Abstract
We have found that daily subcutaneous injection with a maximum tolerated dose (MTD) of the mGluR2/3 agonist LY379268 (20mg/kg) beginning at 4 weeks dramatically improves the phenotype in R6/2 mice. For example, we observed normalization of motor function in distance traveled, speed, the infrequency of pauses, and the ability to locomote in a straight line, and a rescue of a 15–20% striatal neuron loss at 10 weeks. As acute LY379268 treatment is known to increase cortical BDNF production, and BDNF is known to be beneficial for striatal neurons, we investigated if the benefit of daily LY379268 in R6/2 mice for striatal projection neurons was associated with increases in corticostriatal BDNF, with assessments done at 10 weeks of age after daily MTD treatment since the fourth week of life. We found that LY379268 increased BDNF expression in layer 5 neurons in motor cortex, which project to striatum, partly rescued a preferential loss of enkephalinergic striatal neurons, and enhanced substance P (SP) expression by SP striatal projection neurons. The enhanced survival of enkephalinergic striatal neurons was correlated with the cortical BDNF increase, but the enhanced SP expression by SP striatal neurons was not. Thus, LY379268 may protect the two main striatal projection neuron types by different mechanisms, enkephalinergic neurons by the trophic benefit of BDNF, and SP neurons by a mechanism not involving BDNF. The SP neuron benefit may perhaps instead involve the anti-excitotoxic action of mGluR2/3 receptor agonists.
Keywords: Huntington's Disease, Therapy, mGluR2/3, Striatum, BDNF
Introduction
We have found that daily subcutaneous injection with a maximum tolerated dose (MTD) of the mGluR2/3 agonist LY379268 (20mg/kg) beginning at 4 weeks dramatically improves the phenotype in R6/2 mice (Reiner et al., 2012). For example, this regimen of LY379268 administration prevents a 15–20% striatal neuron loss seen at 10 weeks in R6/2 mice. We also observed normalization of motor function at 10 weeks in R6/2 mice treated with daily LY379268 in such parameters as distance traveled, speed, the frequency of pauses, and the ability to locomote in a straight line. The basis of this benefit is uncertain, but the finding that acute LY379268 treatment increases cortical brain-derived neurotrophic factor (BDNF) production (Di Liberto et al., 2010) raises the possibility that the benefit is, at least in part, mediated by BDNF. Four lines of evidence add weight to this possibility.
First, pyramidal neurons of cerebral cortex synthesize BDNF (Altar et al., 1997, Altar and DiStefano, 1998; Cattaneo et al., 2001, 2005; Baquet et al., 2004), and those in layer 5 in particular transport it to striatum, where it is released from terminals and binds to TrkB receptors on striatal neurons (Ivkovic and Ehrlich, 1999; Mizuno et al., 1994; Weiss et al., 1986). Striatal projection neurons are the main targets of this input (Reiner et al., 2003b), and BDNF is a potent trophic and survival factor for these neurons (Mizuno et al., 1994; Widmer and Hefti, 1994; Nakao et al., 1995; Martinez-Serrano and Bjorklund, 1996; Alcantara et al., 1997; Ivkovic and Ehrlich, 1999; Aggerman et al., 2003; Grosse et al., 2005; Ventimiglia et al., 2005). Secondly, wild-type huntingtin promotes BDNF production by cortical neurons but polyglutamine-expanded mutant huntingtin promotes BDNF production less effectively, and accordingly cortical BDNF production is reduced in human Huntington's disease (HD) and in transgenic HD mice (Cattaneo et al., 2001, 2005; Zuccato et al., 2001, 2003, 2005, 2008; Zuccato and Cattaneo, 2007). Depletion of WT huntingtin in cortical neurons by sequestration in mutant protein aggregates and impaired transport of BDNF by cortical neuron axons further exacerbate the striatal and cortical BDNF insufficiency caused by the HD mutation (Gauthier et al., 2004; Cattaneo et al., 2005). Thirdly, studies in various mutant mice indicate that diminished cortical production of BDNF harms striatum (Gorski et al., 2003; Baquet et al., 2004; Canals et al., 2004; Saylor et al., 2006; Strand et al., 2007). Finally, intrastriatal BDNF delivery and selective forebrain overexpression of BDNF reverse cortical and striatal injury, and improve motor performance in transgenic HD mice (Canals et al., 2004; Gharami et al., 2008; Xie et al., 2010; Giralt et al., 2011).
Thus, BDNF deficiency has been implicated in HD pathogenesis, and BDNF overexpression in successful HD treatment. As LY379268 given acutely boosts cortical BDNF production, we investigated if the benefit of daily LY379268 in R6/2 mice was associated with increases in cortical BDNF, and if any BDNF boost was correlated with the improved survival and neurochemistry of either of the two main striatal projection neuron types. Our results show that the LY379268 benefit for ENK but not SP neurons seems linked to a BDNF enhancement.
Results
Behavior
The total distance traveled in a 30-min open field session in vehicle-treated R6/2 (R62V) mice at ten weeks of age was significantly less than in vehicle-treated WT mice (WTV) (57.5% of vehicle-treated WT) (Table 1). By contrast, distance traveled in the LY379268-treated R6/2 mice (R62LY) at ten weeks of age was not significantly different from that in WTV mice (101.3% of WTV), and was significantly more than in R62V mice. Similarly, R62V mice were significantly slower in open field than WTV mice at ten weeks of age (64.8% of vehicle-treated WT), and LY379268 rendered R6/2 speed at ten weeks of age significantly more than in R62V but not significantly different from that in WTV (93.8% of vehicle-treated WT) (Table 1). Thus, daily MTD LY379268 normalized R6/2 mice at 10 weeks on these two motor parameters, which are representative of the motor benefit seen in open field at this age with LY379268 treatment (Reiner et al., 2012).
Table 1.
Effect of LY379268 on activity and speed in open field in WT and R6/2 mice, compared to vehicle treatment, at 10 weeks of age. Numbers of animals per group, and repeat length for the R6/2 groups are shown. R6/2 mice receiving vehicle are significantly impaired in activity and speed, but daily LY379268 treatment normalized them.
| Groups | Distance Traveled in cm | Maximum Speed in cm/sec | Number of Animals | CAG Repeat Length |
|---|---|---|---|---|
| WT - Vehicle | 32026.2±1829.5 | 40.6±1.9 | 15 | |
| WT – LY379268 | 31110.4±1120.1 | 37.0±1.5 | 17 | |
| R62 - Vehicle | 18427.2*#±1920.8 | 26.3*#±2.6 | 12 | 124.7 |
| R62 – LY379268 | 32431.3±3431.2 | 38.1±4.0 | 15 | 124.5 |
significant vs WTV
significant vs R62LY
Quantitative PCR (qPCR)
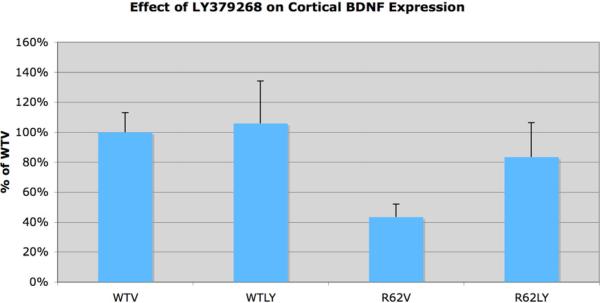
Our qPCR analysis (Fig. 1) showed that daily LY379268 treatment in WT mice (WTLY) had no significant effect (p=0.8345) on overall BDNF expression in the frontal pole of cerebral cortex (WTLY was 105.8% of WTV). BDNF expression in the frontal pole of cerebral cortex of R62V mice was, however, significantly reduced to 43.3% of that in WTV mice (p=0.0429). Daily LY379268 boosted cortical BDNF message in R6/2 mice to 83.4% of that in WTV mice, which was not significantly different than in vehicle-treated WT (p=0.5176). Thus, qPCR shows that the R6/2 mutation causes more than a 50% reduction in overall BDNF expression in frontal cortex at 10 weeks, which was restored to indistinguishable from normal WT levels by daily LY379268.
Figure 1.
Graph showing the effect of LY379268 on BDNF message in frontal cortex in WT and R6/2 mice, compared to vehicle treatment, at 10 weeks of age, as assessed by qPCR. R6/2 mice receiving vehicle have significantly decreased BDNF expression compared to WT mice, but daily LY379268 treatment nearly normalized R6/2 mice. Error bars show SEMs.
