Abstract
DNA methylation is a mechanism of epigenetic regulation that is common to all vertebrates. Functional studies underscore its relevance for tissue homeostasis, but the global dynamics of DNA methylation during in vivo differentiation remain underexplored. Here we report high-resolution DNA methylation maps of adult stem cell differentiation in mouse, focusing on 19 purified cell populations of the blood and skin lineages. DNA methylation changes were locus-specific and relatively modest in magnitude. They frequently overlapped with lineage-associated transcription factors and their binding sites, suggesting that DNA methylation may protect cells from aberrant transcription factor activation. DNA methylation and gene expression provided complementary information, and combining the two enabled us to infer the cellular differentiation hierarchy of the blood lineage directly from genomic data. In summary, these results demonstrate that in vivo differentiation of adult stem cells is associated with small but informative changes in the genomic distribution of DNA methylation.
Keywords: Epigenomics, bioinformatics, stem cells, blood lineage, skin lineage, hematopoietic stem cells, hair follicle stem cells, computational epigenetics
Introduction
DNA methylation provides a mechanism for robust and epigenetically heritable gene silencing. Mouse knockout studies have shown that the mammalian DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b are essential for embryonic development, and that loss of DNA methylation interferes with tissue homeostasis (Bird, 2002). These studies clearly establish the functional relevance of DNA methylation as an epigenetic mark, but they provide only partial insight into the mechanisms by which DNA methylation contributes to the regulation of gene expression and cellular identity. Large-scale DNA methylation mapping has recently emerged as a complementary approach to functional knockout studies, enabling the genome-wide identification of genomic regions that change their DNA methylation states during cellular differentiation. In vitro studies have mapped DNA methylation patterns in pluripotent stem cells (Lister et al., 2009) and in their differentiating progeny (Meissner et al., 2008; Mohn et al., 2008; Stadler et al., 2011), and in vivo studies have started to uncover the DNA methylation dynamics of adult stem cell differentiation and cellular lineage commitment (Hodges et al., 2011; Ji et al., 2010). However, conclusions from in vitro studies tend to be confounded by the presence of cell culture artifacts, while early in vivo studies relied on relatively low-resolution mapping technologies or were based on heterogeneous cell populations.
To dissect the in vivo patterns of epigenetic regulation associated with the differentiation of mammalian cells, we established genomic maps of DNA methylation at single-basepair resolution for two types of adult stem cells (hematopoetic stem cells and hair follicle bulge stem cells) and for a broad selection of blood and skin cell types that are derived from these stem cells. Comparative analysis of 19 highly purified cell types identified DNA methylation changes associated with the differentiation of adult stem cells into progenitor cells, with lymphoid vs. myeloid lineage choice among blood progenitor cells and with the specification of terminally differentiated cells in both lineages. Condensing our observations into a bioinformatic model of differentiation-associated changes, we were able to infer – with good accuracy – the hierarchy of cellular differentiation in the blood lineage based on the combination of DNA methylation and gene expression data.
Results
Single-basepair resolution DNA methylation maps of adult stem cell differentiation
We established genomic DNA methylation maps for 19 highly purified cell populations from the blood and skin lineages of adult mice (Figure 1A). Stem cells, progenitor cells and terminally differentiated cells were purified by subjecting ex vivo cell preparations to fluorescence-activated cell sorting (FACS) under stringent conditions (Table S1). This approach overcomes the heterogeneity of surgically obtained tissues and avoids artifacts that emerge during continued passaging of non-pluripotent cells in vitro (Meissner et al., 2008). For each cell type, two biological replicates were collected from different mice, purified in independent sorting experiments and subjected to genomic DNA methylation mapping (Table S2). In order to provide additional reference points for the identification of blood and skin specific DNA methylation patterns, we also mapped DNA methylation in embryonic stem (ES) cells and in two primary tissues (brain and liver). Furthermore, we obtained gene expression data for the same cell types, in part from public sources (Table S3, Experimental Procedures), allowing us to compare DNA methylation and gene expression differences between cell types (Figure 1B).
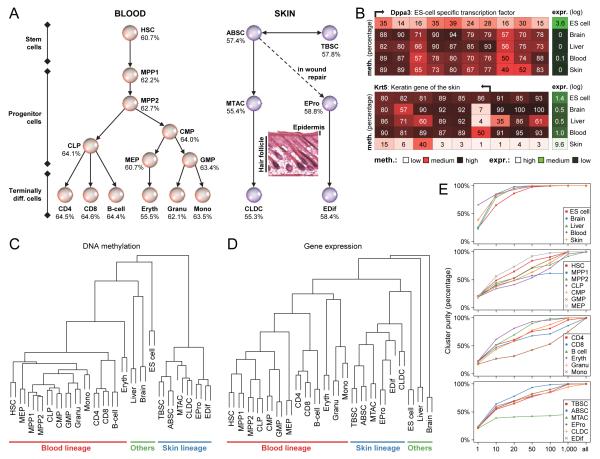
Figure 1.
Genomic DNA methylation maps reflect cellular lineage choice and differentiation stage
(A) Schematic drawing of adult stem cell differentiation in the blood and skin lineages: HSC, Hematopoietic stem cell; MPP1, Multipotent progenitor 1 (Flk2 negative); MPP2, Multipotent progenitor 2 (Flk2 positive); CLP, Common lymphoid progenitor; CMP, Common myeloid progenitor; GMP, Granulocyte-monocyte progenitor; MEP, Megakaryocyte-erythroid progenitor; CD4, T helper cell (CD4 positive); CD8, Cytotoxic T cell (CD8 positive); Eryth, Nucleated erythrocyte; Granu, Granulocyte; Mono, Monocyte; TBSC, Telogen (quiescent) bulge stem cell; ABSC, Anagen (activated) bulge stem cell; MTAC, Matrix/transit-amplifying cell; CLDC, Companion layer differentiated cell; EPro, Epidermis progenitor cell; EDif, Epidermis differentiated cell. Percentage values denote mean DNA methylation levels of 1-kilobase tiling regions with sufficient RRBS coverage across the genome.
(B) High-resolution view of DNA methylation and gene expression for a TF that is specific to ES cells (Dppa3) and a skin-specific keratin gene (Krt5). Red boxes refer to single CpGs (regions are not drawn to scale), with mean DNA methylation levels of single CpGs shown as a percentage values. Green boxes denote expression levels on a truncated log scale ranging from 0 (minimal expression) to 10 (maximal expression). Genomic regions: Dppa3, chr15:101,536,000-101,546,000; Krt5, chr6:122,576,000-122,581,000.
(C) Hierarchical clustering based on the mean DNA methylation levels of 1-kilobase tiling regions throughout the genome. In this type of clustering, the ordering of samples within a cluster or subtree is arbitrary and does not carry biological information. Here, within the constraints imposed by the data-derived clustering tree, cell types were ordered in the same way as in Supplementary Table 1.
(D) Hierarchical clustering based on the expression levels of Ensembl-annotated genes throughput the genome. The ordering of samples was performed in the same way as in Figure 1C.
(E) Percentage of DNA-methylation based clustering analyses in which all biological replicates of the listed cell types cluster more closely with each other than with any other sample. Each data point represents 1,000 hierarchical clustering analyses performed on random subsets of different sizes (x-axis) drawn from the data underlying Figure 1C.
