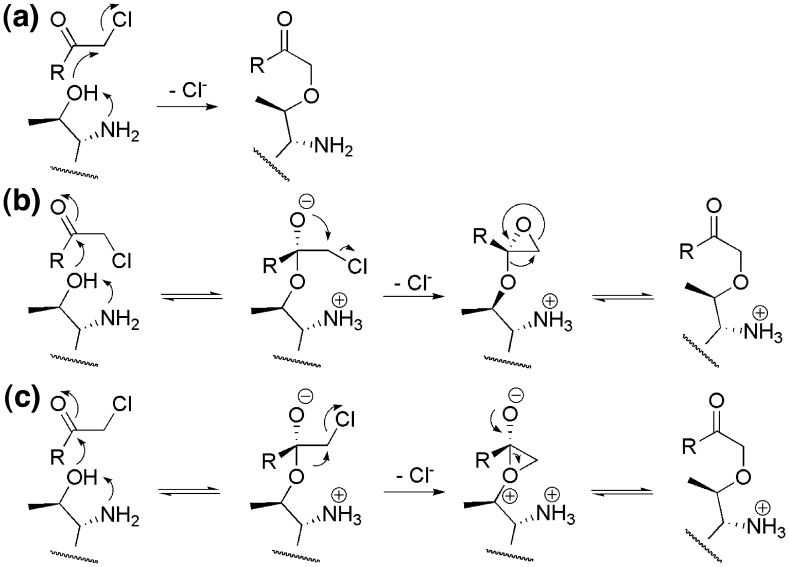
Although a definitive mechanism for inactivation of ThnT is beyond the scope of the present study, the co-crystal structure establishes that ThnT is alkylated at the nucleophilic position and that few alternatives exist with respect to the mechanism of this derivatization. For the Ser proteases, the nucleophile attacks into the carbonyl forming an oxyanion that displaces the adjacent halide forming an oxirane intermediate. This three-membered ring is subsequently opened by attack of the His residue of the catalytic triad, resulting in a second covalent attachment.31 Were a comparable mechanism involving an oxirane to occur in the inactivation of ThnT, its formation would obey the well-known stereoelectronic requirement that attack of the oxyanion into C′ proceeds anti-periplanar to the chloride leaving group (Fig. 6b). The location of the oxyanion is well defined by the location of the oxyanion hole of ThnT. Modeling this species shows that placement of the chloride anti-periplanar to the oxyanion results in a strong steric clash with Ser320 and the carboxylate of Asp321. Because this geometry is intrinsic to oxirane formation, and the ThnT active site harbors features that would significantly impede expulsion of the chloride, we do not believe that inactivation of ThnT occurs through an oxirane intermediate. Instead, the ether linkage reported here is most similar to the thioether formed upon derivatization of the cysteine proteases papain,32 SARS-CoV (Severe Acute Respiratory Syndrome coronavirus) peptidase,33., 34. and hepatitis A viral 3C.29 For the latter two enzymes, an episulfonium species was also trapped, presumably as an intermediate of the inhibition process. A comparable mechanism for ThnT would involve a highly unstable oxonium species that is unlikely to occur (Fig. 6c). Together, these constraints suggest that the mechanism of HMK inhibition in ThnT is distinct from those reported for the serine and cysteine proteases.
Fig. 6.
Potential mechanisms of inhibition of ThnT by the CMK inhibitor. (a) Direct displacement of the chloride follows a traditional SN2 reaction. Evidence suggests that this is the most likely mechanism for ThnT inhibition. (b) The oxyanion of the tetrahedral intermediate expels the chloride in an intramolecular cyclization reaction. This parallels the accepted mechanism of inhibition for traditional Ser proteases, where the imidazole side chain of the catalytic triad subsequently attacks C′ forming a second covalent bond to the inhibitor. Both formation of the oxirane and its collapse to an ether are unlikely to occur within the ThnT active site. (c) After formation of the oxyanion intermediate, a second nucleophilic attack occurs, as has been proposed for thioether formation in Cys proteases. The oxonium species invoked by this mechanism would be too high energy to occur.