In situ Hybridization Histochemistry (ISHH) – Neuron Abundance
Blinded counts of labeled neurons (Fig. 2, Table 2) revealed significantly fewer BDNF neurons in layer 5 of M1 in R62V mice compared to WTV mice (80.6% of WTV) (p=0.0197). The abundance of BDNF neurons in layer 5 of M1 in R62LY mice was, however, not significantly different than in WTV mice (96.8% of WTV), and was significantly more than in R62V mice (p=0.0406). Similarly, relative BDNF message in layer 5 of M1 in R62V mice was also significantly less (p<0.0001) than in WTV mice (47.1% of WTV), and was elevated by LY379268 treatment to 78.9% of WTV mice (Table 2). The layer 5 BDNF message level in R6/2 after daily LY379268 was significantly greater than in R62V mice (p=0.0093), and no longer significantly different from WTV mice (p=0.0782). Thus, LY379268 treatment normalized layer 5 BDNF neuron abundance and boosted BDNF message in layer 5. Note that although our measurements were confined to layer 5 of M1, ISHH supported our qPCR findings that LY379268 had a beneficial effect for all cortical layers and regions (Fig. 2).
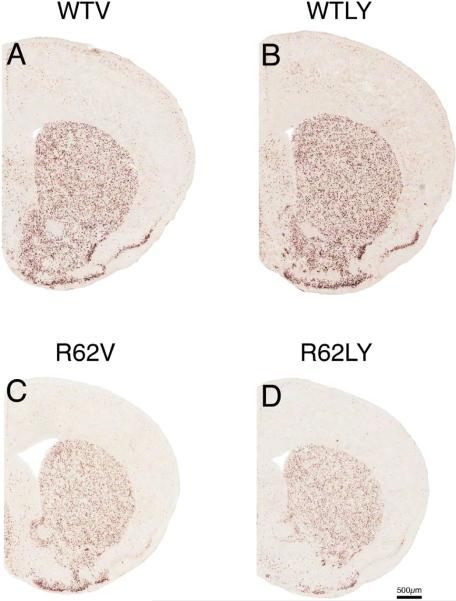
Figure 2.
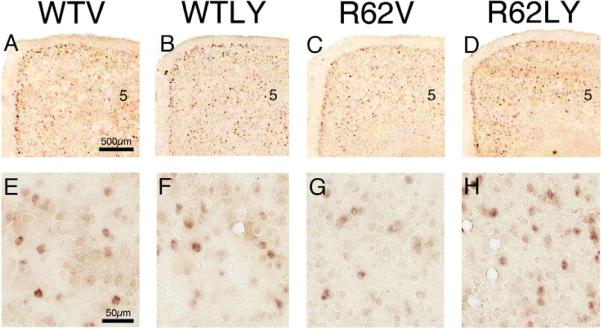
Images of right medial cerebral cortex at the level of M1, at low power (A–D) and high power (E–H), showing ISHH labeling for BDNF message, from 10-week old WT mice treated daily with either vehicle (WTV) or LY379268 (WTLY) compared to 10-week old R6/2 mice treated daily with either vehicle (R62V) or LY379268 (R62LY). Note that cortical BDNF message is somewhat reduced in the R6/2 mice, and LY379268 increases it in R6/2 mice. Images A–D are to the same scale, and images E–H are to the same scale.
Table 2.
Effect of LY379268 on cortical BDNF neuron abundance and message in layer 5 of M1, and on striatal SP and ENK neuron abundance in WT and R6/2 mice, at 10 weeks of age. Numbers of animals per group, and repeat length for the R6/2 groups are shown. R6/2 mice receiving vehicle (R62V) show significantly lower BDNF neuron abundance and message in layer 5 of M1, and significantly fewer ENK neurons in striatum than do WT mice receiving either vehicle (WTV) or LY379268 (WTLY). Daily LY379268 treatment normalized cortical BDNF neuron abundance in R6/2 mice (R62LY), and significantly increased cortical BDNF message and striatal ENK neuron abundance.
| BDNF Neurons in M1 Layer 5 | BDNF Message in M1 Layer 5 | ENK Neurons in Striatum | SP Neurons in Striatum | Number of Animals | CAG Repeat Length | |
|---|---|---|---|---|---|---|
| WTV | 100.0%±6.2 | 100.0%±11.5 | 100.0%±3.6 | 100.0%±3.4 | 11, 11, 12, 12 | |
| WTLY | 107.5%±5.7 | 110.5%±7.9 | 105.7%±4.6 | 90.9%*±3.1 | 13, 13, 13, 13 | |
| R62V | 80.6%*#±3.3 | 47.1%*#±4.7 | 71.2%*#±3.3 | 101.9%±3.7 | 11, 11, 12, 13 | 125.7 |
| R62LY | 96.8%±6.4 | 78.9%±8.9 | 83.1%±3.2* | 101.3%±2.9 | 13, 13, 13, 13 | 126.5 |
significant vs WTV
significant vs R62LY
The abundance of ENK neurons in striatum in R62V mice was also significantly less than in WTV mice (71.2% of WTV) (p<0.0001) (Fig. 3; Table 2). While the abundance of ENK neurons in striatum in LY379268-treated R6/2 mice was significantly more than in R62V mice (p=0.0240), it remained significantly less (83.1% of WTV) than in the WTV mice (p=0.0017). Thus, LY379268 treatment increased striatal ENK neuron abundance in R6/2 mice, but did not normalize it. No shortfall was seen in the abundance of SP striatal neurons in R62V mice, and LY379268 treatment did not alter their abundance in R6/2 mice (Fig. 4; Table 2). Notably, however, LY379268 reduced SP striatal neuron abundance in WT mice to 90.9% of that in WTV mice, and this reduction was significant (p=0.0496).
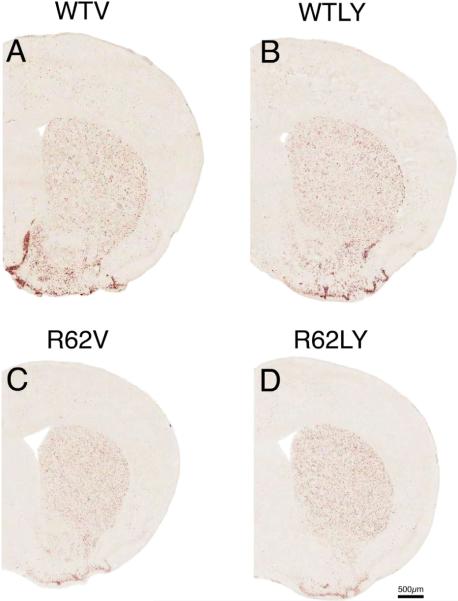
Figure 3.
Images of right half of telencephalon anterior to anterior commissure, showing ISHH labeling for ENK message, from 10-week old WT mice treated daily with either vehicle (WTV) or LY379268 (WTLY) compared to 10-week old R6/2 mice treated daily with either vehicle (R62V) or LY379268 (R62LY). Note that ENK message is clearly reduced in the R6/2 mice, and LY379268 does have not have a benefit that is evident in either WT or R6/2 mice. All images are to the same scale.
Figure 4.
Images of right half of telencephalon anterior to anterior commissure, showing ISHH labeling for SP message, from 10-week old WT mice treated daily with either vehicle (WTV) or LY379268 (WTLY) compared to 10-week old R6/2 mice treated daily with either vehicle (R62V) or LY379268 (R62LY). Note that SP message is slightly reduced in the R6/2 mice, and LY379268 appears to increase message so that it is similar to that in WTV mice. All images are to the same scale.
ISHH – Message per Neuron
In a prior study, we reported that relative striatal signal for ENK and SP message was significantly reduced in R62V mice, with the ENK signal more than halved and the SP signal reduced by about 25% (Reiner et al., 2012). We further found in that study that LY379268 treatment normalized the striatal SP message but not the striatal ENK message in R6/2 mice. In the present study, we report on the effect of the R6/2 mutation and daily LY379268 on ENK and SP message per striatal neuron in these same animals (Table 3). We found that ENK message per ENK neuron was significantly increased (p=0.0268) in WTLY mice (109.2% of vehicle-treated WT), and significantly reduced (p<0.0001) in vehicle-treated R6/2 mice (61.3% of vehicle-treated WT). Despite its beneficial effect on ENK neuron abundance, LY379268 slightly but significantly (p=0.0225) reduced ENK message per neuron below that seen in R62V mice to 50.4% of WTV. By contrast, for SP message per SP neuron, no difference was seen between vehicle-treated and LY37928-treated WT mice. SP message per neuron was, however, significantly reduced (p=0.0010) in R62V mice (77.3% of WTV), and daily LY379268 treatment normalized relative SP message per SP neuron in R6/2 mice so that it was no longer significantly different (p=0.4355) than WTV (94.3% of WTV).