DNA methylation mapping was performed by reduced representation bisulfite sequencing (RRBS), which provides single-basepair resolution and highly quantitative data for a defined subset of cytosines in the genome (Figure S1). We selected RRBS as the most suitable method for this study because it can be applied to very rare cell types (Smith et al., 2012) and because its focus on a defined set of consistently sampled genomic regions confers sensitivity for detecting small differences and minimizes the number of measurements lost due to poor coverage (Bock et al., 2010). Our RRBS analyses covered on average 1.64 million individual CpGs throughout the mouse genome (Table S1), with excellent reproducibility between biological replicates (Pearson’s r > 0.99 for most cell types, Figure S1A). Because RRBS preferentially assays genomic regions with medium to high CpG density, these maps are particularly suitable for studying DNA methylation at putative gene-regulatory elements (Figure S1B, C).
When comparing DNA methylation maps between cell types, we observed high correlations within the blood and skin lineages, with Person’s r ranging from 0.96 to 0.99 for most comparisons (Figure S1A). Nucleated erythrocytes were the only cell type that deviated substantially from all other cell types of the blood and skin lineages (Figure S1A, S1D), as they exhibited the global loss of DNA methylation that has been reported previously (Shearstone et al., 2011). In contrast, the differences between all other cell types among the two line-ages were locus-specific and relatively modest in magnitude. To be able to study these differences on a genomic scale, we developed a bioinformatic method that identifies differentially methylated region and differentially expressed genes within the dataset in a sensitive and robust manner (Supplemental Experimental Procedures). Detailed results of the differential DNA methylation and gene expression analyses are listed in Table 4, and the supplementary website (http://invivomethylation.computational-epigenetics.org/) provides genome browser tracks for visualizing all data at single-basepair resolution.
DNA methylation reflects cellular lineage choice and differentiation stage
Gene expression profiles have been shown to accurately reflect cell type and differentiation stage (Lien et al., 2011; Novershtern et al., 2011), which supports their relevance for understanding cellular differentiation. To assess whether DNA methylation maps reflect cellular identity in similar ways, we performed hierarchical clustering based on DNA methylation data of all biological replicates (Figure 1C). This analysis could accurately distinguish among blood and skin cell types and among the additional reference samples (brain and liver tissue, ES cells). Blood stem and progenitor cells clustered together and separately from lymphocytes, while the skin cell types clustered according to the physiological compartment from which they were derived (hair follicle and interfollicular epidermis).
For comparison, we also performed hierarchical clustering based on gene expression profiles for the same cell types (Figure 1D). Overall, we obtained similar results, while also uncovering one global difference between the two clustering trees. For DNA methylation data (Figure 1C), the branching points within blood and skin lineages were located substantially lower in the tree than for the clustering based on gene expression data (Figure 1D). This observation indicated that between-lineage differences in DNA methylation dominate over within-lineage differences, while the magnitude of change using gene expression is more evenly distributed throughout the tree. To quantify this disparity in the way that epigenetic and transcriptional data reflect cellular identity, we compared the number of differentially methylated regions and the number of differentially expressed genes within and between lineages (Table S4). Indeed, DNA methylation differences were five times as frequent between lineages as they were within the blood and skin lineages, while gene expression differences were only twice as frequent between lineages as they were within lineages.
In order to assess the accuracy with which DNA methylation and gene expression data reflect cellular identity, we repeated the hierarchical clustering several thousand times based on randomly selected region sets of varying sizes (Figure 1E). Ten randomly selected 1-kilobase genomic regions were generally sufficient for achieving 50% correct clustering, while 1,000 or more randomly selected regions were required to achieve near-perfect clustering accuracies. These results were similar to the clustering accuracies observed for gene expression data, although near-perfect accuracy was already achieved based on the expression levels of 100 or more randomly selected genes (Figure S2). To further evaluate the predictive power of DNA methylation, the random selection was restricted to subsets of genomic regions that share a certain characteristic. We found that CpG islands and CpG island shores gave rise to accurate clustering results based on fewer data points than were required for promoter regions (Figure S2), which is consistent with prior reports proposing CpG island shores as hotspots of informative DNA methylation differences between samples (Irizarry et al., 2009). However, genomic regions with DNA methylation levels in the range of 40% to 60% turned out to be even more powerful predictors than CpG island shores (Figure S2). These observations suggest that moderately methylated regions (a large percentage of which overlap with CpG island shores) exhibit DNA methylation levels that accurately reflect cellular identity.
DNA methylation and gene expression changes intersect at cell type specific genes
While DNA methylation maps and gene expression profiles reflected cellular lineage choice and differentiation stage with comparable accuracy (Figures 1, S2), we did not observe strong overlap between cell type specific DNA methylation and expression differences (Figure S3). It is nevertheless believed that DNA methylation contributes to cell type specific repression for a sizable number of genes throughout the genome (Hemberger et al., 2009). The majority of genes, however, need not be regulated by DNA methylation for this epigenetic modification to play a crucial role, and only a small percentage of genes may exhibit coincident changes in DNA methylation and gene expression. In support of this hypothesis, the correlation of DNA methylation and gene expression across cell types was indeed modestly negative (Figure 2A), consistent with DNA methylation’s role as a repressive epigenetic mark. The negative correlation between DNA methylation and gene expression was more pronounced for gene promoters than for the wider gene locus (Figure 2A), suggesting a direct link between DNA methylation at gene-regulatory elements and the expression levels of the associated genes.
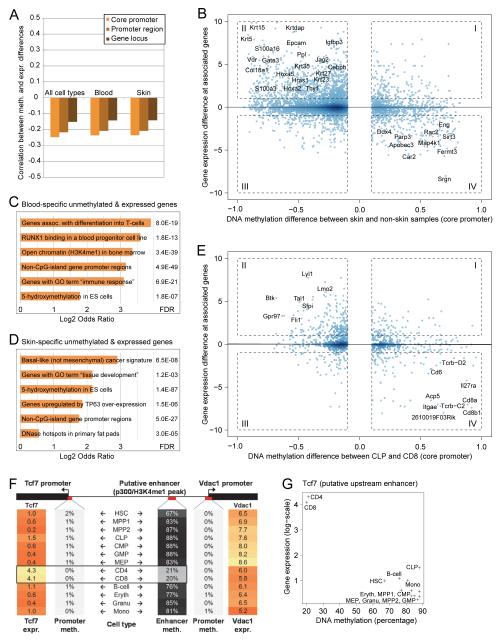
Figure 2.
DNA methylation and gene expression changes intersect at cell type specific genes
(A) Correlation between differences in DNA methylation and in the expression of associated genes, focusing on differentially methylated regions in all cell types (left bars), among blood cells (center bars) and among skin cells (right bars).
(B) Scatterplot of differences in DNA methylation and in the expression of associated genes between skin cells and nonskin samples (i.e. ES cells, brain tissue, liver tissue, blood cells). Genomic regions that exhibit a significant positive correlation (I and III) or negative correlation (II and IV) are shown in boxes and selected genes are highlighted.
(C) Enrichment of genomic properties among regions exhibiting blood-specific reduced DNA methylation and an associated increase in gene expression (box II in Figure S4A). Full results are available from http://invivomethylation.computational-epigenetics.org/.
(D) Enrichment of genomic properties among regions exhibiting skin-specific reduced DNA methylation and an associated increase in gene expression (box II in Figure 2B).
(E) Scatterplot of differences in DNA methylation and in the expression of associated genes between two lymphoid cell types (CLP and CD8).