Because formation of the tetrahedral intermediate is reversible, a competitive and irreversible SN2 attack of T282 into C′ could account for the observed ether linkage. Notably, this reaction would proceed through a trigonal bipyramidal transition state that is intrinsically stabilized by the adjacent carbonyl π-system,35 and the requisite orientation of the chloride for backside attack is reasonable, as indicated by modeling the complex of ThnT and the CMK pantoamide. Consequently, we believe that the simplest, least-motion mechanism is the most likely: derivatization of ThnT occurs by direct displacement of the chloride.
The role of ThnT in thienamycin biosynthesis
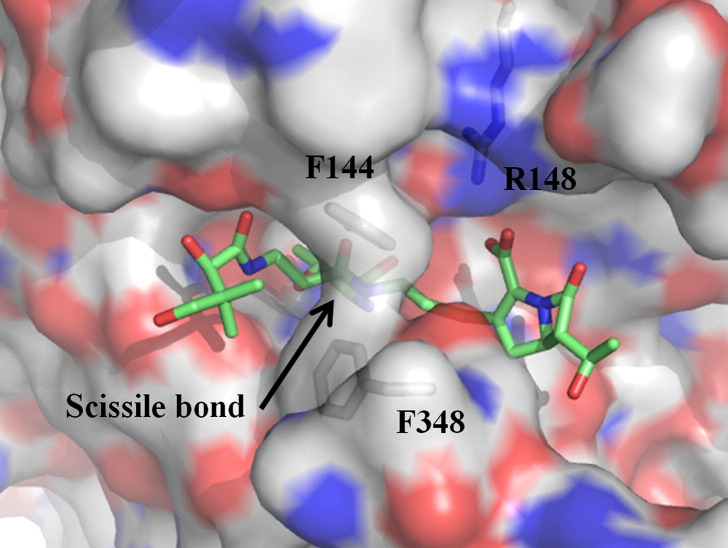
It has been shown that ThnT can hydrolyze a pantetheinylated carbapenem substrate, and circumstantial evidence suggests that it acts late in the biosynthetic pathway.12., 36., 37. Although the data presented here do not provide a ready explanation for the inability of ThnT to cleave phosphopantetheine, the catalytic activity of ThnT is clear. Formation of the Michaelis complex places the carbonyl of the scissile amide into the oxyanion hole (Fig. 7 ). The majority of the enzyme–substrate interactions are in the pantothenate binding region defined by the ThnT–inhibitor structure (Fig. 5b), consistent with the capacity of ThnT to efficiently hydrolyze a variety of pantetheinylated compounds.12 The cysteamine moiety of the substrate provides the ideal distance for the carboxylate of thienamycin to form a salt bridge with Arg148. The catalytic nucleophile is centered within the active site, and its exposure to solvent is minimized by Phe144 and Phe348 from the opposite subunit. Like other N-terminal amidohydrolases,5., 38. the α-amine deprotonates the hydroxyl of Thr282 to generate a reactive nucleophile. Attack into the substrate generates a tetrahedral intermediate whose collapse is concurrent with protonation of the amine leaving group. Studies have shown that amidohydrolases often provide a hydrogen bond acceptor to facilitate inversion of the nitrogen,39 and we hypothesize that Ser320 may satisfy this role for ThnT (Fig. S2). Expulsion of the amino leaving group generates thienamycin and an acyl‐enzyme intermediate. A water molecule in a position similar to Wat1 or Wat2 (Fig. 2b) is then activated by the α-amine, which hydrolyzes the acyl‐enzyme intermediate, regenerating the ThnT active site.
Fig. 7.
Model of the ThnT Michaelis complex with a pantetheinylated carbapenem. This model shows that the substrate binding cavity is well suited to accommodate a carbapenem substrate and that a positively charged residue, R148, is positioned to form a salt bridge with the carboxylate moiety of the substrate.