Table 3.
Effect of LY379268 on cortical BDNF neuron abundance and message in layer 5 of M1, and on relative striatal SP and ENK message per neuron in WT and R6/2 mice, at 10 weeks of age. The BDNF data are shown in this table again to facilitate comparisons to the effects on SP and ENK message. R6/2 mice receiving vehicle (R62V) show significantly lower ENK and SP message per neuron in striatum than do WT mice receiving either vehicle (WTV) or LY379268 (WTLY). Daily LY379268 treatment in R6/2 mice (R62LY) normalized striatal SP message per neuron, but reduced ENK neuron message per neuron.
| BDNF Neurons in M1 Layer 5 | BDNF Message in M1 Layer 5 | ENK Message per ENK Neuron | SP Message per SP Neuron | Number of Animals | CAG Repeat Length | |
|---|---|---|---|---|---|---|
| WTV | 100.0%±6.2 | 100.0%±11.5 | 100.0%±2.7 | 100.0%±5.0 | 11, 11, 12, 12 | |
| WTLY | 107.5%±5.7 | 110.5%±7.9 | 109.2%*±3.5 | 100.5%±4.6 | 13, 13, 13, 13 | |
| R62V | 80.6%*#±3.3 | 47.1%*#±4.7 | 59.9%*#±3.5 | 74.6%*#±6.3 | 11, 11, 12, 13 | 125.7 |
| R62LY | 96.8%±6.4 | 78.9%±8.9 | 50.4%*±2.1 | 94.3%±5.2 | 13, 13, 13, 13 | 126.5 |
significant vs WTV
significant vs R62LY
ISHH – Correlation between Cortical BDNF and ENK Striatal Neuron Abundance
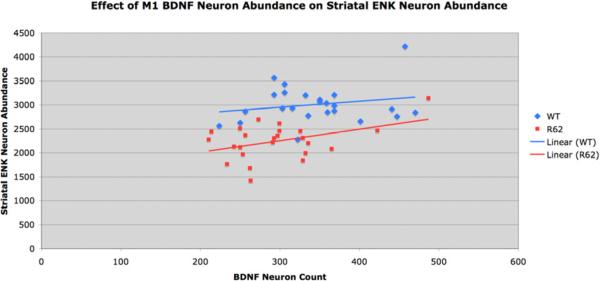
Since cortical neurons provide striatum with trophic support via BDNF, and LY379268 treatment boosted cortical BDNF and rescued striatal ENK neurons in R6/2 mice, we next sought to assess the relationship between cortical layer-5 BDNF neuron abundance and striatal ENK neuron abundance, to determine if variation in striatal ENK neuron abundance was related to variation in cortical BDNF in those same mice. The trends were similar for vehicle-treated and LY379268-treated mice, and thus we combined results per genotype (Fig. 5). For WT mice, we observed no significant correlation of ENK striatal neuron abundance (r=0.2089) with BDNF cortical neuron abundance (p=0.3272). By contrast, although the range of variation in cortical BDNF neuron abundance was similar for WT and R6/2 mice, ENK striatal neuron abundance was significantly correlated (r=0.4299) with cortical layer-5 BDNF neuron abundance in R6/2 mice (p=0.0309). Thus, while ENK striatal neuron abundance does not significantly depend on cortical BDNF neuron abundance in WT mice, such dependence seemingly develops with the mutation in R6/2 mice (Fig. 5). ANCOVA showed that although ENK neuron survival was BDNF-dependent in R6/2 but not WT mice, ENK neuron loss remained significantly greater in R6/2 than WT mice for any given BDNF neuron abundance. A similar effect was evident for layer 5 BDNF message. Thus, as shown by the ANCOVA, the BDNF boost by LY379268 could improve but not entirely normalize ENK neuron survival.
Figure 5.
Graph showing the relationship between M1 layer 5 BDNF neuron abundance and the abundance of striatal ENK neurons. For these graphs, all WT are pooled (i.e. vehicle-treated and LY379268-treated together) as a group, and all R6/2 are pooled (i.e. vehicle-treated and LY379268-treated together) as a group, because trends were similar irrespective of drug treatment. Note that ENK neuron abundance in R6/2 mice is significantly and positively correlated with cortical layer 5 BDNF neuron abundance (r=0.4299).
ISHH – Correlation between Cortical BDNF and Striatal Neuron Message
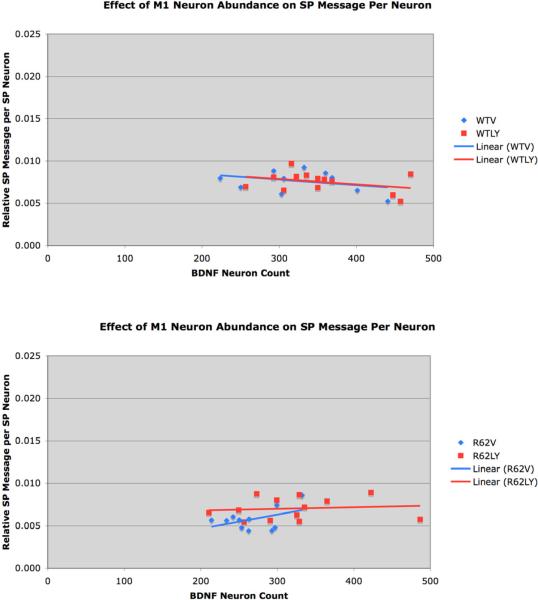
Because the graphed trends appeared to differ for vehicle versus LY379268 treatment in R6/2 mice, we present the correlations of cortical BDNF neuron abundance in M1 with striatal SP message per neuron separately for each group. The striatal SP signal per neuron showed no significant correlation with M1 layer 5 BDNF neuron abundance in WT mice (Fig. 6) and the regression lines were essentially identical for vehicle and LY379268 treated mice (r = −0.3383 for WTV; −0.3467 for WTLY) mice. The results for R6/2 mice were more complex (Fig. 6). R62V mice showed a suggestive but not significant trend toward a correlation (r=0.4503) between cortical BDNF and striatal SP message per neuron (p=0.1646). Although the mean striatal SP signal per neuron was rendered normal by the drug treatment in R62LY mice, the striatal SP signal showed no correlation with M1 layer 5 BDNF neuron abundance (r = 0.1137). Thus, the SP message deficit in R62V mice was not demonstrably attributable to the reduced cortical BDNF, and the LY379268 benefit for striatal neuron SP message in R6/2 mice was unambiguously not attributable to the boosting effect of the drug on cortical BDNF.
Figure 6.
Graphs showing the relationship between M1 layer 5 BDNF neuron abundance and striatal SP message per SP neuron in vehicle-treated (A) and LY379268-treated mice (B). For these graphs, vehicle-treated and LY379268-treated mice are presented separately, because R6/2 showed what might be different trends. Although SP message per neuron appeared possibly correlated with M1 layer 5 BDNF neuron abundance (r=0.4503) in vehicle-treated but not LY379268-treated R6/2 mice (B), this proved not significant. In WT mice with either treatment (A), SP message per neuron was uncorrelated with M1 layer 5 BDNF neuron abundance in vehicle-treated WT mice. Thus, SP message per neuron was independent of cortical BDNF in WT and R6/2 mice, and the benefit of daily LY379268 treatment for SP neuron message was not mediated by BDNF.
Discussion
This study has three major findings. First, our neuron counts indicate that the striatal neurons reported to be lost in symptomatic R6/2 mice in previous studies (Stack et al., 2005, 2007; Reiner et al., 2012) represent ENK neurons but not SP neurons at 10 weeks of age. Secondly, our results show that daily LY379268 treatment boosts BDNF message in layer 5 neurons of cortex in R6/2 mice, which is known to be the main source of the cortical input to the striatum (Reiner et al., 2003b, 2010). A recent study demonstrated that WT mice acutely treated with LY379268 show upregulation of BDNF in cortex and hippocampus (Di Liberto et al., 2010). Our results thus extend this observed BDNF boosting effect of LY379268 administration to chronic treatment in R6/2 HD mutant mice. Our results also suggested a possible slight increase in cortical BDNF expression in WT mice treated daily with MTD LY379268. Finally, our results suggest that LY379268 may yield its neuroprotective effect on striatal ENK neurons in R6/2 mice by its enhancement of the production of BDNF by cortical neurons and its transport to striatum. Consistent with this, our ELISA data indicate that BDNF protein is increased greatly in R6/2 striatum by daily LY379268 administration at an MTD (Reiner et al., 2011).