(F) Interplay of DNA methylation and gene expression at the Tcf7-Vdac1 gene locus. Expression of the Tcf7 gene is specific to T-cells (left orange column), while Vdac1 is a widely expressed house-keeping gene (right orange column). The promoters of Tcf7 and Vdac1 are unmethylated in all cell types (left and right columns in grey). In contrast, a putative enhancer located upstream of Tcf7 and Vdac1 (Figure S1B) is specifically unmethylated in T-cells (black column).
(G) Correlation between DNA methylation levels of the putative Tcf7 enhancer highlighted in Figure 2F and the expression levels of the Tcf7 gene within the blood lineage. Genomic region: chr1:3,027,000-3,028,000.
Given the relatively small number of genes with overlapping DNA methylation and gene expression changes, we reasoned that genes exhibiting consistently negative association between these two properties may constitute strong candidates for a cell type specific functional role. Indeed, we observed well-established marker genes exhibiting lineage-specific decreases in DNA methylation and concomitant increases in gene expression (box II in Figures S4A, 2B). For the blood lineage, this included transcription factors (TFs) involved in hematopoietic regulation (Sfpi, Lmo2), cellular surface antigens (Cd27, Cd93) and cytokines (Il16); and for the skin lineage we detected multiple keratin genes (Krt5, Krt15 Krt23, Krt27, Krt35) as well as several transcription regulators with an established role in skin differentiation (Cebpb, Gata3, Hoxa5). To confirm these observations in a more quantitative manner, we performed an extended gene set enrichment analysis (cf. Supplemental Experimental Procedures) on all genes exhibiting lineage-specifically decrease DNA methylation and concomitantly increased expression levels. This analysis detected significant enrichment of relevant gene sets, epigenetic signatures and gene-regulatory binding events that are characteristic of the blood and skin lineages, respectively (Figure 2C, D), supporting that this combined epigenetic and transcriptional filtering strategy is useful for identifying lineage-specific genes.
The utility of intersecting DNA methylation and gene expression differences was not restricted to the comparison between lineages, but also identified relevant genes associated with cellular differentiation within a lineage. For example, TFs associated with gene regulation in progenitor cells (Fli1, Sfpi) were characterized by lower DNA methylation and higher gene expression levels in common lymphoid progenitors (CLPs), while genes encoding T-cell associated surface markers (Cd6, Cd8a, Cd8b1) were specifically unmethylated and highly expressed in CD8-positive T-cells (Figure 2E). Furthermore, we found that cell type specific correlations between DNA methylation and gene expression frequently extended beyond gene promoters, allowing us to link distal regulatory elements to their target genes. A case in point was the reduced DNA methylation levels found exclusively in T-cells at a putative enhancer element located 60 kilobases upstream of the Tcf7 gene, which encodes a TF that is specifically expressed in T-cells (Figure 2F, G). In a similar way, we identified putative gene-regulatory elements for the T-cell specific surface marker gene Cd8b1 and the lymphoid TF gene encoding Lef1, both of which are specifically unmethylated and expressed in T-cells (Figure S4B). These findings illustrate how locus-specific negative correlations between DNA methylation and gene expression can help identify cell type specific genes and their associated gene-regulatory regions.
Loci and binding sites of myeloid TFs become robustly methylated in lymphoid cells
Within the hematopoietic system, the cell types of the lymphoid lineage control adaptive immunity, while myeloid cells mediate innate immune response and a number of other physiological functions of the blood. On an evolutionary timescale, the myeloid lineage is older and detectable already in primitive vertebrates, whereas the lymphoid lineage is exclusive to vertebrates. Functional evidence suggests that DNA methylation is crucial for lymphoid differentiation but largely dispensable for myeloid differentiation (Broske et al., 2009; Trowbridge et al., 2009); and a substantial number of DNA methylation changes have been identified that associate with lymphoid lineage differentiation (Ji et al., 2010).
Notably, our analysis identified roughly twice as many genomic regions with higher methylation in CLPs than regions with higher methylation in common myeloid progenitors (CMPs) (Figure 3A, Table S5). Regions that were differentially methylated between CLPs and CMPs tended to retain their increased or decreased DNA methylation levels in terminally differentiated lymphocytes (CD4, CD8, B-cells), while the picture was more diverse for myeloid cells (Figure 3A). In terms of gene expression, we observed that lineage-specific downregulation was consistently retained downstream of both lymphoid and myeloid progenitor cells, while lineage-specific upregulation was reversed in terminally differentiated cells of both lineages (Figure S5A). These observations indicate that lymphoid vs. myeloid lineage choice is associated with stable silencing of genes from the alternative lineage at the stage of multipotent progenitor (MPP2) differentiation into CLPs or CMPs. Furthermore, our data indicate that DNA methylation is more robustly used for myeloid gene repression within lymphoid cells than vice versa.
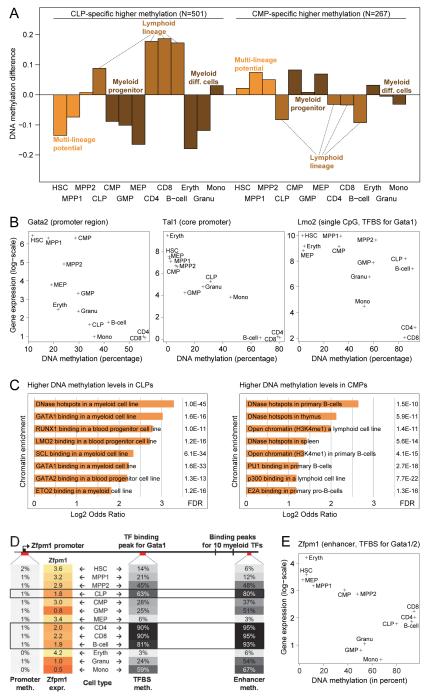
Figure 3.
Myeloid TFs and their binding sites become methylated in lymphoid cells
(A) DNA methylation of genomic regions that are differentially methylated between CLPs and CMPs. Focusing only on regions that are significantly more highly methylated in CLPs than in CMPs (left) or vice versa (right), the diagram shows these regions’ mean DNA methylation levels for all cell types of the blood lineage. To be able to plot multiple regions with different DNA methylation levels on the same scale, DNA methylation levels (y-axis) are shown as absolute differences relative to the mean methylation levels of CLPs and CMPs in each genomic region.
(B) Correlation between DNA methylation and expression levels at myeloid transcription-regulatory genes within the blood lineage. Shown are the mean DNA methylation levels (x-axis) of the 1-kilobase tiling regions that exhibit the highest association with the genes’ expression levels (y-axis). Genomic regions: Gata2, chr6:88,150,000-88,151,000; Tal1, chr4:114,732,000-114,733,000; Lmo2, chr2:103,798,108-103,798,109.
(C) Enrichment of genomic properties among regions that are significantly more highly methylated in CLPs than in CMPs (left) or vice versa (right).
(D) Interplay of DNA methylation and gene expression at the Zfpm1 gene locus. Lymphoid-specific reduced expression of the Zfpm1 gene is associated with increased DNA methylation levels at an intronic Gata1 binding site (center) and also at a putative enhancer element further toward the 3′ end of the gene (right). This putative enhancer overlaps with ChIP-seq binding peaks of multiple myeloid TFs, namely Gata1, Gata2, Lmo2, Lyl1, Runx1, Tal1, Erg, Fli1, Gfi1b and Meis1.