Concluding Remarks
Combined with our initial study of ThnT, the data reported here constitute the first glimpse into the complete “life cycle” of an enzyme from the D/O superfamily (Fig. S3).13 When combined with existing information from the Ntn superfamily, these data enabled an expanded analysis of autoproteolytic activation. In all systems, dissociation of the nascent C-terminus after autoproteolysis is favored by a lack of tertiary contacts. When autoproteolysis is initiated through a si-face attack, the same oxyanion hole is used for both self-cleavage and substrate catalysis. Within the active site, the Thr γ-methyl facilitates this chemistry by directing the nucleophile into a reactive state. Lastly, we identified large structural rearrangements necessary for the formation of the ThnT substrate binding pocket. Although PβS is “insignificantly different”16 before and after autoproteolysis, most other enzymes from the Ntn superfamily (including penicillin acylase,40 CA,41 and γ-glutamyl transpeptidase20) show evidence of a unique conformational rearrangement. These enzymes each contain a high degree of similarity with the ThnT proenzyme active site but are structurally divergent after autoproteolysis. We propose that this is evidence of a more general phenomenon. Autoprocessing systems with shared chemistry, regardless of lineage, will have comparable features prior to autoprocessing that may dissipate as each protein rearranges to its unique mature state. The observation that two markedly different conformations of the Arg74–Thr80 loop of ThnT (Fig. 3b) facilitate each of the hydrolytic chemistries highlights the orchestrated complexity of the structure–function relationship in autoproteolytic enzymes. Not only does sequence encode for an intrinsically reactive folded state but also, as we have shown here for ThnT, the downstream consequences of this self-cleavage result in spontaneous reorganization to the catalytically competent state of the mature enzyme.
Materials and Methods
Crystallization, data collection, and structure determination
ThnT was expressed, purified, and concentrated to 7.7 μg/μL as previously described for the inactive T282C variant.13 The enzyme was incubated at 20 °C for 6 h prior to being flash frozen for storage at − 80 °C. The ThnT–inactivator complex was prepared by adding a 10-fold molar excess of the inactivator and incubating it at 20 °C for 1 h immediately prior to use in crystallization attempts. Crystals were grown by sitting‐drop vapor diffusion against a 1‐mL reservoir solution of 14–18 % polyethylene glycol 3350 and 0.5–0.7 M sodium acetate. Crystals grew overnight and were cryoprotected by immersion into 20% polyethylene glycol 3350, 0.7 M sodium acetate, and 20% glycerol prior to flash freezing in N2. Crystallization and data collection for ThnT were carried out at X6A at the National Synchrotron Light Source. Data for the ThnT–inhibitor complex were collected on BL12-2 at the Stanford Synchrotron Light Source. Both structures were solved by molecular replacement using the Phaser program,42 as implemented in the CCP4 package.43 The structure of the uncleaved T282C variant of ThnT [Protein Data Bank (PDB) ID: 3S3U], with dual occupancies removed, was used as the search model. Iterative rounds of model building were performed in Coot,44 using REFMAC5 for refinement.45 After the protein and solvent atoms were defined, TLS operators were added and further refined in REFMAC5, which yielded significant improvements in R free for both models.45., 46. Crystallographic and refinement statistics are listed in Table 1 . Coordinates were deposited to the PDB with accession numbers 3TM1 (wt-ThnT) and 3TM2 (inactivated complex).
Table 1.