BDNF is produced by cortical neurons and transported axonally to their cortical and striatal projection targets (Altar et al., 1997; Zuccato et al., 2001; Gauthier et al., 2004; Zuccato and Cattaneo, 2007). BDNF promotes development, differentiation, plasticity and survival of neurons in cortex and striatum (Lessmann et al., 2003; Poo 2001; Zuccato and Cattaneo, 2007), and mutant huntingtin reduces cortical BDNF expression and protein transport in HD mice (Zuccato et al., 2001). Placing the R6/1 HD transgene on a hemizygous BDNF knock-out background exacerbates striatal ENK but not SP loss (Canals et al., 2004). Moreover, cortex-specific BDNF knock-out results in cortical and striatal pathology that mimics human HD (Gorski et al., 2003; Baquet et al., 2004; Strand et al., 2007), and embryonic deletion of the BDNF receptor (TrkB) from striatal neurons results in their profound loss, especially ENK neurons (Baydyuk et al., 2011). Conversely, behavioral performance is improved and disease progression slowed in transgenic HD mice overexpressing BDNF in telencephalon (Gharami et al., 2008; Xie et al., 2010; Arregui et al., 2011; Giralt et al., 2011). Similarly, daily intrastriatal administration of BDNF in R6/1 HD mice increases the number of striatal neurons expressing enkephalin and improves behavior (Canals et al., 2004). Thus, a deficit in BDNF may contribute to the pathogenesis of striatal injury in HD, especially for ENK neurons (Deng et al., 2004), and restoration of BDNF in HD mice is therapeutic.
We found that the benefit of LY379268 for ENK striatal neuron survival was correlated with cortical BDNF neuron numbers and message. Because of the evidence summarized above that ENK striatal neurons depend on cortical BDNF for survival, it seems likely that the correlations we observed in R6/2 mice between layer 5 cortical BDNF on one hand and ENK striatal projection neuron abundance on the other reflect a dependence of these striatal projection neurons on BDNF, rather than a shared deleterious effect of the mutation on both cortical BDNF expression and striatal ENK neurons. Note that the benefit of LY379268 for ENK neurons seemed to be specific for their survival and not their message level, since cortical BDNF elevation was not associated with an enhancement of ENK expression by ENK neurons. Daily LY379268 significantly increased SP striatal neuron message in R6/2 mice, but the increase was not correlated with cortical BDNF neuron abundance. Thus, the LY379268 benefit at the level of striatal projection neurons seemingly involves separate mechanisms for ENK and SP neurons – a cortical BDNF boost that enhanced the survival of ENK neurons and some other undetermined mechanism not involving BDNF that increased message level in SP neurons. Consistent with differing LY379268 mechanisms of action on striatal SP and ENK neurons, our results show daily treatment with this mGluR2/3 agonist significantly decreased SP neuron abundance in WT mice but significantly enhanced ENK message in ENK neurons in WT mice.
Two major possibilities exist as to the basis of the SP neuron message benefit in LY379268-treated R6/2 mice. First, SP neurons appear to be more vulnerable to excitotoxic injury to striatum than ENK neurons (Figueredo-Cardenas et al., 1994; Sun et al., 2003), and LY379268 may have opposed HD-mediated excitotoxic striatal injury. One way in which LY379268 may have done this is by acting on mGluR2/3 receptors on the terminals of cortical neurons to reduce glutamate release (Lovinger and McCool, 1995; Battaglia et al., 1997; Cozzi et al., 1997; Kingston et al., 1999; D'Onofrio et al., 2001). Consistent with an anti-excitotoxic mechanism of LY379268 benefit for striatal neurons, one prior study prevented corticostriatal glutamate release in R6/2 mice by ablating cortex, and thereby reversed striatal neuron shrinkage (Stack et al., 2007). Note that LY379268 may have also opposed excitotoxic striatal injury by acting directly on striatal projection neurons that possess mGluR3 receptors. Approximately half of striatal projection neurons express mGluR3 (but none GluR2) (Testa et al., 1994; Allen Brain Atlas), and it is possible this half is predominantly the SP type. Agonist binding to mGluR2/3 receptors, which are negatively coupled to adenylyl cyclase (Nakanishi, 1992; Schoepp et al., 1999), reduces Ca2+ currents via N, L-type Ca2+ channels and increases K+ currents via IRKC channels, and thereby decreases neuronal excitability (Davies et al., 1995; King and Liu, 1996; McCool et al., 1996; Colwell and Levine, 1999). Finally, striatal astrocytes express mGluR3 (Ohishi et al., 1993; Testa et al., 1994; Liu et al., 1998), and LY379268 increases striatal glial production of TGF-β (D'Onofrio et al., 2001). Agonists for mGluR3 counteract neuronal NMDA toxicity in vitro and in vivo, and this protection is mediated by astrocytic release of TGF-β1 (Bruno et al., 1997, 1998), and is absent in mGluR3 knock-out mice and in co-cultures of mGluR3−/− astrocytes and mGluR3+/+ neurons (Corti et al., 2007). Note, however, that TGF-β1 levels are reduced in the brains of human HD victims and in HD mice (R6/2 and YAC128), and acute administration of LY379268 does not elevate cortical or striatal TGF-β1 in R6/2 or YAC128 mice (Battaglia et al., 2011). It is uncertain thus if the beneficial effects of daily LY379268 could have involved boosting cortical and striatal TGF-β1 in our R6/2 mice.
Our results have relevance to the processes mediating striatal projection neuron death in HD. Despite the great variety of processes perturbed by the HD mutation and thus the seeming great variety of possible mechanisms mediating cell death (Reiner et al., 2003a; Zeron et al., 2002; Maglione et al., 2010), several authors have posited on theoretical grounds that a so-called one-hit cell death mechanism is responsible for striatal neuron death in HD (Clarke et al., 2000; Clarke et al., 2001; Clarke and Lumsden, 2005; Miller et al., 2010). In a genetic disease such as HD, the mutant protein is conceived as causing a homeostatic defect that renders the cells vulnerable to single fatal hits by some other stressor(s) that do not cause death in normal cells. Both differences in the fatal stressor and the extent of the mutation-engendered homeostatic defect between cell types can result in differences between cell types in disease vulnerability (Clarke and Lumsden, 2005). Our present evidence suggests that the HD mutation alters SP and ENK striatal neuron homeostasis so as to render ENK neurons much more dependent on BDNF than normal for their survival. Moreover, our results suggest, for two reasons, that the hit that kills ENK neurons in R6/2 striatum (and perhaps in HD itself) might be the single hit of a drop in corticostriatal BDNF below some critical level, while the hit that kills SP neurons must involve some other mechanism, such as excitotoxic injury. First, a drop in corticostriatal BDNF that kills ENK neurons is not adequate to kill SP neurons. Secondly, ENK neurons can be rescued by the boost in BDNF achieved by LY379268, but the rescue of SP message level by LY379268 is independent of its BDNF boost. This interpretation implies that the two cell types require separate drug treatment strategies, or a single strategy (such as mGluR2/3 agonist therapy) that remedies the differing effects of the HD mutation on them.
Experimental Procedure
R6/2 mice were maintained from founders obtained from JAX (Bar Harbor, ME). The repeat length in the mutant transgene had shortened during progressive breeding cycles at JAX from its original 150 CAG to about 120 CAG, unbeknown to JAX at the time we obtained our founders. Our colony was maintained by breeding R6/2 mice with CBA × C57BL/6 F1 (B6CBAF1) mice. Genotype and CAG repeat length were determined by PCR-based amplification using genomic DNA extracted from tail biopsies (Dragatsis et al., 2009). Genotype analysis was carried out by the Laragen Corporation (Culver City, CA). Two sets of mice (all including both males and females) were studied: 1) mutant and wild-type (WT) mice treated with vehicle or LY379268 for behavioral assessment of LY379268 efficacy (open field) (n=59); 2) mutant and WT mice treated with vehicle or LY379268 for qPCR assessment of BDNF expression in frontal cortex (n=24); and 3) mutant and WT mice treated with vehicle or LY379268 for in situ hybridization histochemical (ISHH) assessment of LY379268 efficacy (cortical BDNF, and striatal SP and ENK expression) (n=51). Details on animal numbers per treatment group in the behavioral and histological studies are provided in the Results, but typically about 15 mice were studied per group in the behavioral studies, 6 per group in the qPCR studies, and about 12 per group in the histological studies. Repeat length was about 125 CAG, and mean repeat length per R6/2 study group is also provided in the Results.