(E) Correlation between DNA methylation levels of the putative Zfpm1 enhancer highlighted in Figure 3D and the expression levels of the Zfpm1 gene within the blood lineage. Genomic region: chr8:124,833,000-124,834,000.
Two additional lines of evidence support the hypothesis that DNA methylation plays an important role for silencing myeloid regulatory programs in the lymphoid lineage. First, we observed that lymphoid cells exhibited increased promoter DNA methylation levels at several key regulators of myeloid lineage commitment, which was correlated with robust transcriptional repression in these cell types (Figure 3B). Examples included the TFs Gata2 and Tal1, both of which interfere with normal differentiation when overexpressed in lymphoid cells (Goardon et al., 2002; Tipping et al., 2009). Another interesting case was Lmo2, which blocks T-cell differentiation (Pike-Overzet et al., 2007) and causes T-cell leukemia (McCormack et al., 2003) when aberrantly expressed in lymphoid cells. Although the promoter region of Lmo2 is largely depleted of CpG sites, we observed a significant correlation between Lmo2 expression levels and the DNA methylation levels of a single CpG. This CpG is located 41bp upstream of the transcription start site and overlaps with a Gata1 binding site, suggesting that it may interfere with Gata1 TF binding when methylated (Figure 3B). While this analysis cannot discriminate between causal and consequential changes, DNA methylation levels of a single CpG have been shown to act as a regulatory mechanism for TF binding and gene expression in other systems (Xu et al., 2009). Second, we found that binding sites of important myeloid TFs were strongly enriched among genomic regions that exhibited higher DNA methylation levels in CLPs compared to CMPs (Figure 3C). Previous ChIP-seq studies had identified binding sites for the known myeloid TFs Gata1, Gata2, Lmo2 and Runx1 (Hannah et al., 2011), all of which we found to be enriched among the CLP-specific hypermethylated regions. This trend was statistically significant on the genomic scale and observed for several regulatory elements located near well-characterized myeloid genes. For example, we observed differential DNA methylation at Gata1 and/or Gata2 binding sites in the Zfpm1 gene, which encodes an important cofactor of Gata TFs (Figure 3D, E), for the erythrocyte-specific Rhd and Hpn genes as well as for the myeloid master regulator Sfpi1 (Figure S5B). In summary, these observations suggest that cell type specific DNA methylation of myeloid TFs and their binding sites may provide a checkpoint against accidental activation of myeloid regulatory programs in lymphoid cells.
Blood stem cell differentiation is associated with hypermethylation and downregulation of homeobox genes
Hematopoietic stem cells (HSCs) were the earliest identified adult stem cells and the first that could be purified and characterized with high stringency (Spangrude et al., 1988). The study of HSCs has shaped the functional definition of stem cells, which comprises the capacity for long-term self-renewal and the potency to differentiate into several types of specialized cells. Bona fide HSCs are rare, require complex purification procedures and cannot be cultivated in vitro without losing their stemness, which likely explains why this important cell type has so far eluded comprehensive epigenome characterization.
In our dataset, HSC differentiation into progenitor cells was associated with a moderate number of DNA methylation changes (Figure 4A, Table S5), gain of DNA methylation being more common than its loss. These changes occurred gradually during adult stem cell differentiation into multipotent and lineage-committed progenitor cells and were robustly maintained in terminally differentiated cells. We observed a somewhat more complex picture when analyzing the gene expression changes associated with HSC differentiation (Figure S6A). On the one hand, stem-cell specific genes were gradually downregulated during differentiation and retained their reduced expression in terminally differentiated cells, which resembles the DNA methylation dynamics of HSC differentiation. In contrast, those genes that were specifically upregulated in progenitor cells became downregulated again in terminally differentiated cells (Figure S6A). This progenitor-specific gene cluster was highly enriched for cell cycle associated genes, reflecting the highly proliferative nature of progenitor cells compared to relatively quiescent stem cells and terminally differentiated cells.
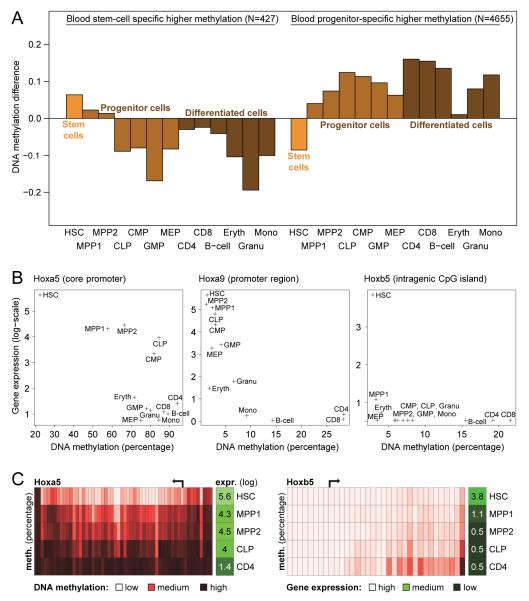
Figure 4.
Blood stem cell differentiation coincides with specific DNA demethylation of hematopoietic regulator genes
(A) DNA methylation of genomic regions that are differentially methylated between blood stem cells (HSCs) and progenitor cells (MPP1, MPP2, CLP, CMP, GMP, MEP). Focusing only on genomic regions that are significantly more highly methylated in stem cells than in progenitor cells (left) or vice versa (right), the diagram shows these regions’ mean DNA methylation levels for all cell types of the blood lineage. DNA methylation levels (y-axis) are shown as absolute differences relative to the mean methylation levels of HSCs and the average of all progenitor cells in each genomic region.
(B) Correlation between DNA methylation and gene expression levels at homeobox genes within the blood lineage. Genomic regions: Hoxa5, chr6:52,154,000-52,155,000; Hoxa9, chr6:52,178,000-52,179,000; Hoxb5, chr11:96,166,000-96,167,000.
(C) Single-basepair resolution view of DNA methylation and gene expression for the Hoxa5 and Hoxb5 genes. The format is the same as in Figure 1B. Genomic regions: Hoxa5, chr6:52,151,500-52,155,000; Hoxb5, chr11:96,164,500-96,168,500.
In order to identify putative HSC-specific regulator genes, we intersected DNA methylation and gene expression data, and we determined those genes that became downregulated and hypermethylated during HSC differentiation into progenitor cells. Examples of such genes included the hormone receptor Lhcg and the TF Smad6 (Figure S6B). Perhaps most remarkably, we identified four HSC-specific homeobox genes that appear to be repressed by DNA methylation in terminally differentiated cells (Figures 4B, S6B). These were the well-characterized oncogenes Hoxa9 and Pbx1, the candidate oncogene Hoxb5 (Bullinger et al., 2004) and the putative tumor suppressor gene Hoxa5, which is aberrantly methylated in some leukemia patients (Strathdee et al., 2007). Each of these genes followed its own trajectory in terms of downregulation and gain of DNA methylation, arguing against a single epigenetic switch that deactivates multiple homeobox genes during HSC differentiation. For example, the Hoxa5 locus was already partially methylated in HSCs and accumulated further DNA methylation while its expression decreased gradually; in contrast, the Hoxb5 locus gained modest levels of DNA methylation only in lymphoid cells, although it was already fully transcriptionally repressed within MPP2 cells (Figure 4C). In summary, these results suggest that certain homeobox genes accumulate DNA methylation during HSC differentiation, which may protect them from aberrant activation in progenitors and terminally differentiated cell types.