Crystallographic statistics for data collection and structure refinement
| Modification | Native | O-(2-Oxo-4-pantoamidobutyl)threonine |
|---|---|---|
| Space group | P21212 | P21212 |
| Cell dimensions | ||
| a, b, c (Å) | 141.6, 67.8, 73.7 | 141.5, 67.3, 73.8 |
| Data processing | ||
| Beamline | BNL X6A | SSRL 12.2 |
| Wavelength (Å) | 0.8001 | 0.9788 |
| Resolution (Å) | 50–1.80 | 50–1.9 |
| Last bin | 1.85–1.80 | 2.05–2.00 |
| No. of observations | 524,211 | 320,826 |
| Completeness (%) | 96.2 (72.0) | 99.7 (98.1) |
| Rmerge (%) | 9.7 (41.5) | 12.1 (65.3) |
| I/σI | 19.9 (2.0) | 16.1 (2.1) |
| Redundancy | 8.2 (3.3) | 6.7 (5.5) |
| Refinement | ||
| Total no. of reflections | 60,481 | 45,361 |
| Total no. of atoms | 5511 | 5471 |
| No. of waters | 430 | 368 |
| Figure of merit | 0.887 | 0.877 |
| Rwork (%) | 17.0 (32.3) | 16.5 (25.3) |
| Rfree (%) | 19.7 (35.0) | 20.2 (30.3) |
| Ramachandran plot allowed (%) | 100.0 | 99.7 |
| Ramachandran plot favored (%) | 97.9 | 97.9 |
| Ramachandran plot outliers (%) | 0.0 | 0.3 |
Values in parenthesis are for the highest‐resolution shell.
Rmerge is ∑|Io − I|/∑Io, where Io is the intensity of an individual reflection, and I is the mean intensity for multiple recorded reflections.
Rwork is ||Fo − Fc||/Fo, where Fo is an observed amplitude, and Fc is a calculated amplitude.
Rfree is the same statistic calculated over a 5% subset of the data that has not been used for refinement.
Characterization of N-terminal hydrolysis
Purified ThnT was diluted to 2 μg/μL into buffer [50 mM Tris (pH 7.5) and 50 mM KCl, with 10% glycerol] with and without 20-fold molar excess of CMK pantoamide inhibitor and incubated at 37 °C. Samples were withdrawn every 12 h for SDS-PAGE analysis and, at 12 h and 48 h, to assay for pantethine hydrolysis activity. Samples were run on 15% SDS-PAGE for 1 h and visualized with Coomassie Blue staining (Fig. S1). Aliquots containing the α-subunit degradation products were rerun on 15% SDS-PAGE for 3 h and electroblotted onto a polyvinylidene fluoride membrane. The smallest α-subunit was sent to the Johns Hopkins Medical Institute Synthesis and Sequencing Facility for N-terminal sequencing. ThnT pantethine hydrolysis activity was monitored by incubation with 10 mM pantethine for 1 h followed by spotting on a TLC plate. Samples without inhibitor present turned purple upon addition of Ninhydrin reagent, consistent with hydrolysis of the pantetheine moiety, whereas those with the CMK pantoamide did not stain.
Molecular modeling of substrate docking and reactive intermediates
All steps were performed with the Discovery Studio 2.1 software suite. The carbapenem substrate and covalent intermediates were generated with the structure drawing functionality of Discovery Studio 2.1 and manually docked in the active site of ThnT. Generally, the system was typed with the CHARMM force field and minimized using an adopted basis NR algorithm.47 Crystallographic waters were explicitly included in all minimizations. Attempts to position the substrate or inhibitor in an orientation substantially different from the observed conformation (e.g., placing the chlorine atom inside the oxyanion hole) resulted in significant active‐site distortion. All rotamers of the CMK C′–Cl bond were independently minimized before and after oxyanion formation in order to assess the capacity of ThnT to accommodate reactive intermediates. All hydrogen bonding interactions defined by the inhibitor-bound structure were constrained to ± 0.2 Å of their measured values. The algorithm exited after achieving a root-mean-squared gradient of < 0.1 kcal/(mol Å). All other settings were default.
Acknowledgements
We gratefully acknowledge Vivian Stojanoff and Jean Jackoncic at the National Synchrotron Light Source for assistance in data collection. N-terminal sequencing was performed by Jodie Franklin at the Johns Hopkins Medical School Sequencing Facility. This work was supported by research grants National Institutes of Health RO1 AI014937 (to C.A.T.) and National Institutes of Health GM61017 (to J.F.S.) and the Dimitri V. d'Arbeloff post-doctoral fellowship (to N.T.W.).