Behavioral Studies
Male and female mice were run in behavioral studies, until death in the case of R6/2 mice or the 18th week in the case of WT mice (by which time all R6/2 mice had died). Note that for concordance to the time point analyzed histologically, only behavioral data at 10 weeks is presented here. All mice were weighed daily, and maintained according to best practice standards for mouse care, which involve feed and environmental enrichment, and group housing (Carter et al., 2000; Hockly et al., 2003). Mutant mice were fed Purina Lab Diet 5001 (as a wet a mash) placed on the cage bottom as symptoms developed. An environmentally enriched cage contains a transparent Mouse Tunnel, (BioServ Product# K3323) and a Petite Gumabone (BioServ Product # K3214), and shredded paper. Behavioral data for open-field testing at the ten-week time point from 52 mice reported in Reiner et al. (2012) are presented here, as well as from 7 additional mice.
Open Field
We conducted weekly automated 30-minute assessment of open field behavior, using a Noldus EthoVision video tracking system (Noldus Information Technology, The Netherlands), and the SEE software of Drai and Golani (2001) to analyze the mouse motor behavior. The SEE software algorithms dichotomize mouse movements into lingering episodes and progression segments, and allow rapid characterization of about 30 endpoints related to locomotion, motivation, navigation, spatial memory and learning (Drai et al., 2000; Drai and Golani, 2001). Each animal was brought from its housing room, introduced into the arena and returned after the 30 min session. The arena was 200 cm in diameter with a non-porous gray floor and a 50 cm high gray wall, which provided a high-contrast background for video tracking of the mice. The arena was illuminated with two ceiling-mounted 40W neon bulbs. We report here on two open field parameters showing a clear R6/2 – WT difference, total distance and maximum speed, to evidence the behavioral benefit of LY37968 at the 10-week time point assessed in the qPCR and ISHH studies.
qPCR Studies
RNA extraction and reverse transcriptase polymerase chain reaction
Total RNA extraction from frontal cortex, and conversion to cDNA were performed as described previously (Jha et al., 2011). Twenty-five ng of cDNA was preamplified in a reaction mix with primers (final concentration, 200 nM) and PreAmp Master Mix (Applied Biosystems, Carlsbad, CA) following manufacture's instructions. The qPCR for BDNF expression was performed using the BioMark 96×96 real-time PCR system (Fluidigm, South San Francisco, CA) following the fabricant instructions. Briefly, for each individual assay, 5 μL of an assay mix that contains 2 μM forward primer, 2 μM reverse primer, and 1 μM UPL probe, and 1× assay reagent (Fluidigm, PN85000736) was loaded into one of the Assay Inlets of the M96 Dynamic Array. In the sample inlets, 5 μL of a sample mix that contains 2.5 μL of preamplified cDNA, 3.25 μL of 2× Universal TaqMan Master Mix (Applied Biosystems), and 0.32 μL of 20× Sample Loading Solution (Fluidigm, PN85000746). The cycling program used consisted of 2 min at 50 °C, 10 min at 95°C, followed by 40 cycles of 95 °C for 15 sec and 1 min at 60 °C. The CT values were obtained by using the BioMark Gene Expression Data Analysis after automatic inspection for quality. CTs higher than 27 and low-quality reactions were excluded and considered as not available. Relative gene expression values were determined by using the 2−ΔΔCT method of Livak and Schmittgen (2001). Six house keeping genes (HGPRT, cyclophilin D, TBP, β-actin, β-tublin, S19) were tested in a pre-assay, and HGPRT and Cyclophilin D were used as reference genes because they showed the minimum deviation among samples. Expression for all individual BDNF exons was determined, summed, and averaged between the two reference genes. Primer design and probes have been described previously (Jha et al., 2011).
Histological Studies
ISHH Methods
We analyzed tissue from vehicle-treated and MTD LY379268-treated 10-week old R6/2 and WT mice (treated daily since the fourth week of life) that had been fresh-frozen processed for ENK, SP and BDNF mRNA detection by ISSH, using previously described methods (Sun et al., 2002; Wang et al., 2006; Reiner et al., 2012). ISHH was performed on 20μm thick fresh frozen cryostat sections through the striatum. The sections were collected onto pre-cleaned Superfrost®/Plus microscope slides, dried on a slide warmer, and stored at −80°C until used for ISHH. To process sections for ISHH, the slides were removed from −80°C, quickly thawed and dried using a hair dryer. After fixation with 2% paraformaldehyde in saline sodium citrate (2× SSC) for 5 minutes, the sections were acetylated with 0.25% acetic anhydride/0.1M triethanolamine hydrochloride (pH 8.0) for 10 minutes, dehydrated through a graded ethanol series, and air-dried. Digoxigenin-UTP labeled cRNA probes (i.e. riboprobes) for preproenkephalin (PPE) and preprotachykinin (PPT), respectively, were transcribed from plasmids with PPE cDNA or PPT cDNA inserts (817bp and 900bp in size, respectively), generated by us using RT-PCR. Primers for PPE PCR were: Sense: 5'-TTCCTGAGGCTTTGCACC-3', and Antisense: 5'-TCACTGCTGGAAAAGGGC-3'. Primers for PPT PCR were: Sense: 5'-TCGAACATGAAAATCCTCGTGGCC-3', and Antisense: 5'-CACATCATACAATGACTGAAGACC-3'. Primers for BDNF PCR were: Sense: 5'-GGCGCCCATGAAAGAAGTAAAC-3', and Antisense: 5'-CGGCAACAAACCACAACATTAT-3'. The PPE riboprobe was directed against nucleotides 312–1128 (GenBank accession number NM_001002927), while the PPT riboprobe was directed against nucleotides 95–994 (GenBank accession number D17584). The BDNF riboprobe was directed against nucleotides 715–1634 (GenBank accession number MN_007540), which includes the protein coding region of BDNF and part of the adjacent 3-prime untranslated sequence. Note that all BDNF transcripts share this sequence, found within exon IX of the BDNF gene, and thus our probe detected all BDNF transcripts (Aid et al., 2007). The sections were incubated with digoxigenin (DIG)-labeled probe in hybridization buffer containing 50% formamide, 4× SSC, 1× Denhardt's solution, 200μg/ml denatured salmon sperm DNA, 250μg/ml yeast tRNA, 10% dextran sulfate, and 20mM dithiothreitol (DTT) at 63°C overnight. After hybridization, the slices were washed at 58°C consecutively in 4× SSC, 50% formamide with 4× SSC, 50% formamide with 2× SSC, and then 2× SSC, followed by treatment with RNase A (20μg/ml) for 30 min at 37°C. Finally, sections were washed at 55°C in 1×SSC, 0.5×SSC, 0.25×SSC, dehydrated through a graded ethanol series, and air-dried. Digoxigenin labeling was detected using anti-digoxigenin Fab fragments conjugated to alkaline phosphatase (AP), as visualized with nitroblue tetrazolium (NBT) histochemistry (Roche, Indianapolis, IN). Sections were coverslipped with a 1% gelatin-based aqueous solution.
Analysis
Open Field
The open field data were analyzed using SAS software and a mixed-model ANOVA, considering genotype, drug, and their interactions in the analysis. Fisher PLSD was used for individual group comparisons.
qPCR
The BDNF expression data from the qPCR were analyzed using SAS software and a mixed-model ANOVA, considering genotype, drug, and their interactions in the analysis. Fisher PLSD was used for individual group comparisons.
ISHH
Blinded neuron counts were performed bilaterally on captured images of cortex and striatum from one section from each animal. Cortical counts were performed for layer 5 of M1, while striatal counts were performed for dorsolateral and central striatum at a level rostral to globus pallidus externus (GPe), which represents a striatal territory to which motor cortex heavily projects (Reiner et al., 2003b, 2010). Neuron abundance reported here for BDNF represents the labeled neuron number per unit area (mm2) of layer 5 of M1. Relative BDNF message was calculated as the product of the area of layer 5 containing BDNF labeled neurons and their mean signal intensity (as measured by ImageJ). Due to striatal shrinkage in R6/2 mice (with compression of surviving neurons), striatal neuron abundance was calculated as the striatal area multiplied by the labeled striatal neuron count per unit area. Thus, the ENK and SP neuron abundance reflected abundance for the entire striatum in the sections analyzed. Densitometric analysis of striatal signal for PPT and PPE mRNA was carried out on the ISHH tissue from vehicle-treated and MTD LY379268-treated WT and R6/2 mice using ImageJ. For the SP and ENK signal, an image of a section from each mouse just anterior to the level of GPe was captured at 4800 dpi using an Epson scanner. Using ImageJ, the area and signal intensity in striatum were measured, and their product taken to reflect striatal message. The striatal message data are presented in Reiner et al., (2012). Striatal message for each animal was then divided by the neuron count to determine message per ENK and per SP neuron, as presented here. Both sides of the brain were analyzed in the case of neuron counts and striatal message per neuron type, and averaged. Note that the optical density measurements reflect relative message signal, and may not be linear with message abundance. Group results were analyzed using SAS software and a mixed-model ANOVA, considering genotype, drug, and their interactions in the analysis. Fisher PLSD was used for individual group comparisons. Linear regression modeling was used to assess the relationship between M1 BDNF neuron abundance and striatal ENK neuron abundance, and between M1 BDNF neuron abundance and SP message per striatal neuron. BDNF neuron abundance and BDNF message were highly correlated, so we only present correlations for BDNF neuron number. Analysis of covariance (ANCOVA) was used to compare fitted regression lines between groups since fitted lines were of similar slope.
Highlights
The striatal neurons lost in R6/2 mice represent ENK neurons but not SP neurons
Daily LY379268 treatment boosts BDNF in cortical layer 5 neurons in R6/2 mice
LY379268 may protect striatal ENK neurons by enhancing BDNF delivery to striatum
LY379268 may protect striatal SP neurons by a presynaptic anti-excitotoxic action
Daily LY379268 protects striatum and delays the R6/2 behavioral phenotype
Acknowledgments
We thank Aminah Henderson, Marion Joni, and Ting Wong for histological assistance, and Michael Piantedosi and Trevon Clark for assistance with behavioral studies and mouse colony maintenance. Supported by the CHDIF (AR), and NIH NS28721 (AR). The authors have no financial interest in the research reported here.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aggerman K, Ernfors P. Differential influence of BDNF and NT3 on the expression of calcium binding proteins and neuropeptide Y in vivo. NeuroReport. 2003;14:2183–2187. doi: 10.1097/00001756-200312020-00010. [DOI] [PubMed] [Google Scholar]
- Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J. Neurosci. Res. 2007;85:525–535. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcantara S, Frisen J, del Rio JA, Soriano E, Barbacid M, Silos-Santiago I. TrkB signaling is required for postnatal survival of CNS neurons and protects hippocampal and motor neurons from axotomy-induced cell death. J. Neurosci. 1997;17:3623–3633. doi: 10.1523/JNEUROSCI.17-10-03623.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen Brain Atlas http, //mouse.brain-map.org/welcome.do.
- Altar CA, Ning CA, Bliven T, Juhasz M, Conner M, Acheson AL, Lindsay RM, Wiegand SJ. Anterograde transport of brain-derived neurotrophic factor and its role in the brain. Nature. 1997;389:856–860. doi: 10.1038/39885. [DOI] [PubMed] [Google Scholar]
- Altar CA, DiStefano PS. Neurotrophin trafficking by anterograde transport. Trends Neurosci. 1998;21:433–437. doi: 10.1016/s0166-2236(98)01273-9. [DOI] [PubMed] [Google Scholar]
- Arregui L, Benítez JA, Razgado LF, Vergara P, Segovia J. Adenoviral astrocyte-specific expression of BDNF in the striata of mice transgenic for Huntington's disease delays the onset of the motor phenotype. Cell. Mol. Neurobiol. 2011;31:1229–1243. doi: 10.1007/s10571-011-9725-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquet ZC, Gorski JA, Jones KR. Early striatal dendrite deficits followed by neuron loss with advanced age in the absence of anterograde cortical brain-derived neurotrophic factor. J. Neurosci. 2004;24:4250–4258. doi: 10.1523/JNEUROSCI.3920-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia G, Monn JA, Schoepp DD. In vivo inhibition of veratridine-evoked release of striatal excitatory amino acids by the group II metabotropic glutamate receptor agonist LY354740. Neurosci. Lett. 1997;229:161–164. doi: 10.1016/s0304-3940(97)00442-4. [DOI] [PubMed] [Google Scholar]
- Battaglia G, Cannella M, Riozzi B, Orobello S, Maat-Schieman ML, Aronica E, Busceti CL, Ciarmiello A, Alberti S, Amico E, Sassone J, Sipione S, Bruno V, Frati L, Nicoletti F, Squitieri F. Early defect of transforming growth factor β1 formation in Huntington's disease. J. Cell. Mol. Med. 2011;15:555–571. doi: 10.1111/j.1582-4934.2010.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baydyuk M, Russell T, Liao GY, Zang K, An JJ, Reichardt LF, Xu B. TrkB receptor controls striatal formation by regulating the number of newborn striatal neurons. Proc Natl Acad Sci U.S.A. 2011;108:1669–1674. doi: 10.1073/pnas.1004744108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno V, Battaglia G, Casabona G, Copani A, Caciagli F, Nicoletti F. Neuroprotection by glial metabotropic glutamate receptors is mediated by transforming growth factor-beta. J. Neurosci. 1998;18:9594–9600. doi: 10.1523/JNEUROSCI.18-23-09594.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno V, Sureda FX, Storto M, Casabona G, Caruso A, Knopfel T, Kuhn R, Nicoletti F. The neuroprotective activity of group-II metabotropic glutamate receptors requires new protein synthesis and involves a glial-neuronal signaling. J. Neurosci. 1997;17:1891–1897. doi: 10.1523/JNEUROSCI.17-06-01891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canals JM, Pineda JR, Torres-Peraza JF, Bosch M, Martin-Ibanez R, Munoz MT, Mengod G, Ernfors P, Alberch J. Brain-derived neurotrophic factor regulates the onset and severity of motor dysfunction associated with enkephalinergic neuronal degeneration in Huntington's disease. J. Neurosci. 2004;24:7727–7739. doi: 10.1523/JNEUROSCI.1197-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter RJ, Hunt MJ, Morton AJ. Environmental stimulation increases survival in mice transgenic for exon 1 of the Huntington's disease gene. Mov. Disord. 2000;15:925–937. doi: 10.1002/1531-8257(200009)15:5<925::aid-mds1025>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Cattaneo E, Rigamonti D, Goffredo D, Zuccato C, Squitieri F, Sipione S. Loss of normal huntingtin function: new developments in Huntington's disease research. Trends Neurosci. 2001;24:182–188. doi: 10.1016/s0166-2236(00)01721-5. [DOI] [PubMed] [Google Scholar]
- Cattaneo E, Zuccato C, Tartari M. Normal Huntingtin function: An alternative approach to Huntington's disease. Nat. Rev. Neurosci. 2005;6:919–930. doi: 10.1038/nrn1806. [DOI] [PubMed] [Google Scholar]
- Clarke G, Collins RA, Leavitt BR, Andrews DF, Hayden MR, Lumsden CJ, McInnes RR. A one-hit model of cell death in inherited neuronal degenerations. Nature. 2000;406:195–199. doi: 10.1038/35018098. [DOI] [PubMed] [Google Scholar]
- Clarke G, Lumsden CJ, McInnes RR. Inherited neurodegenerative diseases: the one-hit model of neurodegeneration. Hum. Mol. Genet. 2001;10:2269–2275. doi: 10.1093/hmg/10.20.2269. [DOI] [PubMed] [Google Scholar]
- Clarke G, Lumsden CJ. Heterogeneous cellular environments modulate one-hit neuronal death kinetics. Brain Res. Bull. 2005;65:59–67. doi: 10.1016/j.brainresbull.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Colwell CS, Levine MS. Metabotropic glutamate receptor modulation of excitotoxicity in the neostriatum: role of calcium channels. Brain Res. 1999;833:234–241. doi: 10.1016/s0006-8993(99)01545-0. [DOI] [PubMed] [Google Scholar]
- Corti C, Battaglia G, Molinaro G, Riozzi B, Pittaluga A, Corsi M, Mugnaini M, Nicoletti F, Bruno V. The use of knock-out mice unravels distinct roles for mGlu2 and mGlu3 metabotropic glutamate receptors in mechanisms of neurodegeneration/neuroprotection. J. Neurosci. 2007;27:8297–8308. doi: 10.1523/JNEUROSCI.1889-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzi A, Attucci S, Peruginelli F, Marinozzi M, Luneia R, Pellicciari R, Moroni F. Type 2 metabotropic glutamate (mGlu) receptors tonically inhibit transmitter release in rat caudate nucleus: in vivo studies with (2S,1'S,2'S,3'R)-2-(2'-carboxy-3'-phenylcyclopropyl)glycine, a new potent and selective antagonist. Eur. J. Neurosci. 1997;9:1350–1355. doi: 10.1111/j.1460-9568.1997.tb01489.x. [DOI] [PubMed] [Google Scholar]
- Davies CH, Clarke VR, Jane DE, Collingridge GL. Pharmacology of postsynaptic metabotropic glutamate receptors in rat hippocampal CA1 pyramidal neurones. Br. J. Pharmacol. 1995;116:1859–1869. doi: 10.1111/j.1476-5381.1995.tb16674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng YP, Penney JB, Young AB, Albin RL, Anderson KD, Reiner A. Differential loss of striatal projection neurons in Huntington's disease: A quantitative immunohistochemical study. J. Chem. Neuroanat. 2004;27:143–164. doi: 10.1016/j.jchemneu.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Di Liberto V, Bonomo A, Frinchi M, Belluardo N, Mudo G. Group II metabotropic glutamate receptor activation by agonist LY379268 treatment increases the expression of brain derived neurotrophic factor in the mouse brain. Neuroscience. 2010;165:863–73. doi: 10.1016/j.neuroscience.2009.11.012. [DOI] [PubMed] [Google Scholar]
- D'Onofrio M, Cuomo L, Battaglia G, Ngomba RT, Storto M, Kingston AE, Orzi F, De Blasi A, Di Iorio P, Nicoletti F, Bruno V. Neuroprotection mediated by glial group-II metabotropic glutamate receptors requires the activation of the MAP kinase and the phosphatidylinositol-3-kinase pathways. J. Neurochem. 2001;78:435–445. doi: 10.1046/j.1471-4159.2001.00435.x. [DOI] [PubMed] [Google Scholar]
- Dragatsis I, Goldowitz D, Del Mar N, Deng YP, Meade CA, Liu L, Sun Z, Dietrich P, Yue J, Reiner A. CAG repeat lengths ≥335 attenuate the phenotype in the R6/2 Huntington's disease transgenic mouse. Neurobiol. Dis. 2009;33:315–330. doi: 10.1016/j.nbd.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drai D, Golani I. SEE: a tool for the visualization and analysis of rodent exploratory behavior. Neurosci. Biobehav. Rev. 2001;25:409–426. doi: 10.1016/s0149-7634(01)00022-7. [DOI] [PubMed] [Google Scholar]
- Drai D, Benjamini Y, Golani I. Statistical discrimination of natural modes of motion in rat exploratory behavior. J. Neurosci. Methods. 2000;96:119–131. doi: 10.1016/s0165-0270(99)00194-6. [DOI] [PubMed] [Google Scholar]
- Figueredo-Cardenas G, Anderson KD, Chen Q, Veenman CL, Reiner A. Relative survival of striatal projection neurons and interneurons after intrastriatal injection of quinolinic acid in rats. Exper. Neurol. 1994;129:37–56. doi: 10.1006/exnr.1994.1145. [DOI] [PubMed] [Google Scholar]
- Gauthier LR, Charrin B, Borrell-Pages M, Dompierre J, Rangone H, Cordelieres F, De Mey J, MacDonald M, Lessmann HS. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Gharami K, Xie Y, An JJ, Tonegawa S, Xu B. BDNF overexpression in the forebrain ameliorates huntington's disease phenotypes in mice. J. Neurochem. 2008;105:369–379. doi: 10.1111/j.1471-4159.2007.05137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giralt A, Carretón O, Lao-Peregrin C, Martín ED, Alberch J. Conditional BDNF release under pathological conditions improves Huntington's disease pathology by delaying neuronal dysfunction. Molec. Neurodegen. 2011;6:71–86. doi: 10.1186/1750-1326-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski JA, Zeiler SR, Tamowski S, Jones KR. Brain-derived neurotrophic factor is required for the maintenance of cortical dendrites. J. Neurosci. 2003;23:6856–6865. doi: 10.1523/JNEUROSCI.23-17-06856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse G, Djalali S, Deng DR, Holtje M, Hinz B, Schwartzkopff K, Cygon M, Rothe T, Stroh T, Hellweg R, Ahnert-Hilger G, Hortnagl H. Area-specific effects of brain-derived neurotrophic factor (BDNF) genetic ablation on various neuronal subtypes of the mouse brain. Dev. Brain Res. 2005;156:111–126. doi: 10.1016/j.devbrainres.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Hockly E, Woodman B, Mahal A, Lewis CM, Bates G. Standardization and statistical approaches to therapeutic trials in the R6/2 mouse. Brain Res. Bull. 2003;61:469–479. doi: 10.1016/s0361-9230(03)00185-0. [DOI] [PubMed] [Google Scholar]
- Ivkovic S, Ehrlich ME. Expression of the striatal DARPP-32/ARPP-21 phenotype in GABAergic neurons requires neurotrophins in vivo and in vitro. J. Neurosci. 1999;19:5409–5419. doi: 10.1523/JNEUROSCI.19-13-05409.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S, Dong B, Sakata K. Enriched environment treatment reverses depression-like behavior and restores reduced hippocampal neurogenesis and protein levels of brain-derived neurotrophic factor in mice lacking its expression through promoter IV. Transl. Psychiatry. 2011;1:e40. doi: 10.1038/tp.2011.33. doi:10.1038/tp.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AE, Liu XH. Dual action of metabotropic glutamate receptor agonists on neuronal excitability and synaptic transmission in spinal ventral horn neurons in vitro. Neuropharmacol. 1996;35:1673–1680. doi: 10.1016/s0028-3908(96)00140-2. [DOI] [PubMed] [Google Scholar]
- Kingston AE, O'Neill MJ, Lam A, Bales KR, Monn JA, Schoepp DD. Neuroprotection by metabotropic glutamate receptor agonists: LY2354740, LY379268 and LY389795. Eur. J. Pharmacol. 1999;377:155–165. doi: 10.1016/s0014-2999(99)00397-0. [DOI] [PubMed] [Google Scholar]
- Lessmann V, Gottmann K, Malcangio M. Neurotrophin secretion: current facts and future prospects. Prog. Neurobiol. 2003;69:341–374. doi: 10.1016/s0301-0082(03)00019-4. [DOI] [PubMed] [Google Scholar]
- Liu XB, Munoz A, Jones EG. Changes in subcellular localization of metabotropic glutamate receptor subtypes during postnatal development of mouse thalamus. J. Comp. Neurol. 1998;395:450–465. doi: 10.1002/(sici)1096-9861(19980615)395:4<450::aid-cne3>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;4:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lovinger DM, McCool BA. Metabotropic glutamate receptor-mediated presynaptic depression at corticostriatal synapses involves mGLuR2 or 3. J. Neurophys. 1995;73:1076–1083. doi: 10.1152/jn.1995.73.3.1076. [DOI] [PubMed] [Google Scholar]
- Maglione V, Marchi P, Di Pardo A, Lingrell S, Horkey M, Tidmarsh E, Sipione S. Impaired ganglioside metabolism in Huntington's Disease and neuroprotective role of GM1. J. Neurosci. 2010;30:4072–4080. doi: 10.1523/JNEUROSCI.6348-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Serrano A, Bjorklund A. Protection of the neostriatum against excitotoxic damage by neurotrophin-producing, genetically modified neural stem cells. J. Neurosci. 1996;16:4604–4616. doi: 10.1523/JNEUROSCI.16-15-04604.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCool BA, Pin JP, Brust PF, Harpold MM, Lovinger DM. Functional coupling of rat group II metabotropic glutamate receptors to an omega-conotoxin GVIA-sensitive calcium channel in human embryonic kidney 293 cells. Mol. Pharmacol. 1996;50:912–922. [PubMed] [Google Scholar]
- Miller J, Arrasate M, Shaby BA, Mitra S, Masliah E, Finkbeiner S. Quantitative relationships between huntingtin levels, polyglutamine length, inclusion body formation, and neuronal death provide novel insight into huntington's disease molecular pathogenesis. J. Neurosci. 2010;30:10541–10550. doi: 10.1523/JNEUROSCI.0146-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K, Carnahan J, Nawa H. Brain-derived neurotrophic factor promotes differentiation of striatal GABAergic neurons. Dev. Biol. 1994;165:243–256. doi: 10.1006/dbio.1994.1250. [DOI] [PubMed] [Google Scholar]
- Nakanishi S. Molecular diversity of glutamate receptors and implications for brain function. Science. 1992;258:597–603. doi: 10.1126/science.1329206. [DOI] [PubMed] [Google Scholar]
- Nakao N, Brundin P, Funa K, Lindvall O, Odin P. Trophic and protective actions of brain-derived neurotrophic factor on striatal DARPP-32-containing neurons in vitro. Dev. Brain Res. 1995;90:92–101. doi: 10.1016/0165-3806(96)83489-4. [DOI] [PubMed] [Google Scholar]
- Ohishi H, Shigemoto R, Nakanishi S, Mizuno N. Distribution of the mRNA for a metabotropic glutamate receptor (mGluR3) in the rat brain: an in situ hybridization study. J. Comp. Neurol. 1993;335:252–266. doi: 10.1002/cne.903350209. [DOI] [PubMed] [Google Scholar]
- Poo MM. Neurotrophins as synaptic modulators. Nat. Rev. Neurosci. 2001;2:24–32. doi: 10.1038/35049004. [DOI] [PubMed] [Google Scholar]
- Reiner A, Dragatsis I, Zeitlin SO, Goldowitz D. Wild-type huntingtin plays a role in brain development and neuronal survival. Mol. Neurobiol. 2003a;28:259–275. doi: 10.1385/MN:28:3:259. [DOI] [PubMed] [Google Scholar]
- Reiner A, Jiao Y, Del Mar N, Laverghetta AV, Lei WL. Differential morphology of pyramidal-tract type and intratelencephalically-projecting type corticostriatal neurons and their intrastriatal terminals in rats. J. Comp. Neurol. 2003b;457:420–440. doi: 10.1002/cne.10541. [DOI] [PubMed] [Google Scholar]
- Reiner A, Hart NM, Lei WL, Deng YP. Corticostriatal projection neurons – Dichotomous types and dichotomous functions. Frontiers in Neuroanatomy. 2010;4 doi: 10.3389/fnana.2010.00142. article 142; doi: 10.3389/fnana.2010.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A, Deng YP, Wang HB, Lafferty DC, Del Mar N, Sakata K, Wang B, Liao F. Striatal neuroprotection in R6/2 mice by the group 2 metabotropic glutamate receptor agonist LY379268 may be mediated by the BDNF-Akt pathway. Soc. Neurosci. Abst. 2011 #557.15. [Google Scholar]
- Reiner A, Lafferty DC, Wang HB, Del Mar N, Deng YP. The group 2 metabotropic glutamate receptor agonist LY379268 rescues neuronal, neurochemical and motor abnormalities in R6/2 Huntington's disease mice. Neurobiol. Dis. 2012;47:75–91. doi: 10.1016/j.nbd.2012.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saylor AJ, Meredith GE, Vercillo MS, Zahm DS, McGinty JF. BDNF heterozygous mice demonstrate age-related changes in striatal and nigral gene expression. Exp. Neur. 2006;199:362–372. doi: 10.1016/j.expneurol.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Schoepp DD, Jane DE, Monn JA. Pharmacological agents acting at subtypes of metabotropic glutamate receptors. Neuropharmacol. 1999;38:1431–1476. doi: 10.1016/s0028-3908(99)00092-1. [DOI] [PubMed] [Google Scholar]
- Stack EC, Kubilus JK, Smith K, Cormier K, Del Signore SJ, Guelin E, Ryu H, Hersch SM, Ferrante RJ. Chronology of behavioral symptoms and neuropathological sequela in R6/2 Huntington's disease transgenic mice. J. Comp. Neurol. 2005;490:354–370. doi: 10.1002/cne.20680. [DOI] [PubMed] [Google Scholar]
- Stack EC, Dedeoglu A, Smith KM, Cormier K, Kubilus JK, Bogdanov M, Matson WR, Yang L, Jenkins BG, Luthi-Carter R, Kowall NW, Hersch SM, Beal MF, Ferrante RJ. Neuroprotective effects of synaptic modulation in Huntington's disease R6/2 mice. J. Neurosci. 2007;27:12908–12915. doi: 10.1523/JNEUROSCI.4318-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand AD, Baquet ZC, Aragaki AK, Holmans P, Yang L, Cleren C, Beal MF, Jones L, Kooperberg C, Olson JM, Jones KR. Expression profiling of Huntington's disease models suggests that brain-derived neurotrophic factor depletion plays a major role in striatal degeneration. J. Neurosci. 2007;27:11758–11768. doi: 10.1523/JNEUROSCI.2461-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Del Mar N, Meade C, Goldowitz D, Reiner A. Differential changes in striatal projection neurons in R6/2 mice transgenic for Huntington's disease. Neurobiol. Dis. 2002;11:369–385. doi: 10.1006/nbdi.2002.0554. [DOI] [PubMed] [Google Scholar]
- Sun Z, Chen Q, Reiner A. Enkephalinergic but not substance P-containing striatal projection neurons lose vulnerability to quinolinic acid with age in rats. Exp. Neurol. 2003;184:1034–1042. doi: 10.1016/j.expneurol.2003.08.016. [DOI] [PubMed] [Google Scholar]
- Testa CM, Standaert DG, Young AB, Penney JB., Jr. Metabotropic glutamate receptor mRNA expression in the basal ganglia of the rat. J. Neurosci. 1994;14:3005–3008. doi: 10.1523/JNEUROSCI.14-05-03005.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventimiglia R, Mather PE, Jones BE, Lindsay RM. The neurotrophins BDNF, NT-3 and NT-4/5 promote survival and morphological and biochemical differentiation of striatal neurons in vitro. Eur. J. Neurosci. 1995;7:213–222. doi: 10.1111/j.1460-9568.1995.tb01057.x. [DOI] [PubMed] [Google Scholar]
- Wang HB, Laverghetta AV, Foehring RF, Deng YP, Sun Z, Yamamoto K, Lei WL, Jiao Y, Reiner A. Single-Cell RT-PCR, in situ hybridization histochemical, and immunohistochemical studies of substance P and enkephalin co-occurrence in striatal projection neurons in rats. J. Chem. Neur. 2006;31:178–199. doi: 10.1016/j.jchemneu.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Weiss S, Pin JP, Sebben M, Kemp DE, Sladeczek F, Gabrion J, Bockaert J. Synatogenesis of cultured striatal neurons in serum-free medium: A morphological and biochemical study. Proc. Natl. Acad. Sci. U.S.A. 1986;83:2238–2243. doi: 10.1073/pnas.83.7.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmer HR, Hefti F. Neurotrophin-4/5 promotes survival and differentiation of rat striatal neurons developing in culture. Eur. J. Neurosci. 1994;6:1669–1679. doi: 10.1111/j.1460-9568.1994.tb00559.x. [DOI] [PubMed] [Google Scholar]
- Xie Y, Hayden MR, Xu B. BDNF Overexpression in the forebrain rescues Huntington's disease phenotypes in YAC128 mice. J. Neurosci. 2010;30:14708–14718. doi: 10.1523/JNEUROSCI.1637-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeron MM, Hansson O, Chen N, Wellington CL, Leavitt BR, Brundin P, Hayden MR, Raymond LA. Increased sensitivity to N-methyl-D-aspartate receptor-mediated excitotoxicity in a mouse model of Huntington's disease. Neuron. 2002;33:849–860. doi: 10.1016/s0896-6273(02)00615-3. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L, MacDonald ME, Friedlander RM, Silani V, Hayden MR, Timmusk T, Sipione S, Cattaneo E. Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Tartari M, Crotti A, Goffredo D, Valenza M, Conti L, Cataudella T, Leavitt BR, Hayden MR, Timmusk T, Rigamonti D, Cattaneo E. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat. Gen. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Liber D, Ramos C, Tarditi A, Rigamonti D, Tartari M, Valenza M, Cattaneo E. Progressive loss of BDNF in a mouse model of Huntington's disease and rescue by BDNF delivery. Pharmacol. Res. 2005;52:133–139. doi: 10.1016/j.phrs.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Marullo M, Conforti P, McDonald ME, Tartari M, Cattaneo E. Systematic assessment of BDNF and its receptor levels in human cortices affected by Huntington's disease. Brain Path. 2008;18:225–238. doi: 10.1111/j.1750-3639.2007.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C, Cattaneo E. Role of brain-derived neurotrophic factor in Huntington's disease. Prog. Neurobiol. 2007;81:294–330. doi: 10.1016/j.pneurobio.2007.01.003. [DOI] [PubMed] [Google Scholar]