Skin stem cells remodel their epigenome in response to cell cycle activation and cellular differentiation
Two distinct populations of skin stem cells can be purified from the hair follicle: quiescent telogen bulge stem cells (TBSCs) and activated anagen bulge stem cells (ABSCs). Both cell types are capable of long-term self-renewal and have the potency to differentiate into several types of specialized cells, thus fulfilling the defining criteria of stem cells (Blanpain et al., 2004). During the normal hair cycle, TBSCs and ABSCs give rise to short-lived matrix transit-amplifying cells (MTACs), from which the seven differentiated lineages of the hair follicle are derived (including companion layer differentiated cells, CLDCs). Hair follicle bulge stem cells can also regenerate the epidermis, but only under the special conditions of wound repair. A recent study mapped three histone modifications (histone H3K4me3, H3K27me3, H3K79me2) during stem cell maintenance and differentiation (Lien et al., 2011). This provided us with the opportunity to compare the DNA methylation changes observed in our dataset with a catalog of chromatin states in the same cell types.
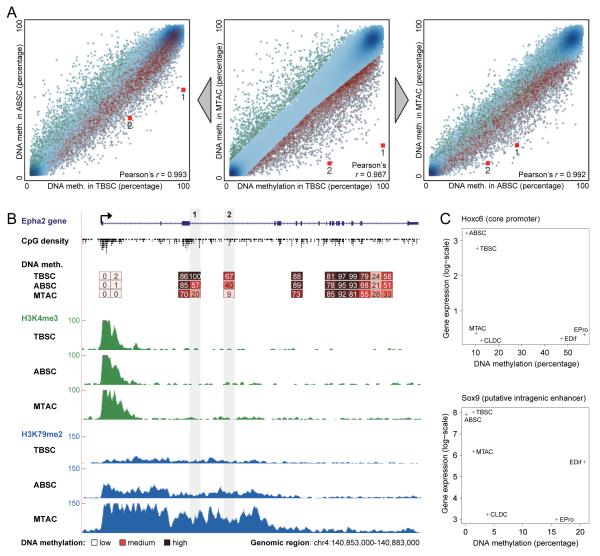
In contrast to HSCs, hair follicle stem cells can be purified during their quiescent resting stage (TBSCs) and following their activation (ABSCs), when follicles are growing hair and stem cells undergo self-renewal. This allowed us to assess the prevalence of DNA methylation changes associated with stemness versus those associated with cell proliferation. In pairwise comparisons of DNA methylation maps (Figures 5A, S7A), we observed marginally higher correlation between the two stem cell populations (ABSC vs. TBSC, Pearson’s r = 0.993) than between the proliferating stem and progenitor cell populations of the hair follicle (ABSC vs. MTAC, Pearson’s r = 0.992). Direct comparison between quiescent stem cells and proliferating progenitor cells resulted in a lower correlation (TBSC vs. MTAC, Pearson’s r = 0.987). An even lower correlation was observed when comparing hair follicle stem cells with epidermal progenitor cells (TBSC vs. EPro, Pearson’s r = 0.974) and with terminally differentiated cells of the epidermis (TBSC vs. EDif, Pearson’s r = 0.982), consistent with the fact that the interfollicular epidermis is maintained by a separate stem cell population under conditions of normal tissue homeostasis and may therefore constitute a distinct cellular lineage.
Figure 5.
Skin stem cells undergo epigenetic remodeling upon cell cycle activation and upon differentiation
(A) Pairwise scatterplots of DNA methylation observed in skin stem cells (TBSC, ABSC) and skin progenitor cells (MTAC). Genomic regions that were significantly more highly methylated in TBSCs than in MTACs are highlighted in red across all three diagrams, and genomic regions that were significantly less methylated in TBSCs than in MTACs are highlighted in green. Numbered red squares highlight two specific genomic regions that are displayed in detail in Figure 5B.
(B) DNA methylation and histone modifications at the Epha2 gene locus. Red boxes refer to 1-kilobase genomic regions, with the mean DNA methylation level of each region shown as a percentage value. The frequency plots display ChIP-seq read counts of the promoter-associated histone modification H3K4me3 (green) and the transcription-associated histone modification H3K79me2 (blue) across the locus, based on published ChIP-seq data (Lien et al., 2011). Genomic region: chr4:140,853,000-140,883,000.
(C) Correlation between DNA methylation and gene expression levels at the Hoxc6 and Sox9 genes within the skin lineage. Genomic regions: Hoxc6, chr15:102842000-102843000; Sox9, chr11:112,646,000-112,647,000.
To test whether skin stem cell differentiation is associated with similarly consistent DNA methylation changes as we observed for HSC differentiation, we identified all genomic regions that were differentially methylated between TBSCs and MTACs (Table S5) and highlighted them in each of the scatterplots (Figures 5A, S7A). Genomic regions that were more highly methylated in TBSCs than in MTACs (red) were on average also more highly methylated in TBSCs than in ABSCs or in epidermis cells (EPro, EDif). Similarly, genomic regions that were more highly methylated in MTACs than in TBSCs (green) were on average also more highly methylated in MTACs than in ABSCs. These observations confirmed that the DNA methylation levels of a significant number of genomic regions were robustly associated either with the stem cell compartment or with the progenitor cell population. An illustrative example is the Epha2 gene, which encodes a receptor tyrosine kinase and has been shown to contribute to terminal differentiation in skin cells (Lin et al., 2010). Two putative regulatory elements downstream of the Epha2 promoter gradually lose DNA methylation during skin stem cell differentiation (red squares in Figure 5A). In parallel, this gene locus acquires a chromatin structure indicative of active transcription (Figure 5B).
Finally, we identified putative skin stem cell specific genes in the same way as we did for the blood lineage, by intersecting DNA methylation and gene expression differences associated with skin stem cell differentiation (Figures 5C and S7B). Among these candidate regulator genes was an additional homeobox TF, the putative oncogene Hoxc6 (Ramachandran et al., 2005). The list also included several other TFs with an established regulatory role in skin stem cells, such as Sox9 (Nowak et al., 2008), Tcf7l2 (Nguyen et al., 2009) and Runx1 (Osorio et al., 2008). In summary, our data for the skin lineage suggest that stemness and quiescence contribute to the characteristic DNA methylation signatures observed among TBSCs, and they provide an additional example for differentiation-associated gain of DNA methylation and loss of expression of a homeobox TF.
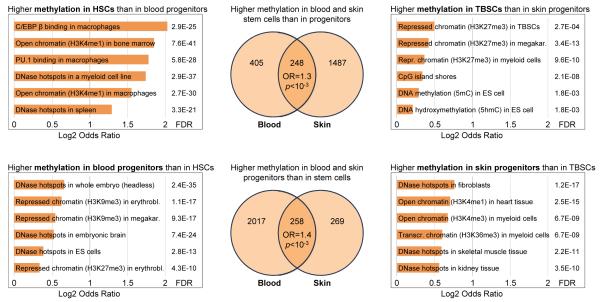
Stem cells exhibit incomplete programming of DNA methylation toward the blood and skin lineages
In order to identify common themes associated with adult stem cell differentiation, we performed a systematic comparison between stem cells and committed progenitor cell types in the blood and skin lineages (Figures 6, S8). A total of 248 genomic regions were more highly methylated in stem cells of both lineages when compared to the corresponding progenitor cells, which constitutes a modest enrichment over random chance (odds ratio = 1.3, p < 10−3). Similarly, 258 genomic regions were less methylated in stem cells than in progenitor cells (odds ratio = 1.4, p < 10−3). These shared signatures were statistically significant but small in absolute terms, indicating that the DNA methylation changes in adult stem cell differentiation are to a large degree lineage-specific.
Figure 6.
Stem cells and progenitors exhibit characteristic DNA methylation signatures Magnitude of overlap and enrichment of genomic properties among regions that are consistently more highly methylated in stem cells (top row) or in progenitor cells (bottom row) of the blood and skin lineages. Differences between blood stem cells (HSCs) and progenitor cells (MPP1, MPP2, CLP, CMP, GMP, MEP) were compared to differences between quiescent skin stem cells (TBSCs) and progenitor cells (MTACs), and the significance of overlap was assessed by the odds ratio (OR) and Fisher’s exact test (p). The total numbers of genomic regions in the Venn diagrams are lower than in the separate analyses of the blood and skin lineages because fewer regions fulfill the minimum coverage filtering across all cell types.
When we performed an extended gene set enrichment analysis separately on the blood and skin data and compared enriched properties, we observed more pronounced similarities (Figure 6A). For both lineages, genomic regions characterized by lower DNA methylation in stem cell than in progenitor cells were significantly enriched for open chromatin and DNase hypersensitive sites associated with other somatic lineages (brain, fibroblasts, heart, skeletal muscle, kidney) and with ES cells. We also observed enrichment of blood-specific TF binding, open chromatin and putative gene-regulatory elements among those genomic regions that were more highly methylated in blood stem cells than in progenitor cells. These results suggest that blood and skin stem cells retain open chromatin at gene-regulatory elements associated with other lineages, which is lost upon differentiation. At the same time, lineage-specific regulatory regions become increasingly unmethylated when adult stem cells differentiate into progenitor cells.
We performed a similar comparison for gene expression data and obtained quite different results (Figure S8), highlighting that DNA methylation and gene expression reflect complementary aspects of adult stem cell differentiation. Stem cell specific genes of the blood and skin lineage exhibited considerable overlap (odds ratio = 3.3, p < 10−99), and an even stronger overlap was observed among genes that were specifically expressed in committed progenitor cell types as opposed to stem cells (odds ratio = 4.7, p < 10−99). Stem cell specific genes frequently coded for TFs carrying zinc finger domains, overlapped with published gene signatures of stem cells, and exhibited an active chromatin state in the two adult stem cell populations for which epigenome data was available (TBSC, ABSC). In contrast, progenitor specific genes exhibited strong enrichment for cell cycle, proliferation and aggressive cancer gene signatures, consistent with the highly proliferative nature of most progenitor cell populations.
Combining DNA methylation and gene expression data enables inference of cellular lineage hierarchies
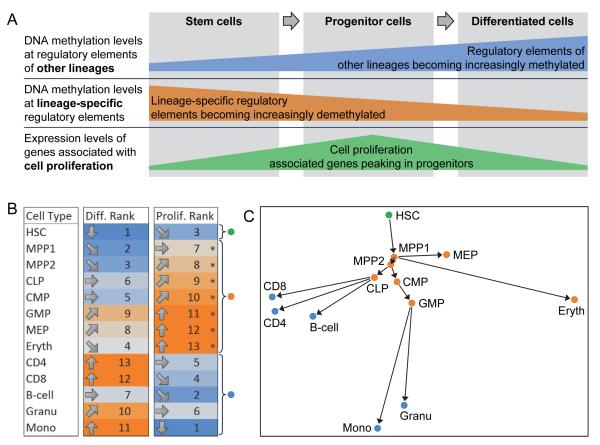
Our efforts to dissect the role of DNA methylation in adult stem cell differentiation uncovered certain properties of cellular differentiation that were consistently observed among the blood and skin lineages (Figure 7A). First, gene-regulatory regions associated with other lineages and with ES cells became increasingly methylated during adult stem cell differentiation. Second, gene-regulatory elements of the chosen lineage were only partially demethylated in stem cells and exhibited significantly reduced DNA methylation levels in committed progenitors and in terminally differentiated cells. Third, committed progenitor cells were characterized by a strong proliferative gene expression signature, which was essentially absent from both stem cells and terminally differentiated cells. Finally, all cell types clustered by their known biological similarity in an unsupervised analysis of DNA methylation and gene expression data (Figure 1).
Figure 7.
Cellular lineage hierarchies can be inferred from DNA methylation maps and gene expression profiles
(A) Schematic drawing summarizing trends in the DNA methylation and gene expression data that were observed among the blood and skin lineages. The size of the blue, orange and red bars are not drawn to scale, i.e. they do not attempt to quantify what percentage of regulatory regions and genes are methylated or expressed in each cell type.
(B) Differentiation and proliferation ranking of all analyzed blood cell types. The cell type with the lowest differentiation rank was tagged in green (“stem cell”), all cell types with proliferation scores above the mean (indicated by asterisks) were tagged in orange (“progenitor cells”), and all remaining cell types were tagged in blue (“terminally differentiated cells”).
(C) Blood lineage hierarchy inferred from genomic data. Similarities and differences between the DNA methylation and gene expression profiles among cell types were projected onto a two-dimensional map, each cell type was colored following the classification in Figure 7B, and the points were connected by arrows according to a method for data-driven lineage inference described in the Experimental Procedures.
We reasoned that these observations could in aggregate provide the basis for bioinformatic inference of cellular differentiation hierarchies, combining measures of differentiation directionality and proliferation state with global estimates of similarity among genome-scale DNA methylation maps and gene expression profiles. To that end, we calculated differentiation and proliferation ranks for all cell types of the blood lineage relative to one another (cf. Experimental Procedures), and each cell type was classified as stem cells, progenitor cells or terminally differentiated cells based on these ranks (Figure 7B). Specifically, we identified all cell types whose proliferation scores exceeded the observed mean value across samples and classified them as progenitor cells; and we designated the cell type with the lowest differentiation rank as stem cells, while also verifying that these stem cells exhibited a low proliferation rank. Next, we performed an unsupervised multidimensional scaling analysis of the combined genome-scale DNA methylation maps and gene expression profiles. This method uses the projection of the complex datasets onto a two-dimensional map to depict the similarities and differences of cell types as spatial distances (Figure 7C). In this map, we tagged the most undifferentiated cell type as stem cell (green), highly proliferating cell types as progenitors (orange) and all remaining cell types as terminally differentiated cells (blue). Finally, we inferred a cellular differentiation tree by starting from the stem cells and iteratively connecting each cell type to the spatially closest stem cell or progenitor cell type that was already part of the tree.
This analysis was conducted without entering any prior knowledge on cell type specific marker genes, functional similarities between cell types or observations from lineage tracing studies. Instead it was solely based on a combination of two genomic datasets (DNA methylation and gene expression), well-established statistical methods and a small set of rules that we derived from the observations made in the current study. The resulting predictions closely recapitulate our current understanding of hematopoiesis with its hierarchical differentiation tree rooted on the HSC (Figure 7C). For example, the inferred map accurately reflects the establishment of lymphoid and myeloid lineages downstream of MPP2 cells, and it correctly positions lymphocytes downstream of CLP, while placing monocytes and granulocytes downstream of GMPs. On the other hand, the predictions for the erythrocyte lineage are more debatable. While it is reasonable that the model classified nucleated erythrocytes as progenitor cells (they are a highly proliferating cell type that retains the potency for differentiation into enucleated erythrocytes), it would be consistent with widely accepted models of hematopoietic differentiation to place erythrocytes downstream of MEPs, and MEPs downstream of CMPs; although some evidence exists that is supporting the same relationship of MPPs and MEPs that we inferred from the data (Adolfsson et al., 2005).
When we applied the same approach to the skin lineage the proliferation ranks accurately reflected increased proliferation of progenitor cells compared stem cells and terminally differentiated cells (Figure S9A). Furthermore, the two-dimensional map identified biologically plausible properties of the skin lineage hierarchy, such as strong separation between cell types of the hair follicle and epidermis, and stepwise changes among the hair follicle cells (Figure S9B). However, all cell types scored equally in terms of their differentiation rank (Figure S9A), which precluded us from performing a confident classification of skin cell types into stem cells, progenitors and terminally differentiated cell types. For this reason, we could not add directional arrows denoting differentiation trajectories into an otherwise highly accurate map of the skin lineage (Figure S9B). It is possible that higher plasticity within the skin lineage constitutes the biological reason for our inability to infer skin differentiation hierarchies, but technical reasons related to the smaller number of cell types, less developed cell sorting strategies or insufficient robustness of the bioinformatic methods could also explain this result. In summary, cellular lineage inference based on the combination of DNA methylation and gene expression data can be applied to the cell types of the blood lineage and may provide a useful complement to more classical approaches. However, further validation on additional cell types both within and beyond the blood lineage will be required to validate and refine the proposed method.
Discussion
In order to foster our understanding of DNA methylation in cellular lineage commitment, we established high-resolution genomic maps of DNA methylation for 19 cell types of the blood and skin lineages that were purified directly from the mouse, without exposing them to cell culture. These data constitute a single-basepair resolution resource of the DNA methylation dynamics of adult stem cell differentiation in vivo. Through integrated bioinformatic analyses, we discovered epigenetic regulatory patterns that were in part shared between the blood and skin lineages, raising the possibility that these patterns may constitute general principles of adult stem cell differentiation and epigenetic regulation of cellular lineage commitment. For example, we found that adult stem cells were characterized by reduced levels of DNA methylation at gene-regulatory elements associated with other lineages and with ES cells, while lineage-specific gene-regulatory elements (such as the binding sites of hematopoietic TFs) were more highly methylated in stem cells than in progenitor cells. This observation suggests that it is more feasible and biologically meaningful to establish an epigenetic signature of stemness based on enrichment of certain chromatin features than by trying to identify a single set of stem-cell associated genomic regions that would be unmethylated in all stem cell populations.
We also observed that gain of DNA methylation and loss of gene expression were robustly maintained downstream of adult stem cells, indicating that cellular identity is in part defined by epigenetic switches that change their state only once in the course of the differentiation. Such switches might act as gatekeepers and prevent differentiated cells from aberrantly expressing stem-cell associated genes. In a similar way, DNA methylation appears to provide a two-tier epigenetic barrier against spurious expression of myeloid TFs in lymphoid cells. We observed that both the gene loci and the binding sites of key myeloid specification factors (Gata2, Tal1 and Lmo2) became robustly methylated during lymphoid differentiation, suggesting that DNA methylation may interfere not only with their transcription but also with the DNA binding of these TFs in case they are spuriously transcribed in a lymphoid cell. Given that all of these TFs are well-characterized oncogenes, it seems plausible to speculate that simultaneous epigenetic repression of TF gene loci and their binding sites across the genome may help protect lineage-committed cells from TF-induced oncogenesis.
Across all cell types analyzed, we found that DNA methylation and gene expression data reflected cellular lineage choice and differentiation stage with similar accuracy, although DNA methylation differences were comparatively rare among the blood and skin lineages, modest in magnitude and not strongly correlated with differences in gene expression. From a practical perspective, these results support the value of DNA methylation as a fingerprint of cellular identity, which may prove valuable for inferring the tissue of origin of samples that lack RNA of sufficient quality and quantity (e.g. archival tumors, forensic samples, museum collections). Furthermore, we could show that a relatively straightforward bioinformatic method was able to accurately infer the lineage hierarchy among blood cell types based on the combination of DNA methylation and gene expression profiles. While this method requires further validation and refinement based on larger datasets and additional cellular lineages, it may ultimately provide a fast and unbiased method for mapping the topology of cellular differentiation hierarchies.
Experimental Procedures
DNA methylation
Blood and skin cells were obtained from adult mice and purified by fluorescence-activated cell sorting (FACS) using stringent sorting criteria (cf. Supplemental Experimental Procedures and Table S1). RRBS libraries for DNA methylation analysis were prepared from 30ng of input DNA per biological replicate following a published protocol (Gu et al., 2011) and sequenced by the Broad Institute’s Genome Sequencing Platform on Illumina Genome Analyzer II or HiSeq 2000 machines. Bioinformatic data processing and quality control were performed as described previously (Bock et al., 2011).
Gene expression
All gene expression data were based on the Affymetrix GeneChip Mouse Genome 430 2.0 Array. Microarray profiles for a subset of the cell types used in this study have been published previously (Chambers et al., 2007; Ji et al., 2010; Lien et al., 2011); and the corresponding data are publicly available from the Gene Expression Omnibus (GEO) repository (accession numbers: GSE20244, GSE6506 and GSE31028). Microarray data that were not publicly available have been submitted to GEO (GSE38557). All microarray data were obtained as CEL files, and they have been quality-controlled and normalized in the same way in order to minimize batch effects (cf. Supplemental Experimental Procedures).
Data analysis
DNA methylation data were mapped to a 1-kilobase tiling region set of the mouse genome, giving rise to a total of 95,086 genomic regions for which highly quantitative DNA methylation levels were available throughout the dataset. Similarly, gene expression data were mapped to Ensembl gene identifiers, resulting in 20,666 genes with robust data across the entire dataset. Data from biological replicates were integrated and cell type specific differences detected using a bioinformatic method that makes it possible to treat DNA methylation and gene expression data in an equivalent way (cf. Supplemental Experimental Procedures). The extended gene set enrichment analysis combined two gene-based approaches – functional enrichment analysis using DAVID (http://david.abcc.ncifcrf.gov/) and a parametric gene set enrichment analysis based on MSigDB (http://www.broadinstitute.org/gsea/msigdb/) – with two region-based approaches, namely an enrichment analyses from chromatin annotations collected from Cistrome (http://cistrome.org/) and other sources, and a genomic feature analyzing using EpiGRAPH (http://epigraph.mpi-inf.mpg.de/). With the exception of the DAVID and EpiGRAPH analyses, which were performing using the publicly available web servers, all data analyses were performed with the R statistics package (http://www.r-project.org/).
Lineage inference
The differentiation rank was calculated as the mean rank of DNA methylation for gene-regulatory elements specific to ES cells (higher levels resulted in higher ranks) and for gene-regulatory elements specific to the blood or skin lineage (lower levels resulted in higher ranks). In both cases, DNase hypersensitivity hotspot data and ChIP-seq peaks for transcription factor binding as well as chromatin modifications were obtained from public sources, and the DNA methylation levels at 1-kilobase tiling regions overlapping with these peaks were compared to the background of 1-kilobase tiling regions that did not overlap. Similarly, the proliferation rank was calculated based on the expression levels of genes included in proliferation-associated gene signatures obtained from MSigDB. Based on these scores, cell types were classified as stem cells (lowest differentiation rank), progenitor cells (proliferation score above the mean) and terminally differentiated cells (all remaining cell types). In the next step, multidimensional scaling was performed on the combination of the DNA methylation and gene expression profiles, using Pearson’s r as distance function (higher correlations corresponding to smaller distances) and averaging the normalized distance matrix between DNA methylation and gene expression (in order to give both data types equal weight). Finally, cell types were iteratively connected by arrows, starting from the stem cells and in the order of increasing differentiation scores. With each step, the next cell type in the list was connected to the spatially closest cell type that had already been selected and had been classified as either stem cell or progenitor cell.
Supplementary website
Additional material is available from http://invivomethylation.computational-epigenetics.org/, including raw DNA methylation data (BAM and BED files), preprocessed DNA methylation and gene expression data tables and genome browser tracks for interactive visualization of the DNA methylation data. All genomic coordinates in this paper and on the supplementary website refer to the mm9 (NCBI37) assembly of the mouse genome.
Supplementary Material
Highlights.
Genomic DNA methylation maps for 19 purified cell types of blood and skin lineages
Myeloid transcription factors and binding sites become methylated in lymphoid cells
Adult stem cells retain low DNA methylation at regulatory regions of other lineages
Epigenome and transcriptome enable inference of the blood differentiation hierarchy
Acknowledgements
We would like to thank other members of the Meissner, Rossi and Fuchs labs for their support. From Rockefeller University, we also thank S. Maizel at the flow cytometry facility as well as Nicole Stokes and the Comparative Biology Center animal facility. Furthermore, we thank Tarjei S. Mikkelsen for his contributions to the early parts of the project and Fontina Kelley for supporting the sequencing efforts. C. Bock was supported by a Feodor Ly-nen Fellowship (Alexander von Humboldt Foundation) and a Charles A. King Trust Postdoctoral Fellowship (Charles A. King Trust, N.A., Bank of America, Co-Trustee). W.-H. Lien was supported by a Harvey L. Karp Postdoctoral Fellowship and a Jane Coffin Child Postdoctoral Fellowship. E. Fuchs is a Howard Hughes Medical Institute Investigator. A. Meissner was supported by the Massachusetts Life Science Center and the Pew Charitable Trusts. D. Rossi was supported by the New York Stem Cell Foundation. This work was funded by the Harvard Stem Cell Institute (to D. Rossi and A. Meissner) and partially supported by a grant (to E. Fuchs) from the NIH/NIAMS (R01AR31737).
Footnotes
Accession numbers GSE38557
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adolfsson J, Mansson R, Buza-Vidas N, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Blanpain C, Lowry WE, Geoghegan A, et al. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Bock C, Kiskinis E, Verstappen G, et al. Reference Maps of Human ES and iPS Cell Variation Enable High-Throughput Characterization of Pluripotent Cell Lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C, Tomazou EM, Brinkman AB, et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol. 2010;28:1106–1114. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broske AM, Vockentanz L, Kharazi S, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009;41:1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- Bullinger L, Dohner K, Bair E, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004;350:1605–1616. doi: 10.1056/NEJMoa031046. [DOI] [PubMed] [Google Scholar]
- Chambers SM, Boles NC, Lin KY, et al. Hematopoietic fingerprints: an expression database of stem cells and their progeny. Cell Stem Cell. 2007;1:578–591. doi: 10.1016/j.stem.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goardon N, Schuh A, Hajar I, et al. Ectopic expression of TAL-1 protein in Ly-6E.1-htal-1 transgenic mice induces defects in B- and T-lymphoid differentiation. Blood. 2002;100:491–500. doi: 10.1182/blood.v100.2.491. [DOI] [PubMed] [Google Scholar]
- Gu H, Smith ZD, Bock C, et al. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc. 2011;6:468–481. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- Hannah R, Joshi A, Wilson NK, et al. A compendium of genome-wide hematopoietic transcription factor maps supports the identification of gene regulatory control mechanisms. Exp Hematol. 2011;39:531–541. doi: 10.1016/j.exphem.2011.02.009. [DOI] [PubMed] [Google Scholar]
- Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington’s canal. Nat Rev Mol Cell Biol. 2009;10:526–537. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- Hodges E, Molaro A, Dos Santos CO, et al. Directional DNA methylation changes and complex intermediate states accompany lineage specificity in the adult hematopoietic compartment. Mol Cell. 2011;44:17–28. doi: 10.1016/j.molcel.2011.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Ehrlich LI, Seita J, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien WH, Guo X, Polak L, et al. Genome-wide maps of histone modifications unwind in vivo chromatin states of the hair follicle lineage. Cell Stem Cell. 2011;9:219–232. doi: 10.1016/j.stem.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Gordon K, Kaplan N, et al. Ligand targeting of EphA2 enhances keratinocyte adhesion and differentiation via desmoglein 1. Mol Biol Cell. 2010;21:3902–3914. doi: 10.1091/mbc.E10-03-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack MP, Forster A, Drynan L, et al. The LMO2 T-cell oncogene is activated via chromosomal translocations or retroviral insertion during gene therapy but has no mandatory role in normal T-cell development. Mol Cell Biol. 2003;23:9003–9013. doi: 10.1128/MCB.23.24.9003-9013.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Nguyen H, Merrill BJ, Polak L, et al. Tcf3 and Tcf4 are essential for long-term homeostasis of skin epithelia. Nat Genet. 2009;41:1068–1075. doi: 10.1038/ng.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novershtern N, Subramanian A, Lawton L, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak JA, Polak L, Pasolli HA, et al. Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell. 2008;3:33–43. doi: 10.1016/j.stem.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio KM, Lee SE, McDermitt DJ, et al. Runx1 modulates developmental, but not injury-driven, hair follicle stem cell activation. Development. 2008;135:1059–1068. doi: 10.1242/dev.012799. [DOI] [PubMed] [Google Scholar]
- Pike-Overzet K, de Ridder D, Weerkamp F, et al. Ectopic retroviral expression of LMO2, but not IL2Rgamma, blocks human T-cell development from CD34+ cells: implications for leukemogenesis in gene therapy. Leukemia. 2007;21:754–763. doi: 10.1038/sj.leu.2404563. [DOI] [PubMed] [Google Scholar]
- Ramachandran S, Liu P, Young AN, et al. Loss of HOXC6 expression induces apoptosis in prostate cancer cells. Oncogene. 2005;24:188–198. doi: 10.1038/sj.onc.1207906. [DOI] [PubMed] [Google Scholar]
- Shearstone JR, Pop R, Bock C, et al. Global DNA demethylation during mouse erythropoiesis in vivo. Science. 2011;334:799–802. doi: 10.1126/science.1207306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012 doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- Strathdee G, Holyoake TL, Sim A, et al. Inactivation of HOXA genes by hypermethylation in myeloid and lymphoid malignancy is frequent and associated with poor prognosis. Clin Cancer Res. 2007;13:5048–5055. doi: 10.1158/1078-0432.CCR-07-0919. [DOI] [PubMed] [Google Scholar]
- Tipping AJ, Pina C, Castor A, et al. High GATA-2 expression inhibits human hematopoietic stem and progenitor cell function by effects on cell cycle. Blood. 2009;113:2661–2672. doi: 10.1182/blood-2008-06-161117. [DOI] [PubMed] [Google Scholar]
- Trowbridge JJ, Snow JW, Kim J, et al. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell. 2009;5:442–449. doi: 10.1016/j.stem.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Watts JA, Pope SD, et al. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 2009;23:2824–2838. doi: 10.1101/gad.1861209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.