Edited by G. Schulz
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2012.06.012
Appendix A. Supplementary Data
Supplementary materials
References
- 1.Macao B., Johansson D.G.A., Hansson G.C., Hard T. Autoproteolysis coupled to protein folding in the SEA domain of the membrane-bound MUC1 mucin. Nat. Struct. Mol. Biol. 2006;13:71–76. doi: 10.1038/nsmb1035. [DOI] [PubMed] [Google Scholar]
- 2.Lin H.H., Chang G.W., Davies J.Q., Stacey M., Harris J., Gordon S. Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. J. Biol. Chem. 2004;279:31823–31832. doi: 10.1074/jbc.M402974200. [DOI] [PubMed] [Google Scholar]
- 3.Rosenblum J.S., Blobel G. Autoproteolysis in nucleoporin biogenesis. Proc. Natl Acad. Sci. USA. 1999;96:11370–11375. doi: 10.1073/pnas.96.20.11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tinel A., Janssens S., Lippens S., Cuenin S., Logette E., Jaccard B. Autoproteolysis of PIDD marks the bifurcation between pro-death caspase-2 and pro-survival NF-κB pathway. EMBO J. 2007;26:197–208. doi: 10.1038/sj.emboj.7601473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brannigan J.A., Dodson G., Duggleby H.J., Moody P.C.E., Smith J.L., Tomchick D.R., Murzin A.G. A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature. 1995;378:416–419. doi: 10.1038/378416a0. [DOI] [PubMed] [Google Scholar]
- 6.Perler F.B., Xu M.Q., Paulus H. Protein splicing and autoproteolysis mechanisms. Curr. Opin. Chem. Biol. 1997;1:292–299. doi: 10.1016/s1367-5931(97)80065-8. [DOI] [PubMed] [Google Scholar]
- 7.Xu Q.A., Buckley D., Guan C.D., Guo H.C. Structural insights into the mechanism of intramolecular proteolysis. Cell. 1999;98:651–661. doi: 10.1016/s0092-8674(00)80052-5. [DOI] [PubMed] [Google Scholar]
- 8.Guan C.D., Cui T., Rao V., Liao W., Benner J., Lin C.L., Comb D. Activation of glycosylasparaginase. Formation of active N-terminal threonine by intramolecular autoproteolysis. J. Biol. Chem. 1996;271:1732–1737. doi: 10.1074/jbc.271.3.1732. [DOI] [PubMed] [Google Scholar]
- 9.Seemuller E., Lupas A., Stock D., Lowe J., Huber R., Baumeister W. Proteasome from Thermoplasma acidophilum: a threonine protease. Science. 1995;268:579–582. doi: 10.1126/science.7725107. [DOI] [PubMed] [Google Scholar]
- 10.Bompard-Gilles C., Villeret V., Davies G.J., Fanuel L., Joris B., Frere J.M., Van Beeumen J. A new variant of the Ntn hydrolase fold revealed by the crystal structure of l-aminopeptidase d-Ala-esterase/amidase from Ochrobactrum anthropi. Structure. 2000;8:153–162. doi: 10.1016/s0969-2126(00)00091-5. [DOI] [PubMed] [Google Scholar]
- 11.Cheng H., Grishin N.V. DOM-fold: a structure with crossing loops found in DmpA, ornithine acetyltransferase, and molybdenum cofactor-binding domain. Protein Sci. 2005;14:1902–1910. [Google Scholar]
- 12.Freeman M.F., McIshos K.A., Bodner M.J., Li R.F., Townsend C.A. Four enzymes define the incorporation of coenzyme A in thienamycin biosynthesis. Proc. Natl Acad. Sci. USA. 2008;105:11128–11133. doi: 10.1073/pnas.0804500105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buller A.R., Freeman M.F., Wright N.T., Schildbach J.F., Townsend C.A. Insights into cis-autoproteolysis reveal a reactive state formed through conformational rearrangement. Proc. Natl Acad. Sci. USA. 2012;109:2308–2313. doi: 10.1073/pnas.1113633109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y.M., Guo H.C. Crystallographic snapshot of glycosylasparaginase precursor poised for autoprocessing. J. Mol. Biol. 2010;403:120–130. doi: 10.1016/j.jmb.2010.08.038. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y.M., Guo H.C. Crystallographic snapshot of a productive glycosylasparaginase–substrate complex. J. Mol. Biol. 2007;366:82–92. doi: 10.1016/j.jmb.2006.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ditzel L., Huber R., Mann K., Heinemeyer W., Wolf D.H., Groll M. Conformational constraints for protein self-cleavage in the proteasome. J. Mol. Biol. 1998;279:1187–1191. doi: 10.1006/jmbi.1998.1818. [DOI] [PubMed] [Google Scholar]
- 17.Volpato G., Rodrigues R.C., Fernandez-Lafuente R. Use of enzymes in the production of semi-synthetic penicillins and cephalosporins: drawbacks and perspectives. Curr. Med. Chem. 2010;17:3855–3873. doi: 10.2174/092986710793205435. [DOI] [PubMed] [Google Scholar]
- 18.Sio C.F., Quax W.J. Improved β-lactam acylases and their use as industrial biocatalysts. Curr. Opin. Biotechnol. 2004;15:349–355. doi: 10.1016/j.copbio.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 19.Oinonen C., Rouvinen J. Structural comparison of Ntn-hydrolases. Protein Sci. 2000;9:2329–2337. doi: 10.1110/ps.9.12.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boanca G., Sand A., Okada T., Suzuki H., Kumagai H., Fukuyama K., Barycki J.J. Autoprocessing of Helicobacter pylori γ-glutamyltranspeptidase leads to the formation of a threonine–threonine catalytic dyad. J. Biol. Chem. 2007;282:534–541. doi: 10.1074/jbc.M607694200. [DOI] [PubMed] [Google Scholar]
- 21.Dunitz J.D. The entropic cost of bound water in crystals and biomolecules. Science. 1994;264:670. doi: 10.1126/science.264.5159.670. [DOI] [PubMed] [Google Scholar]
- 22.Duggleby H.J., Tolley S.P., Hill C.P., Dodson E.J., Dodson G., Moody P.C.E. Penicillin acylase has a single-amino-acid catalytic center. Nature. 1995;373:264–268. doi: 10.1038/373264a0. [DOI] [PubMed] [Google Scholar]
- 23.Zhiryakova D., Ivanov I., Ilieva S., Guncheva M., Galunsky B., Stambolieva N. Do N-terminal nucleophile hydrolases indeed have a single amino acid catalytic center? FEBS J. 2009;276:2589–2598. doi: 10.1111/j.1742-4658.2009.06987.x. [DOI] [PubMed] [Google Scholar]
- 24.Bond C.S. TopDraw: a sketchpad for protein structure topology cartoons. Bioinformatics. 2003;19:311–312. doi: 10.1093/bioinformatics/19.2.311. [DOI] [PubMed] [Google Scholar]
- 25.Jung M.E., Gervay J. gem-Dialkyl effect in the intramolecular Diels–Alder reaction of 2-furfuryl methyl fumarates: the reactive rotamer effect, enthalpic basis for acceleration, and evidence for a polar transition state. J. Am. Chem. Soc. 1991;113:224–232. [Google Scholar]
- 26.Lqbal A., Clifton I.J., Bagonis M., Kershaw N.J., Domene C., Claridge T.D.W. Anatomy of a simple acyl intermediate in enzyme catalysis: combined biophysical and modeling studies on ornithine acetyl transferase. J. Am. Chem. Soc. 2009;131:749–757. doi: 10.1021/ja807215u. [DOI] [PubMed] [Google Scholar]
- 27.Yin J., Deng Z., Zhao G., Huang X. The N-terminal nucleophile serine of cephalosporin acylase executes the second autoproteolytic cleavage and acylpeptide hydrolysis. J. Biol. Chem. 2011;286:24476–24486. doi: 10.1074/jbc.M111.242313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Powers J.C., Asgian J.L., Ekici O.D., James K.E. Irreversible inhibitors of serine, cysteine, and threonine proteases. Chem. Rev. 2002;102:4639–4750. doi: 10.1021/cr010182v. [DOI] [PubMed] [Google Scholar]
- 29.Yin J., Cherney M.M., Bergmann E.M., Zhang J., Huitema C., Pettersson H. An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors. J. Mol. Biol. 2006;361:673–686. doi: 10.1016/j.jmb.2006.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savory P.J., Djaballah H., Angliker H., Shaw E., Rivett A.J. Reaction of proteasomes with peptidylchloromethanes and peptidyldiazomethanes. Biochem. J. 1993;296:601–605. doi: 10.1042/bj2960601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prorok M., Albeck A., Foxman B.M., Abeles R.H. Chloroketone hydrolysis by chymotrypsin and N-methylhistidyl-57-chymotrypsin: implications for the mechanism of chymotrypsin inactivation by chloroketones. Biochemistry. 1994;33:9784–9790. doi: 10.1021/bi00198a050. [DOI] [PubMed] [Google Scholar]
- 32.Drenth J., Kalk K.H., Swen H.M. Binding of chloromethyl ketone substrate analogs to crystalline papain. Biochemistry. 1976;15:3731–3738. doi: 10.1021/bi00662a014. [DOI] [PubMed] [Google Scholar]
- 33.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 34.Yin J.A., Niu C.G., Cherney M.M., Zhang J.M., Huitema C., Eltis L.D. A mechanistic view of enzyme inhibition and peptide hydrolysis in the active site of the SARS-CoV 3C-like peptidase. J. Mol. Biol. 2007;371:1060–1074. doi: 10.1016/j.jmb.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bordwell F.G., Brannen W.T. The effect of the carbonyl and related groups on the reactivity of halides in SN2 reactions. J. Am. Chem. Soc. 1964;86:4645. [Google Scholar]
- 36.Bodner M.J., Phelan R.M., Freeman M.F., Li R., Townsend C.A. Non-heme iron oxygenases generate natural structural diversity in carbapenem antibiotics. J. Am. Chem. Soc. 2010;132 doi: 10.1021/ja907320n. 12- + [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bodner M.J., Li R.F., Phelan R.M., Freeman M.F., Moshos K.A., Lloyd E.P., Townsend C.A. Definition of the common and divergent steps in carbapenem β-lactam antibiotic biosynthesis. ChemBioChem. 2011;12:2159–2165. doi: 10.1002/cbic.201100366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fanuel L., Goffin C., Cheggour A., Devreese B., Van Driessche G., Joris B. The DmpA aminopeptidase from Ochrobactrum anthropi LMG7991 is the prototype of a new terminal nucleophile hydrolase family. Biochem. J. 1999;341:147–155. [PMC free article] [PubMed] [Google Scholar]
- 39.Syrén P., Hult K. Amidases have a hydrogen bond that facilitates nitrogen inversion, but esterases have not. ChemCatChem. 2011;3:853–860. [Google Scholar]
- 40.Bokhove M., Yoshida H., Hensgens C.M.H., van der Laan J.M., Sutherland J.D., Dijkstra B.W. Structures of an isopenicillin N converting Ntn-hydrolase reveal different catalytic roles for the active site residues of precursor and mature enzyme. Structure. 2010;18:301–308. doi: 10.1016/j.str.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 41.Kim Y., Kim S., Earnest T.N., Hol W.G.J. Precursor structure of cephalosporin acylase: insights into autoproteolytic activation in a new N-terminal hydrolase family. J. Biol. Chem. 2002;277:2823–2829. doi: 10.1074/jbc.M108888200. [DOI] [PubMed] [Google Scholar]
- 42.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bailey S. The CCP4 suite: programs for protein crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 44.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Winn M.D., Murshudov G.N., Papiz M.Z. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 46.Painter J., Merritt E.A. TLSMD web server for the generation of multi-group TLS models. J. Appl. Crystallogr. 2006;39:109–111. [Google Scholar]
- 47.Brooks B.R., Bruccoleri R.E., Olafson B.D., States D.J., Swaminathan S., Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials