Abstract
Retroviruses are the original source of oncogenes. The discovery and characterization of these genes were made possible by the introduction of quantitative cell biological and molecular techniques for the study of tumor viruses. Key features of all retroviral oncogenes were first identified in src, the oncogene of Rous sarcoma virus. These include non-involvement in viral replication, coding for a single protein, and cellular origin. The myc, ras and erbB oncogenes quickly followed src, and these together with pi3k are now recognized as critical driving forces in human cancer.
Introduction
Most oncogenes that play predominant roles in human cancer were first recognized in retroviruses. This includes the receptor tyrosine kinase epidermal growth factor receptor (EGFR), the small GTPase Ras, the lipid kinase PI3K and the transcriptional regulator Myc. The discovery of retroviral oncogenes during the last four decades has set in motion an era of progress that culminated in our current view of cancer as a genetic disease (Timeline). This view guides and inspires efforts of therapeutic innovation. At this time it appears attractive to look back to the origins of the oncogene field, because they illustrate first principles that are still valid and applicable to the legions of oncogenes encountered today.
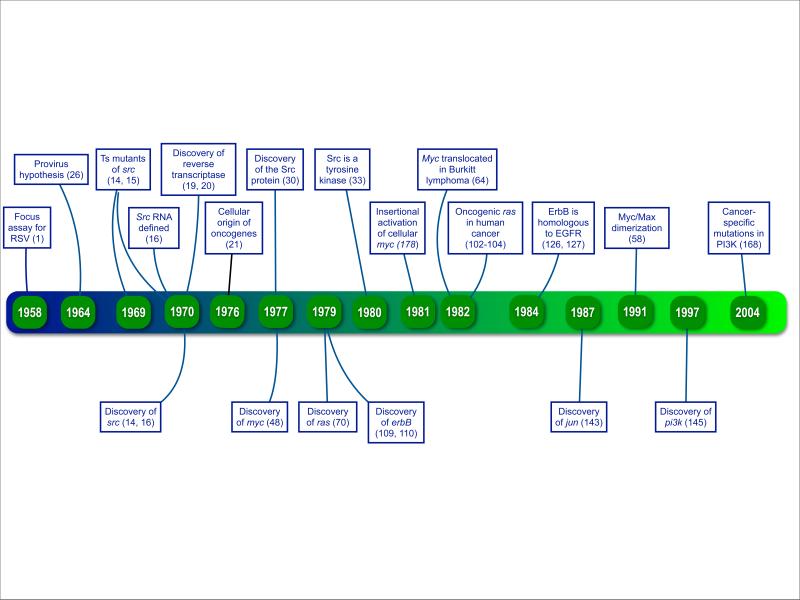
Timeline. Retroviral oncogenes: 50 years of discovery.
The top banners refer to transforming discoveries that have shaped the development of the field. The bottom banners mark the years in which the important oncogenes highlighted in this article were identified.
There are slightly over 30 retroviral oncogenes, identified almost exclusively in avian and rodent viruses. Their products can be grouped into eight functional classes (Table 1). The unifying functional assignment of these genes and proteins is signaling in the control of cellular replication. From this list, I will discuss a few oncogenes that best illustrate the history of experimental and theoretical breakthroughs but also play critical roles in human disease.
Table 1.
Functional classes of retroviral oncoproteins
| Functional class | Representative example | Source virus |
|---|---|---|
| Growth factor | Sis, platelet-derived growth factor | Simian sarcoma virus |
| Receptor tyrosine kinase | ErbB, epidermal growth factor receptor | Avian erythroblastosis virus |
| Hormone receptor | ErbA, thyroid hormone receptor | Avian erythroblastosis virus |
| G-protein | H-Ras, GTPase K-Ras, GTPase |
Harvey sarcoma virus Kirsten sarcoma virus |
| Adaptor protein | Crk, modular signaling link | CT10 avian sarcoma virus |
| Non-receptor tyrosine kinase | Src, signaling protein kinase Abl, signaling protein kinase |
Rous sarcoma virus Abelson murine leukemia virus |
| Serine-threonine kinase | Akt, signaling protein kinase Mos, signaling protein kinase |
Akt8 murine thymoma virus Moloney murine sarcoma virus |
| Transcriptional regulator | Jun, component of the AP-1 (activator protein 1) complex Fos, component of the AP-1 (activator protein 1) complex Myc, transcription factor |
Avian sarcoma virus 17 Finkel-Biskis-Jinkins murine sarcoma virus Avian myelocytomatosis virus MC29 |
| Lipid kinase | Pi3k, phosphatidylinositol 3-kinase | Avian sarcoma virus 16 |
The src paradigm
Src was the first retroviral oncogene discovered. This was no accident. Preparations of Rous sarcoma virus (RSV), the avian sarcoma virus carrying the src gene, induce readily visible oncogenic transformation within a few days in primary fibroblasts. RSV can be accurately titrated in cell culture with a focus assay developed in 19581. In this assay, the focus number is directly proportional to the amount of virus, hence a single RSV particle can fully transform a host cell, and no cooperation between complementing viruses is required. Soon methods for the biological cloning of RSV particles were developed, the fruit of extensive studies devoted to a replication-defective variant of RSV2, 3. A procedure for assaying non-oncogenic but actively replicating avian retroviruses by interference with RSV focus formation had also been devised4. In the 1960ies, these were powerful quantitative cell biological tools, and the avian sarcoma viruses were the only retroviruses for which such tools were available. This technological advantage was decisive in the discovery of the first oncogene.
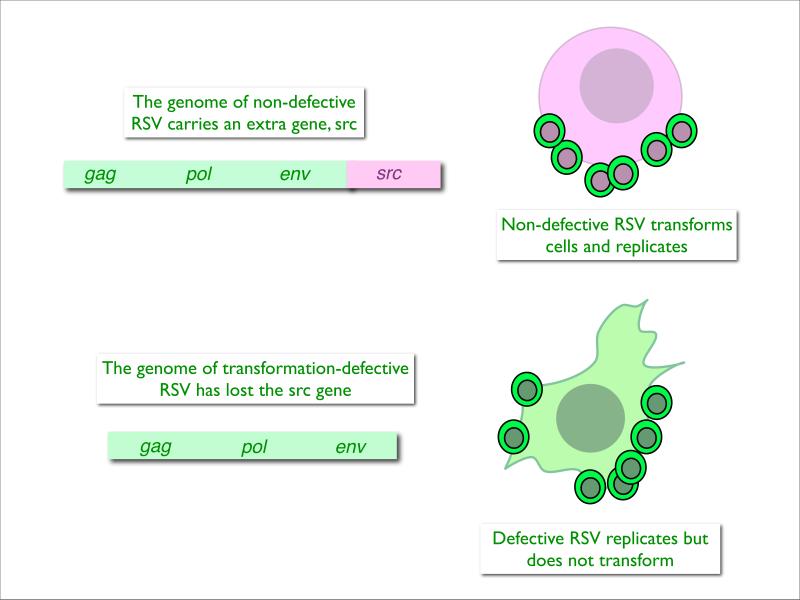
Our knowledge of src and of its protein product is the culmination of a long and complex evolution with stepwise, successive contributions from genetics, biochemistry, immunology, and structural biology5. Each of these steps built on and complemented the preceding one. Three early genetic observations helped define the problem: First, there are mutants of RSV that instead of transforming the fibroblast host into a rounded cell, induce an elongated, fusiform cell shape6,7. Therefore, the phenotype of the transformed cell is under the control of the viral genome. Second, a replication-defective variant of RSV transforms cells without producing infectious progeny, which indicated that the generation of progeny virus is not a prerequisite for oncogenicity8, 9. Third, most strains of RSV are non-defective10, 11 (meaning that they carry all viral replicative genes and the oncogene in the same RNA molecule) (Fig. 1) but spontaneously segregate deletion mutants that still replicate but can no longer transform cells12, 13. Reproduction and oncogenicity are separate and distinct functions.
Fig. 1. The biochemical definition of src.
The protein-coding regions of non-defective RSV encompass the complete information required for virus reproduction (gag, pol, env) and the information needed for oncogenic transformation (src). The RSV-infected cell produces progeny virus and is transformed.
During the replication of RSV, mutant viruses are generated that are no longer oncogenic but contain all the essential viral genes and are fully capable of producing progeny virus that fails to transform cells in culture.
A comparison of the genome sizes of parental RSV and transformation-defective mutant shows that loss of oncogenicity is correlated with loss of about 20% of the genome. The lost sequences represent the src gene which is not essential for virus replication. Using DNA transcripts of these two viral genomes, src sequences can be purified by subtractive hybridization.
The critical proof for the existence of a viral gene that initiates and maintains the transformed cellular phenotype came from temperature-sensitive mutants. In 1970, a groundbreaking paper in Nature described a mutant of the replication-competent Schmidt-Ruppin strain of RSV that transforms cells at a low, permissive temperature but fails to transform at an elevated, non-permissive temperature14. However, production of progeny virus is unaffected by temperature. The mutant pointed to the existence of a viral gene that directs oncogenicity but is dispensable for virus replication. An earlier report of temperature-sensitive mutants of RSV had also shown this temperature-dependence of transformation, but the temperature effect extended to virus replication as well, probably due to multiple mutations15.
Biochemistry then provided the physical underpinning for the existence of a specific oncogene in RSV. This work depended on a unique property of the RSV genome, its non-defectiveness, as discussed above. All other oncogene-carrying retroviruses are replication-defective, the oncogene having displaced one or several of the viral replicative genes. Mutant RSVs that are transformation-defective, but replication-competent, contain a smaller RNA than the parental virus, suggesting that the lost sequences represent the oncogene16 (Fig. 1). This hypothesis was supported by genetic mapping experiments. Temperature-sensitive mutations that affect the ability to transform cells were located to the region of the RSV genome that is deleted in the transformation-defective viruses17. Biochemical mapping with RNA fingerprinting showed that the deleted RNA was a contiguous fragment, located at the 3’ terminus of the viral RNA genome18. Here then was a piece of the retroviral genome, not required for virus survival but essential for oncogenic transformation. The fact that this gene was readily lost from the viral genome showed that it did not convey an evolutionary advantage to the virus.
Where did this accessory piece of information come from? The biochemical experiments had defined a distinct nucleic acid segment of the retroviral RNA genome as the oncogene. This definition then paved the way to a physical isolation of src. The discovery of reverse transcriptase in 1970 had shifted the biochemistry of retroviruses from RNA to DNA for which there existed better and more versatile tools of experimentation19, 20. One of these tools, subtractive hybridization, was applied to DNA transcripts of non-defective RSV and its replication-defective deletion mutant and resulted in the isolation of src-specific DNA sequences. With these sequences it was possible to explore the origin of src, using hybridization as a measure of relatedness. These experiments showed that src originated from the cellular genome and that it was a cellular, not a viral gene21. This fundamental insight, at first ridiculed, was soon extended to other retroviral oncogenes that had been discovered in the meantime, and it changed the landscape of tumor virology22. Retroviruses were no longer originators of oncogenic information; they were demoted to mere carriers of oncogenes that are part of the host genome. This discovery resulted in a huge expansion of the oncogene concept. Any cellular gene with an oncogenic potential that could be activated by a gain of function qualified as an oncogene. Most of these activating genetic events do not involve viruses, but retroviruses that lack an oncogene in their genome can still activate cellular oncogenes by insertional mutagenesis (Box 1).
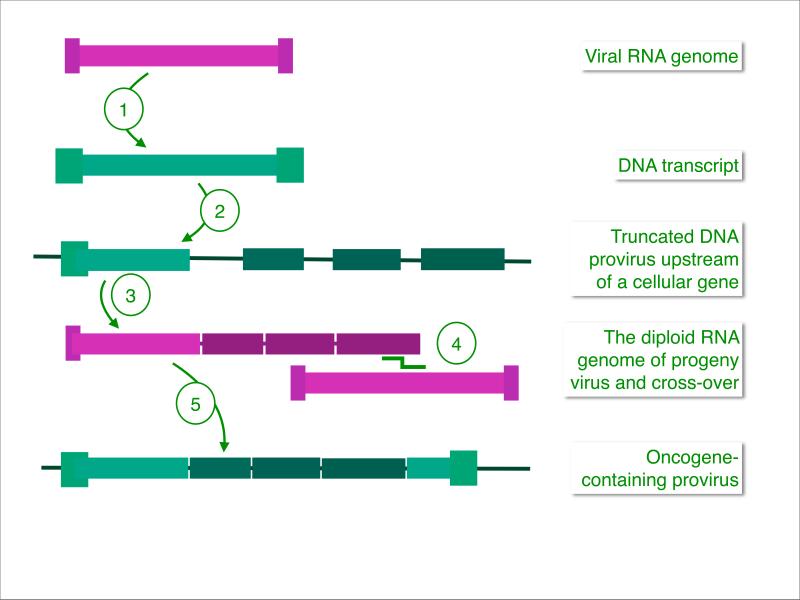
The essential foundation for the genetics of src and of other retroviral oncogenes is the unique life cycle of retroviruses that involves reverse transcription of the virion RNA into DNA and integration of this DNA into the host genome23-25. The genetic stability of the oncogenic phenotype induced by RSV had prompted Temin to propose the main elements of such a life cycle as the provirus hypothesis26, 27. At the time, this seemed a preposterous idea, because RNA-dependent synthesis of DNA overturned the central dogma and its unidirectional flow of genetic information from DNA to RNA to protein. The sensitivity of retrovirus replication to inhibitors of DNA synthesis supported Temin's claim, but the evidence was far from compelling until the discovery of reverse transcriptase provided firm proof for the provirus hypothesis19, 20. Today reverse transcriptase is a routine tool for copying genetic information, so it is important to remember that the generation of a double-stranded DNA copy from virion RNA and the integration of the provirus into the cellular genome are at the root of our understanding of retroviral oncogenes. Proviral integrations are genetic recombination events that can result in the incorporation of a cellular oncogene into the viral genome (Fig. 2.) Such acquisitions are rare. They can occur during viral passage in an animal but are almost never seen in cell culture. There is no experimental system that predictably reproduces spontaneous oncogene acquisition, therefore the molecular details remain hypothetical28, 29.
Fig 2. Acquisition of a cellular oncogene by a retroviral genome.
(1) Virion RNA is transcribed into double-stranded DNA. (2) An accidentally truncated provirus is located upstream of a cellular gene. (Cellular exons indicated in dark green.) (3) A spliced fusion transcript of viral and cellular sequences is packaged into a progeny virion together with a wild type viral genome. (Retroviruses are diploid.) (4) During next-generation reverse transcription, recombination between the two genomes generates a DNA provirus that encodes the cellular oncogene fused to viral sequences (5). As a result of acquiring cellular sequences, viral information essential for the production of progeny has been lost, and such highly oncogenic viruses are replication-defective, with the exception of most strains of RSV which can reproduce and are oncogenic. This mechanism for the acquisition of cellular sequences is hypothetical but in agreement with available experimental data.
The data on the src gene had left an important question unanswered: what is the product of this oncogene? Considering the technical arsenal available at the time, it was not an easy question to answer. The phenomenal breakthrough was achieved in 1977 with a Src-specific antibody raised by a technique that was as ingenious as it was non-obvious: injecting a mammalian-adapted RSV into young rabbits30. This antibody identified the src product as a 60 kD protein which soon was found to have protein kinase activity31, 32. The critical insight that differentiated the Src kinase from other protein kinases known at the time came with the discovery of its target amino acid: it is not serine or threonine but tyrosine33. The Src protein was the first representative of this new class of tyrosine protein kinases, rapidly followed by EGFR34. Today the members of this class are actively studied; they perform key regulatory functions in the cell35.
In the early 1980ies, the cellular and viral src genes were sequenced36-39. The viral Src protein differs from its cellular progenitor by a C-terminal deletion that includes a critical regulatory phosphorylation site and by several point mutations. A comparison of the two proteins showed that the cellular Src had a lower kinase and negligible oncogenic activity compared to viral Src40-42. The explanation of this difference evolved from the discovery that Src carries two modular protein-protein interaction domains, a phosphotyrosine binding SH2 and a poly-proline binding SH3 domain43, 44. Both are crucial for the regulation of Src kinase activity. The molecular details of this regulation were revealed by the crystal structure of Src and of the Src family kinase Hck45, 46. Cellular Src requires activation that opens the catalytic domain by disrupting intramolecular interactions involving both the SH2 and SH3 domains. In viral Src, these inhibitory interactions are absent because of the C-terminal deletion and point mutations in the SH3 domain, making viral Src constitutively active.
The kinase activity of Src invited a search for target proteins that would shed light on the normal and oncogenic functions of the enzyme. Multiple direct and indirect Src targets have been identified, but the search for cancer-relevant functions is far from complete and remains an active area of cancer research47.
Discovering diversity
For the discovery and characterization of other retroviral oncogenes, some lessons from src could be transferred, but there were also new and unique challenges. Other retroviruses that carry an oncogene are replication-defective, in contrast with non-defective RSV. Replication-defective viruses require a helper virus that supplies the missing viral functions in trans. These viruses always occur as mixtures of transforming and of non-transforming helper virus. Because of this dependence on a helper, the genetic experiments are less straightforward than with RSV. However, the structure of the genomes of replication-defective viruses can also offer an advantage: the displacement of viral replicative genes by an oncogene can generate a fusion gene, combining cell-derived and viral sequences and resulting in the production of an oncogenic fusion protein. Such viral-cellular fusion products are readily identifiable with available viral antibodies.
A standard succession of events characterizes the history of most retroviral oncogenes. It starts with the identification of the gene in the virus. Here two criteria first established for src have become signature traits of virtually all retroviral oncogenes: cellular origin and non-identity with viral replicative genes. Identification of the protein, cloning and sequencing are the next steps and are extended to the cellular counterpart of the gene. Questions of oncogenic and normal functions are then addressed; such studies build on pre-existing knowledge of the cellular protein. In the early days of oncogene discovery, temperature sensitive mutants played an important role. With the advances in cloning and sequencing, identifying such mutants became less critical. The discovery of oncogenes in DNA viruses also started with temperature-sensitive mutants31. However the genetic origins and molecular mechanisms of these oncogenes and oncoproteins stand in contrast to those of retroviruses. The critical differences are summarized in Box 2.
The potent trio in human cancer
Myc
Myc was one of the first oncogenes that emerged after src. An RNA fingerprint analysis of the genome of the avian myelocytomatosis virus MC29 had revealed oligonucleotides unrelated to viral replicative genes or to src48. The same sequences were also identified in the avian retroviruses CMII, OK10 and MH249-51. The sequences were shown not to be scattered over the genome but to form a contiguous stretch of RNA, indicating that they were derived from a distinct gene. A fusion protein combining viral Gag sequences of MC29 with the presumptive new oncoprotein was rapidly identified with viral antibodies52. DNA sequencing had just been invented53, 54 and within a few years was applied to the viral and the human myc genes55, 56.
A first important insight into the functions of the Myc protein came with the discovery that it is localized in the cell nucleus57. One of the possible roles for this protein was to act as a transcriptional regulator. However, the failure of Myc to bind DNA under physiological conditions could not be easily reconciled with this idea. The impasse was broken with the discovery of the Max protein as an obligatory dimerization partner of Myc. Only the Myc-Max heterodimer can bind DNA with high affinity and affect transcription58. Max is the required partner of several Myc-related proteins, forming the central component of a regulatory network that can stimulate as well as repress transcription59. The workings of this network are based on selective dimerization. Max forms DNA-binding homodimers, but none of its partners have that ability, so they depend on dimerization with Max to bind DNA and to regulate transcription.
The identification of Myc target genes has been challenging, because thousands of copies of its short DNA target sequence, the E-box CACGTG, are present in vertebrate genomes. A recent study using a combination of chromatin immunoprecipitation and deep sequencing identified over 7000 genomic binding sites in a cell that overexpresses Myc60. Cellular levels of Myc are tightly regulated, and overexpression leads to uncontrolled cell replication or to apoptosis, depending on contextual factors that are not completely understood.
There are three MYC genes in the human genome, c-MYC L-MYC and N-MYC55,61, 62, The cellular homolog of the retroviral myc gene is c-MYC. N-MYC and L-MYC were discovered later in human cells; they play important roles in diverse human cancers63. The two representative mechanisms for the involvement of MYC in human disease came to light from studies of Burkitt lymphoma and neuroblastoma. Burkitt lymphoma cells always carry a chromosomal translocation that places c-MYC under the control of an immunoglobulin enhancer64. The result is increased transcription of c-MYC driven by the immunoglobulin-regulatory sequences. The discovery of c-MYC rearrangements in a human lymphoma was the first indication that cellular counterparts of retroviral oncogenes are involved in the pathogenesis of human disease. In neuroblastoma, the N-MYC gene is frequently amplified, and the expression of N-MYC is correspondingly elevated65. Upregulated transcription and amplification are the two mechanisms for the oncogenic gain of function in the MYC genes. Mutations in the coding region of MYC do not play a significant role in human cancer.
Recent studies indicate that the role of MYC in cancer goes beyond the situations where it appears to be the primary driver. c-MYC has emerged as the mediator of resistance to inhibitors of PI3-kinase, and dominant negative Myc causes regression of Ras-induced tumors in mice66, 67.
Ras
The discovery of ras faced a different set of challenges. The two principal viruses carrying this oncogene, Harvey sarcoma virus and Kirsten sarcoma virus, arose by recombination with the host genome during passage of murine leukemia virus in rats68, 69. The rat-derived oncogene in these replication-defective viruses is not fused to viral genes, and in the absence of such viral markers, the Ras protein could not be identified with viral antibodies. However, animals bearing Kirsten or Harvey sarcomas generated antibodies that interacted with the 21 kD product of the ras gene70. The Ras protein was also obtained by in vitro translation of the viral genome71. Cloning and sequencing of the Harvey and Kirsten sarcoma viruses defined the viral and rat-derived contributions to these recombinant genomes and completed the molecular information on the viral ras gene72-74. The two ras genes, Kras and Hras, do not differ in the properties we consider here.
A first clue about biochemical functions came from the observation that Ras has guanine nucleotide binding activity, a finding that quickly culminated in the discovery that Ras is a GTPase75-78. In its active form, Ras is bound to GTP, and binding could be enhanced by activated EGFR79. How could Ras be integrated into cellular signaling and what was responsible for the oncogenic activity of that protein? Half of the answer came from linking an adaptor protein and a guanine nucleotide exchange factor to the activity of Ras80, 81. The SH2 domain of the adaptor, GRB2, binds to phosphorylated tyrosine, typically in a receptor tyrosine kinase (such as EGFR) and with its SH3 domain GRB2 can recruit the guanine nucleotide exchange factor (GEF). This GEF stimulates the release of GDP from Ras and thus enhances loading with GTP. This sequence of interactions established the upstream signaling path that leads to Ras activation82.
The other half of the answer, outlining the downstream activities of Ras, was initiated by the discovery of the raf oncogene in a murine sarcoma virus 83, 84 and of its avian homolog mil in the chicken tumor virus MH285, 86. The Raf protein binds to GTP-loaded Ras and connects it to the MAP kinase pathway87-89. Activated Ras also binds to the catalytic subunit of PI3K, and this interaction is important for PI3K signaling90. Although numerous somatic mutations occur within the catalytic subunit of human PI3K, no mutations have been found in the Ras binding domain, suggesting that interaction with Ras is essential for the function of PI3K. The oncogenic activities of PI3K are discussed in greater detail below.
The GTPase activity of Ras is stimulated by association with a GTPase-activating protein (GAP)91. Ras acquires oncogenic potency by point mutations affecting residues 12 and 61. These mutations disturb the interaction with GAPs. They reduce the rate of GTP hydrolysis and result in elevated levels of the active, GTP-bound Ras78, 91-96. An important aspect of all Ras activity is cellular localization. Ras is positioned at the inner side of the plasma membrane, and this location is essential for activity97. The interaction with membrane lipids is mediated by an obligatory posttranslational isoprenylation of the protein98, 99.
A series of exciting and dramatic experiments linked Ras directly to human cancer. Initially, transfer of DNA from human cancer cells was found to transform recipient mouse cells. Integration of the source DNA into the genome of the recipient cells was verified by the presence of readily identifiable repetitive human sequences100, 101. The breakthrough came with the discovery that the transforming DNA derived from human cancer cells is homologous to ras102-105. This discovery also linked a retroviral oncogene that in experimental systems induces sarcomas to epithelial cancers in humans. The activity of retroviral oncogenes is clearly not restricted to fibroblasts or hematopoietic cells but includes epithelial cells. Oncogenic activity in DNA transfection experiments also revealed the existence of a third ras gene, Nras, in the human genome106.
Within the span of two years, 1982 to 1984, the findings of c-MYC in Burkitt lymphoma, N-MYC in neuroblastoma and oncogenic RAS in diverse human cancers, linked the two retroviral oncogenes unequivocally to human disease as probable causative agents. These connections between retroviral model systems and human cancer could have been predicted from the cellular origin of retroviral oncogenes, but they came as a surprise nonetheless. The discoveries with MYC and RAS have special historical significance, because they consolidated the view of cancer as a genetic disease.
ErbB
The story of ErbB takes us back to the early days of retrovirology. ErbB is the oncogene of avian retroviruses that induce an acute form of erythroid leukemia called erythroblastosis. One of these viruses, referred to as strain R, dates from 1935107. It contains two cell-derived oncogenes, erbA, a hormone receptor, and erbB22, 108-110. For the induction of oncogenic growth, erbA is auxiliary but dispensable, whereas erbB is both necessary and sufficient, because a separate isolate of erythroblastosis virus, strain H, carries only the erbB oncogene yet does not differ significantly from strain R in tumor spectrum or pathogenic potency111. Studies on additional independent isolates of avian erythroblastosis virus have supported this dominant role of erbB in oncogenesis112. Analyses of the cloned genomes and of in vitro translated proteins from strain R and H viruses suggested that the viral ErbB protein is produced by a fused mRNA consisting of a very short N-terminal viral sequence and part of the cellular erbB113-116. In addition, specific antibodies detected a 74 kD transformation-specific protein in cells infected by avian erythroblastosis virus117, 118.
At the time, this information on chicken viruses seemed almost esoteric, but it has acquired great relevance for human disease. Within a period of a few months in 1984, the viral ErbB protein was found to be glycosylated and phosphorylated and structurally related to tyrosine kinases. It showed sequence features of tyrosine kinases and was localized as an integral membrane protein at the cell surface115, 119-125. Finally and most importantly, sequence analysis revealed both close homology to EGFR and a large deletion in the extracellular domain of viral ErbB126, 127. In addition to the N-terminal truncation, viral ErbB proteins also show mutations in the kinase domain located in the C-terminal cytoplasmic portion of the protein. Viral ErbB functions as a constitutively active receptor tyrosine kinase; the activity is ligand-independent and also requires the kinase domain mutations. The mutations in viral ErbB do not cause a mere quantitative enhancement of the same signaling pathways that are controlled by cellular EGFR but induce qualitative changes in the spectrum of signaling targets. These changes are critical to the oncogenic potency of the protein112.
EGFR can function as an oncogenic “driver” in diverse human cancers. Mutations that mechanistically resemble those seen in viral ErbB occur in the EGFR of glioblastoma multiforme and non-small cell lung cancer. About 50% of glioblastomas carry the EGFRvIII mutant which has lost a large portion of the extracellular domain and no longer binds ligand but signals constitutively, addressing targets that are different from those of wild-type EGFR128. Such a cancer-specific mutation in a kinase would appear to be an ideal therapeutic target. However, the clinical experience with inhibitors of EGFR in glioblastoma has been uneven, with tumor shrinkage linked to the co-expression of EGFRvIII and of the tumor suppressor PTEN129. In non-small cell lung cancer, EGFR mutations are located in the kinase domain and lead to constitutive autophosphorylation and activation. Such cancers, seen mostly in non-smokers, are uniquely sensitive to EGFR inhibitors but regularly develop resistance to these drugs130-133.
The human genome contains three additional genes that are closely related to EGFR, HER2, HER3 and HER4134-136. The oncogenic potential of HER2 was discovered in transfection experiments with DNA from human neuroblastoma cells. The cell-transforming gene in these experiments was identified as EGFR-related with an activating mutation in the transmembrane domain137. HER2 is frequently amplified in breast cancer138. A humanized monoclonal antibody that inhibits HER2 signaling (trastuzumab) shows substantial clinical benefit and is now part of standard therapy for HER2+ breast cancers139. HER3 is unusual in that it has extremely low kinase activity and functions predominantly as a dimerization partner of other EGFR family members140. HER4 differs from the other EGFR-related genes in that it mediates cellular differentiation and inhibits replication141.
Oncogenes from slaughterhouse viruses
In the 1980ies, it became clear that avian retroviruses are a particularly rich source of oncogenes. In chickens, retrovirus infection is common and widespread. Most of these viruses are replication-competent, do not carry an oncogene, and induce tumors (mostly lymphoid leukosis) by insertional activation of a cellular oncogene142. But occasionally the genetic recombination between virus and host results in the incorporation of an oncogene into the viral genome. Such an acquisition converts the virus from slowly oncogenic to rapidly oncogenic, resulting in solid tumors that are distinct from the endemic leukosis (Fig. 2). Chicken slaughterhouses process up to 30,000 birds a day; each of these is inspected for signs of disease. At these numbers, even rare viral-cellular recombination events that result in aggressive cancers can be found.
Slaughterhouse veterinarians have been the source of several new, rapidly oncogenic retroviruses, and from these viruses, three new oncogenes were isolated: jun, qin and pi3k143-145. The discovery of the retroviral Jun, the finding of its cellular counterpart in the transcription factor complex AP1, and the identification of the tight partnership with the oncoprotein Fos (discovered separately in the Finkel-Biskis-Jinkins murine sarcoma virus146), marked an exciting period in the history of oncogenes. The story of these events has been told elsewhere147, 148. Qin (also referred to as FOXG1) is a representative of the winged helix or FOX family of DNA-binding proteins which function as developmental and metabolic transcriptional regulators144. Although Qin has not been implicated in human cancer, the FOX protein family is linked to human disease by the involvement of FOXO1 in a chromosomal translocation that contributes to the development of alveolar rhabdomyosarcoma, an aggressive childhood tumor. Another member of the family, FOXA1 controls the sexual dimorphism seen with hepatocellular carcinoma149. A broader survey of the association of FOX proteins with cancer has been presented in a recent review150.
Among the oncogenes derived from these recently isolated avian retroviruses, pi3k stands out, because its cellular counterpart controls signaling pathways that show aberrant activation in most human tumors and contain promising drug targets. The retroviral pi3k has served as an important model for the oncogenic activities of human PI3K. There has long been a suspicion that the lipid kinase PI3K may have oncogenic potential. In early work, the oncoproteins of DNA viruses as well as Src were shown to be associated with a cellular lipid kinase activity151-155. The interaction was essential for the oncogenicity of these viral proteins. The transformation-associated lipid kinase activity was then found to catalyze the phosphorylation of the D3 position of the inositol ring, defining a novel enzymatic activity that generates phosphatidylinositol 3 phosphates156. The fundamental importance of this finding was not realized until much later when it became clear that this PI3K was at the center of an extensive and versatile cellular signaling network that becomes corrupted in most cancers157, 158. Direct evidence for the oncogenicity of PI3K came with the discovery of an avian hemangiosarcoma virus, ASV16, in a tumor obtained from a chicken processing plant. ASV16 is a replication-defective virus with a genome that encodes a single protein encompassing the p110α isoform of the catalytic subunit of chicken PI3K fused N-terminally to viral gag sequences 145.
The viral pi3k harbors several mutations in the p110α coding sequence, but these do not induce a gain of function and are irrelevant for oncogenic activity. Oncogenicity depends on the N-terminal Gag sequences159. The function of the gag sequences was first thought to facilitate membrane localization and to bring the enzyme in direct contact with its substrate. Support for this idea comes from the observation that a myristylation signal added to the N-terminus of cellular p110α also has a strongly activating effect and makes the protein oncogenic. However, recent data cast doubt on this interpretation. Even random amino acid sequences, added to the N-terminus of p110α, are activating, there is no requirement for a membrane localizing function in these sequences. Rather, these N-terminal additions appear to induce a conformational change that mimics activation of p110α by upstream signals160. A similar mechanism for constitutive activity is seen with cancer-specific mutations that carry an amino acid substitution in the helical domain of p110α 161-164. In such mutants, the inhibitory interaction with the regulatory subunit p85 is disrupted.
PI3K has moved into the limelight as a cancer target because of frequent cancer-specific genetic and epigenetic changes that result in enhanced activity. These include loss of function in the PI3K antagonist and tumor suppressor PTEN, elevated activity and amplification of PI3K and gain-of-function mutations in the catalytic subunit p110α165-170. Enhanced PI3K signaling is a driving force in cancer development. Academic laboratories and the pharmaceutical industry have responded to this situation by generating small molecule inhibitors of PI3K, and several of these are currently in advanced clinical trial171.
From simplicity to complexity
As we look at the history of oncogenes and their significance for human disease, two developmental trends unfold (see Timeline). One is a steady increase in relevance, the other a broadening of the concept of cancer as genetic disease. Rapidly tumorigenic retroviruses that carry oncogenes have been found mostly in chickens and in mice. Early work with these viruses focused on cancer as an infectious condition. But the concepts and mechanisms uncovered with readily transmissible animal tumors appeared not applicable to the human situation. Therefore the significance of identifying specific oncogenes in viruses was at first exclusively experimental and theoretical. These discoveries showed that normal vertebrate cells could be transformed into cancer cells by the action of a single gene. This was a revolutionary insight, offering simplicity and the prospect of complete molecular understanding.
Retroviral oncogenes remained mainly experimental tools with uncertain ties to human cancer until 1976, when oncogene sequences were found in cellular genomes21. This discovery transformed the field. Retroviruses with their ability to acquire and transduce host genes became just one out of several possible ways by which a cellular oncogene can be activated. In principle, any genetic change in the cellular oncogene is potentially activating. The next transformative step on the way to relevance established the direct connection between the cellular versions of retroviral oncogenes and human cancer. The key discoveries were finding transcriptional activation of c-MYC by chromosomal translocation in Burkitt lymphoma, amplification of N-Myc in neuroblastoma and identification of activated RAS in DNA from human cancer cells (see also Table 2)64, 65, 102-104. These findings were fundamental in revealing cancer as a genetic disease. They also appeared like a reductionist triumph, explaining cancer with changes in one or at most a few genes that would generate novel and highly specific therapeutic targets.
Table 2.
Oncogenes first identified in retroviruses can function as drivers in human cancer
| Oncogene | Mechanism of activation | Cancer type | References |
|---|---|---|---|
| MYC | Increased transcription | Burkitt lymphoma | 64, 186 |
| Increased transcription | B-cell lymphoma | 187, 188 | |
| Amplification | Neuroblastoma | 65, 189 | |
| Amplification | Medulloblastoma | 190 - 192 | |
| EGFR | Mutation | Glioblastoma | 128, 193 |
| Mutation | Non-small cell lung cancer | 130 - 133 | |
| RAS | Mutation | Pancreatic cancer | 194 - 196 |
| RAF | Mutation | Melanoma | 197 |
This development took retroviral oncogenes from obscurity to prominence. But in subsequent years, genetic changes that affect the oncogenic cellular phenotype have steadily increased in type and in number. If we define an oncogene as a replication-promoting gene that encodes a protein and shows gain of function in cancer, then the number of such genes is probably in excess of a thousand and growing. A comprehensive view of cell-autonomous genetic alterations in cancer further includes tumor suppressors which contribute to the oncogenic phenotype by a loss of function, often as a result of epigenetic changes172. Micro RNAs have added another layer of complexity with both pro- and anti-oncogenic effects173. The vast extend of the non-coding transcriptome including large antisense transcripts and pseudogenes is beginning to be functionally explored and likely holds even more surprises174. The cancer genome project has uncovered an unexpected multitude of genetic changes in all cancers, revealing mutational landscapes that are characteristic of tumor origin and histology. A similar trend toward complexity can be seen in our understanding of oncoprotein functions. All these proteins show multiple activities, generating diverse signals. A complete molecular understanding of how these activities initiate and maintain cancer remains a challenge.
The complexity of genetic alterations becomes irrelevant in certain cancers that show a striking and apparently irreversible dependency on a single, dominant genetic change. Such oncogene addiction can be the basis for stunning clinical successes with targeted therapy175. However, it is questionable whether the model of cellular addiction to a single oncoprotein is applicable to a broad spectrum of cancers. In the more common scenario, complexity rules and dictates a therapeutic strategy that relies on targeting a few critical drivers of the oncogenic cellular phenotype. Success depends on the identification and validation of these drivers as cancer targets176, 177. These efforts are guided by the general principle that it is easier to correct a gain of function than to restore a loss of function. Oncogenes remain very much in the line of fire.
Text Box 1.
Activation of cellular oncogenes by insertional mutagenesis. Retroviruses of the subfamily oncovirus that lack an oncogene in their genome are able to induce cancer by insertional mutagenesis178. In this process, a provirus integrating in the vicinity of a cellular oncogene functions as positive transcriptional regulator and thus activates the latent tumorigenic potential of the cellular gene. Insertional, retrovirus-mediated mutagenesis is a slow process that occurs only in the vertebrate host and typically requires prolonged and extensive viral replication and integration. It has been widely used to reveal the oncogenic potential of cellular genes that are never transduced by viruses179.
Text Box 2.
Contrasting mechanisms in viral oncogenicity: RNA versus DNA viruses. The oncogenes of retroviruses are cell-derived; they deregulate cellular signaling and transcriptional controls. In contrast, oncogenic DNA viruses, including the papilloma viruses, polyoma virus, simian virus 40 (SV40) and some tumor-inducing adeno- and herpesviruses carry their own oncogenes. Some of the best understood among these disrupt the retinoblastoma (Rb) protein-mediated control of the cell cycle180-184. During the G1 phase of the cell cycle, the Rb protein is hypophosphorylated and bound to E2F transcription factors, forming transcriptional repressor complexes. These are essential components of the restriction point that prevents entry into the S phase of the cell cycle. Upon mitogenic stimulation, cyclin-dependent kinases phosphorylate Rb, thus releasing the E2F proteins which then initiate a transcriptional program that marks the entry into the S phase. Several DNA viral proteins bind to hypophosphorylated Rb: the E1A protein of adenoviruses, the large T antigen of SV40 virus and the E7 proteins of oncogenic human papilloma viruses. These interactions free the E2F proteins without a requirement for mitogenic signals and start the S phase of the cell cycle. Proteins of oncogenic DNA viruses can also operate as constitutive signaling receptors or can interfere with the functions of inhibitors of cyclin-dependent kinases185.
Acknowledgments
My sincere thanks go to Klaus Bister and Anja Zembrzycki for their help with this paper. Klaus Bister read several iterations of this review, offering insightful comments and valuable suggestions. Anja Zembrzycki transformed multiple disorganized drafts into a formatted manuscript.
Work of the author is supported by National Institutes of Health grants R01CA078230, R01CA153124 and R01CA151574.
This review is dedicated to Harry Rubin. His pioneering work started the field.
References
- 1.Temin HM, Rubin H. Characteristics of an assay for Rous sarcoma virus and Rous sarcoma cells in tissue culture. Virology. 1958;6:669–88. doi: 10.1016/0042-6822(58)90114-4. [DOI] [PubMed] [Google Scholar]
- 2.Hanafusa H, Hanafusa T, Rubin H. Analysis of the Defectiveness of Rous Sarcoma Virus. I. Characterization of the Helper Virus. Virology. 1964;22:591–601. doi: 10.1016/0042-6822(64)90081-9. [DOI] [PubMed] [Google Scholar]
- 3.Hanafusa H, Hanafusa T, Rubin H. Analysis of the Defectiveness of Rous Sarcoma Virus, Ii. Specification of Rsv Antigenicity by Helper Virus. Proc Natl Acad Sci U S A. 1964;51:41–8. doi: 10.1073/pnas.51.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubin H, Vogt PK. An avian leukosis virus associated with stocks of Rous sarcoma virus. Virology. 1962;17:184–94. doi: 10.1016/0042-6822(62)90096-x. [DOI] [PubMed] [Google Scholar]
- 5.Martin GS. The road to Src. Oncogene. 2004;23:7910–7. doi: 10.1038/sj.onc.1208077. [DOI] [PubMed] [Google Scholar]
- 6.Temin HM. The control of cellular morphology in embryonic cells infected with rous sarcoma virus in vitro. Virology. 1960;10:182–97. doi: 10.1016/0042-6822(60)90038-6. [DOI] [PubMed] [Google Scholar]
- 7.Yoshii S, Vogt PK. A mutant of rous sarcoma virus (type O) causing fusiform cell transformation. Proceedings of the Society for Experimental Biology and Medicine. Society for Experimental Biology and Medicine. 1970;135:297–301. doi: 10.3181/00379727-135-35039. [DOI] [PubMed] [Google Scholar]
- 8.Hanafusa H, Hanafusa T, Rubin H. The defectiveness of Rous sarcoma virus. Proc Natl Acad Sci U S A. 1963;49:572–80. doi: 10.1073/pnas.49.4.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Temin HM. Separation of morphological conversion and virus production in Rous sarcoma virus infection. Cold Spring Harbor symposia on quantitative biology. 1962;27:407–14. doi: 10.1101/sqb.1962.027.001.038. [DOI] [PubMed] [Google Scholar]
- 10.Goldé A, Lacassagne M. Non-défectivité de la souche de virus de Rouse de Schmidt-Ruppin. C. R. Acad. Sci. Paris. 1966;262:329–331. [PubMed] [Google Scholar]
- 11.Duff RG, Vogt PK. Characteristics of two new avian tumor virus subgroups. Virology. 1969;39:18–30. doi: 10.1016/0042-6822(69)90344-4. [DOI] [PubMed] [Google Scholar]
- 12.Martin GS, Duesberg PH. The a subunit in the RNA of transforming avian tumor viruses. I. Occurrence in different virus strains. II. Spontaneous loss resulting in nontransforming variants. Virology. 1972;47:494–7. doi: 10.1016/0042-6822(72)90287-5. [DOI] [PubMed] [Google Scholar]
- 13.Vogt PK. Spontaneous segregation of nontransforming viruses from cloned sarcoma viruses. Virology. 1971;46:939–46. doi: 10.1016/0042-6822(71)90092-4. [DOI] [PubMed] [Google Scholar]
- 14.Martin GS. Rous sarcoma virus: a function required for the maintenance of the transformed state. Nature. 1970;227:1021–3. doi: 10.1038/2271021a0. [DOI] [PubMed] [Google Scholar]
- 15.Toyoshima K, Vogt PK. Temperature sensitive mutants of an avian sarcoma virus. Virology. 1969;39:930–1. doi: 10.1016/0042-6822(69)90030-0. [DOI] [PubMed] [Google Scholar]
- 16.Duesberg PH, Vogt PK. Differences between the ribonucleic acids of transforming and nontransforming avian tumor viruses. Proc Natl Acad Sci U S A. 1970;67:1673–80. doi: 10.1073/pnas.67.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernstein A, MacCormick R, Martin GS. Transformation-defective mutants of avian sarcoma viruses: the genetic relationship between conditional and nonconditional mutants. Virology. 1976;70:206–9. doi: 10.1016/0042-6822(76)90254-3. [DOI] [PubMed] [Google Scholar]
- 18.Wang LH, Duesberg PH, Kawai S, Hanafusa H. Location of envelope-specific and sarcoma-specific oligonucleotides on RNA of Schmidt-Ruppin Rous sarcoma virus. Proc Natl Acad Sci U S A. 1976;73:447–51. doi: 10.1073/pnas.73.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970;226:1209–11. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- 20.Temin HM, Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970;226:1211–3. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- 21.Stehelin D, Varmus HE, Bishop JM, Vogt PK. DNA related to the transforming gene(s) of avian sarcoma viruses is present in normal avian DNA. Nature. 1976;260:170–3. doi: 10.1038/260170a0. [DOI] [PubMed] [Google Scholar]
- 22.Roussel M, et al. Three new types of viral oncogene of cellular origin specific for haematopoietic cell transformation. Nature. 1979;281:452–5. doi: 10.1038/281452a0. [DOI] [PubMed] [Google Scholar]
- 23.Coffin J, Hughes S, Varmus H, editors. Retroviruses. Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 24.Strauss J, Strauss E. Viruses and Human Disease. Academic Press; 2007. [Google Scholar]
- 25.Flint S, Enquist L, Racaniello V. Principles of Virology. ASM Press; 2009. [Google Scholar]
- 26.Temin H. Nature of the Provirus of Rous Sarcoma. National Cancer Institute Monograph. 1964;17:557–570. [Google Scholar]
- 27.Temin HM. The Participation of DNA in Rous Sarcoma Virus Production. Virology. 1964;23:486–94. doi: 10.1016/0042-6822(64)90232-6. [DOI] [PubMed] [Google Scholar]
- 28.Vogt P, Bader A. In: Encyclopedia of Virology. BWJ M, MHV VR, editors. Elsevier Publishing; Oxford: 2008. pp. 445–450. [Google Scholar]
- 29.Varmus HE. Form and function of retroviral proviruses. Science. 1982;216:812–20. doi: 10.1126/science.6177038. [DOI] [PubMed] [Google Scholar]
- 30.Brugge JS, Erikson RL. Identification of a transformation-specific antigen induced by an avian sarcoma virus. Nature. 1977;269:346–8. doi: 10.1038/269346a0. [DOI] [PubMed] [Google Scholar]
- 31.Collett MS, Erikson RL. Protein kinase activity associated with the avian sarcoma virus src gene product. Proc Natl Acad Sci U S A. 1978;75:2021–4. doi: 10.1073/pnas.75.4.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levinson AD, Oppermann H, Levintow L, Varmus HE, Bishop JM. Evidence that the transforming gene of avian sarcoma virus encodes a protein kinase associated with a phosphoprotein. Cell. 1978;15:561–72. doi: 10.1016/0092-8674(78)90024-7. [DOI] [PubMed] [Google Scholar]
- 33.Hunter T, Sefton BM. Transforming gene product of Rous sarcoma virus phosphorylates tyrosine. Proc Natl Acad Sci U S A. 1980;77:1311–5. doi: 10.1073/pnas.77.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ushiro H, Cohen S. Identification of phosphotyrosine as a product of epidermal growth factor-activated protein kinase in A-431 cell membranes. J Biol Chem. 1980;255:8363–5. [PubMed] [Google Scholar]
- 35.Hunter T. Tyrosine phosphorylation: thirty years and counting. Current opinion in cell biology. 2009;21:140–6. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Czernilofsky AP, et al. Corrections to the nucleotide sequence of the src gene of Rous sarcoma virus. Nature. 1983;301:736–8. doi: 10.1038/301736b0. [DOI] [PubMed] [Google Scholar]
- 37.Czernilofsky AP, et al. Nucleotide sequence of an avian sarcoma virus oncogene (src) and proposed amino acid sequence for gene product. Nature. 1980;287:198–203. doi: 10.1038/287198a0. [DOI] [PubMed] [Google Scholar]
- 38.Schwartz DE, Tizard R, Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983;32:853–69. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- 39.Takeya T, Feldman RA, Hanafusa H. DNA sequence of the viral and cellular src gene of chickens. 1. Complete nucleotide sequence of an EcoRI fragment of recovered avian sarcoma virus which codes for gp37 and pp60src. J Virol. 1982;44:1–11. doi: 10.1128/jvi.44.1.1-11.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coussens PM, Cooper JA, Hunter T, Shalloway D. Restriction of the in vitro and in vivo tyrosine protein kinase activities of pp60c-src relative to pp60v-src. Mol Cell Biol. 1985;5:2753–63. doi: 10.1128/mcb.5.10.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iba H, Cross FR, Garber EA, Hanafusa H. Low level of cellular protein phosphorylation by nontransforming overproduced p60c-src. Mol Cell Biol. 1985;5:1058–66. doi: 10.1128/mcb.5.5.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iba H, Takeya T, Cross FR, Hanafusa T, Hanafusa H. Rous sarcoma virus variants that carry the cellular src gene instead of the viral src gene cannot transform chicken embryo fibroblasts. Proc Natl Acad Sci U S A. 1984;81:4424–8. doi: 10.1073/pnas.81.14.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sadowski I, Stone JC, Pawson T. A noncatalytic domain conserved among cytoplasmic protein-tyrosine kinases modifies the kinase function and transforming activity of Fujinami sarcoma virus P130gag-fps. Mol Cell Biol. 1986;6:4396–408. doi: 10.1128/mcb.6.12.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pawson T. Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell. 2004;116:191–203. doi: 10.1016/s0092-8674(03)01077-8. [DOI] [PubMed] [Google Scholar]
- 45.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–9. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 46.Xu W, Doshi A, Lei M, Eck MJ, Harrison SC. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Molecular cell. 1999;3:629–38. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 47.Martin GS. The hunting of the Src. Nature reviews. Molecular cell biology. 2001;2:467–75. doi: 10.1038/35073094. [DOI] [PubMed] [Google Scholar]
- 48.Duesberg PH, Bister K, Vogt PK. The RNA of avian acute leukemia virus MC29. Proc Natl Acad Sci U S A. 1977;74:4320–4. doi: 10.1073/pnas.74.10.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bister K, Ramsay G, Hayman MJ, Duesberg PH. OK10, an avian acute leukemia virus of the MC 29 subgroup with a unique genetic structure. Proc Natl Acad Sci U S A. 1980;77:7142–6. doi: 10.1073/pnas.77.12.7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duesberg PH, Vogt PK. Avian acute leukemia viruses MC29 and MH2 share specific RNA sequences: evidence for a second class of transforming genes. Proc Natl Acad Sci U S A. 1979;76:1633–7. doi: 10.1073/pnas.76.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kan NC, Flordellis CS, Garon CF, Duesberg PH, Papas TS. Avian carcinoma virus MH2 contains a transformation-specific sequence, mht, and shares the myc sequence with MC29, CMII, and OK10 viruses. Proc Natl Acad Sci U S A. 1983;80:6566–70. doi: 10.1073/pnas.80.21.6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bister K, Hayman MJ, Vogt PK. Defectiveness of avian myelocytomatosis virus MC29: isolation of long-term nonproducer cultures and analysis of virus-specific polypeptide synthesis. Virology. 1977;82:431–48. doi: 10.1016/0042-6822(77)90017-4. [DOI] [PubMed] [Google Scholar]
- 53.Maxam AM, Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977;74:560–4. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. Journal of molecular biology. 1975;94:441–8. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- 55.Dalla-Favera R, et al. Cloning and characterization of different human sequences related to the onc gene (v-myc) of avian myelocytomatosis virus (MC29). Proc Natl Acad Sci U S A. 1982;79:6497–501. doi: 10.1073/pnas.79.21.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alitalo K, et al. Nucleotide sequence to the v-myc oncogene of avian retrovirus MC29. Proc Natl Acad Sci U S A. 1983;80:100–4. doi: 10.1073/pnas.80.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abrams HD, Rohrschneider LR, Eisenman RN. Nuclear location of the putative transforming protein of avian myelocytomatosis virus. Cell. 1982;29:427–39. doi: 10.1016/0092-8674(82)90159-3. [DOI] [PubMed] [Google Scholar]
- 58.Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–7. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 59.Eisenman RN. The Max network: coordinated transcriptional regulation of cell growth and proliferation. Harvey Lect. 2000;96:1–32. [PubMed] [Google Scholar]
- 60.Seitz V, et al. Deep sequencing of MYC DNA-binding sites in Burkitt lymphoma. PLoS One. 2011;6:e26837. doi: 10.1371/journal.pone.0026837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ikegaki N, Minna J, Kennett RH. The human L-myc gene is expressed as two forms of protein in small cell lung carcinoma cell lines: detection by monoclonal antibodies specific to two myc homology box sequences. The EMBO journal. 1989;8:1793–9. doi: 10.1002/j.1460-2075.1989.tb03573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schwab M, et al. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature. 1983;305:245–8. doi: 10.1038/305245a0. [DOI] [PubMed] [Google Scholar]
- 63.Depinho RA, et al. Myc family genes: a dispersed multi-gene family. Annals of clinical research. 1986;18:284–9. [PubMed] [Google Scholar]
- 64.Dalla-Favera R, et al. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci U S A. 1982;79:7824–7. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schwab M, et al. Enhanced expression of the human gene N-myc consequent to amplification of DNA may contribute to malignant progression of neuroblastoma. Proc Natl Acad Sci U S A. 1984;81:4940–4. doi: 10.1073/pnas.81.15.4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu P, et al. Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nature medicine. 2011;17:1116–20. doi: 10.1038/nm.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Soucek L, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–83. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harvey JJ. An Unidentified Virus Which Causes the Rapid Production of Tumours in Mice. Nature. 1964;204:1104–5. doi: 10.1038/2041104b0. [DOI] [PubMed] [Google Scholar]
- 69.Kirsten WH, Mayer LA. Morphologic responses to a murine erythroblastosis virus. J Natl Cancer Inst. 1967;39:311–35. [PubMed] [Google Scholar]
- 70.Shih TY, Weeks MO, Young HA, Scolnick EM. Identification of a sarcoma virus-coded phosphoprotein in nonproducer cells transformed by Kirsten or Harvey murine sarcoma virus. Virology. 1979;96:64–79. doi: 10.1016/0042-6822(79)90173-9. [DOI] [PubMed] [Google Scholar]
- 71.Parks WP, Scolnick EM. In vitro translation of Harvey murine sarcoma virus RNA. J Virol. 1977;22:711–9. doi: 10.1128/jvi.22.3.711-719.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hager GL, et al. Molecular cloning of the Harvey sarcoma virus closed circular DNA intermediates: initial structural and biological characterization. J Virol. 1979;31:795–809. doi: 10.1128/jvi.31.3.795-809.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsuchida N, Ryder T, Ohtsubo E. Nucleotide sequence of the oncogene encoding the p21 transforming protein of Kirsten murine sarcoma virus. Science. 1982;217:937–9. doi: 10.1126/science.6287573. [DOI] [PubMed] [Google Scholar]
- 74.Tsuchida N, Uesugi S. Structure and functions of the Kirsten murine sarcoma virus genome: molecular cloning of biologically active Kirsten murine sarcoma virus DNA. J Virol. 1981;38:720–7. doi: 10.1128/jvi.38.2.720-727.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gibbs JB, Sigal IS, Poe M, Scolnick EM. Intrinsic GTPase activity distinguishes normal and oncogenic ras p21 molecules. Proc Natl Acad Sci U S A. 1984;81:5704–8. doi: 10.1073/pnas.81.18.5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McGrath JP, Capon DJ, Goeddel DV, Levinson AD. Comparative biochemical properties of normal and activated human ras p21 protein. Nature. 1984;310:644–9. doi: 10.1038/310644a0. [DOI] [PubMed] [Google Scholar]
- 77.Scolnick EM, Papageorge AG, Shih TY. Guanine nucleotide-binding activity as an assay for src protein of rat-derived murine sarcoma viruses. Proc Natl Acad Sci U S A. 1979;76:5355–9. doi: 10.1073/pnas.76.10.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sweet RW, et al. The product of ras is a GTPase and the T24 oncogenic mutant is deficient in this activity. Nature. 1984;311:273–5. doi: 10.1038/311273a0. [DOI] [PubMed] [Google Scholar]
- 79.Kamata T, Feramisco JR. Epidermal growth factor stimulates guanine nucleotide binding activity and phosphorylation of ras oncogene proteins. Nature. 1984;310:147–50. doi: 10.1038/310147a0. [DOI] [PubMed] [Google Scholar]
- 80.Downward J, Riehl R, Wu L, Weinberg RA. Identification of a nucleotide exchange-promoting activity for p21ras. Proc Natl Acad Sci U S A. 1990;87:5998–6002. doi: 10.1073/pnas.87.15.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wolfman A, Macara IG. A cytosolic protein catalyzes the release of GDP from p21ras. Science. 1990;248:67–9. doi: 10.1126/science.2181667. [DOI] [PubMed] [Google Scholar]
- 82.McCormick F. Signal transduction. How receptors turn Ras on. Nature. 1993;363:15–6. doi: 10.1038/363015a0. [DOI] [PubMed] [Google Scholar]
- 83.Kan NC, Flordellis CS, Mark GE, Duesberg PH, Papas TS. A common onc gene sequence transduced by avian carcinoma virus MH2 and by murine sarcoma virus 3611. Science. 1984;223:813–6. doi: 10.1126/science.6320371. [DOI] [PubMed] [Google Scholar]
- 84.Rapp UR, Todaro GJ. Generation of oncogenic mouse type C viruses: in vitro selection of carcinoma-inducing variants. Proc Natl Acad Sci U S A. 1980;77:624–8. doi: 10.1073/pnas.77.1.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jansen HW, et al. Homologous cell-derived oncogenes in avian carcinoma virus MH2 and murine sarcoma virus 3611. Nature. 1984;307:281–4. doi: 10.1038/307281a0. [DOI] [PubMed] [Google Scholar]
- 86.Jansen HW, Ruckert B, Lurz R, Bister K. Two unrelated cell-derived sequences in the genome of avian leukemia and carcinoma inducing retrovirus MH2. The EMBO journal. 1983;2:1969–75. doi: 10.1002/j.1460-2075.1983.tb01686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moodie SA, Willumsen BM, Weber MJ, Wolfman A. Complexes of Ras.GTP with Raf-1 and mitogen-activated protein kinase kinase. Science. 1993;260:1658–61. doi: 10.1126/science.8503013. [DOI] [PubMed] [Google Scholar]
- 88.Vojtek AB, Hollenberg SM, Cooper JA. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–14. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 89.Warne PH, Viciana PR, Downward J. Direct interaction of Ras and the amino-terminal region of Raf-1 in vitro. Nature. 1993;364:352–5. doi: 10.1038/364352a0. [DOI] [PubMed] [Google Scholar]
- 90.Rodriguez-Viciana P, et al. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature. 1994;370:527–32. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- 91.Adari H, Lowy DR, Willumsen BM, Der CJ, McCormick F. Guanosine triphosphatase activating protein (GAP) interacts with the p21 ras effector binding domain. Science. 1988;240:518–21. doi: 10.1126/science.2833817. [DOI] [PubMed] [Google Scholar]
- 92.Scheffzek K, et al. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–8. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 93.Reddy EP, Reynolds RK, Santos E, Barbacid M. A point mutation is responsible for the acquisition of transforming properties by the T24 human bladder carcinoma oncogene. Nature. 1982;300:149–52. doi: 10.1038/300149a0. [DOI] [PubMed] [Google Scholar]
- 94.Tabin CJ, et al. Mechanism of activation of a human oncogene. Nature. 1982;300:143–9. doi: 10.1038/300143a0. [DOI] [PubMed] [Google Scholar]
- 95.Taparowsky E, et al. Activation of the T24 bladder carcinoma transforming gene is linked to a single amino acid change. Nature. 1982;300:762–5. doi: 10.1038/300762a0. [DOI] [PubMed] [Google Scholar]
- 96.Trahey M, McCormick F. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. 1987;238:542–5. doi: 10.1126/science.2821624. [DOI] [PubMed] [Google Scholar]
- 97.Willingham MC, Pastan I, Shih TY, Scolnick EM. Localization of the src gene product of the Harvey strain of MSV to plasma membrane of transformed cells by electron microscopic immunocytochemistry. Cell. 1980;19:1005–14. doi: 10.1016/0092-8674(80)90091-4. [DOI] [PubMed] [Google Scholar]
- 98.Finegold AA, Schafer WR, Rine J, Whiteway M, Tamanoi F. Common modifications of trimeric G proteins and ras protein: involvement of polyisoprenylation. Science. 1990;249:165–9. doi: 10.1126/science.1695391. [DOI] [PubMed] [Google Scholar]
- 99.Schafer WR, et al. Genetic and pharmacological suppression of oncogenic mutations in ras genes of yeast and humans. Science. 1989;245:379–85. doi: 10.1126/science.2569235. [DOI] [PubMed] [Google Scholar]
- 100.Perucho M, et al. Human-tumor-derived cell lines contain common and different transforming genes. Cell. 1981;27:467–76. doi: 10.1016/0092-8674(81)90388-3. [DOI] [PubMed] [Google Scholar]
- 101.Shih C, Padhy LC, Murray M, Weinberg RA. Transforming genes of carcinomas and neuroblastomas introduced into mouse fibroblasts. Nature. 1981;290:261–4. doi: 10.1038/290261a0. [DOI] [PubMed] [Google Scholar]
- 102.Der CJ, Krontiris TG, Cooper GM. Transforming genes of human bladder and lung carcinoma cell lines are homologous to the ras genes of Harvey and Kirsten sarcoma viruses. Proc Natl Acad Sci U S A. 1982;79:3637–40. doi: 10.1073/pnas.79.11.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Parada LF, Tabin CJ, Shih C, Weinberg RA. Human EJ bladder carcinoma oncogene is homologue of Harvey sarcoma virus ras gene. Nature. 1982;297:474–8. doi: 10.1038/297474a0. [DOI] [PubMed] [Google Scholar]
- 104.Santos E, et al. Malignant activation of a K-ras oncogene in lung carcinoma but not in normal tissue of the same patient. Science. 1984;223:661–4. doi: 10.1126/science.6695174. [DOI] [PubMed] [Google Scholar]
- 105.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nature reviews. Cancer. 2003;3:459–65. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 106.Hall A, Marshall CJ, Spurr NK, Weiss RA. Identification of transforming gene in two human sarcoma cell lines as a new member of the ras gene family located on chromosome 1. Nature. 1983;303:396–400. doi: 10.1038/303396a0. [DOI] [PubMed] [Google Scholar]
- 107.Engelbreth-Holm J, Rothe Meyer A. On the Connection between Erythroblastosis (Hæmocytoblastosis), Myelosis and Sarcoma in Chicken. Acto Pathol. Microbiol. Scand. 1935;12:352–365. [Google Scholar]
- 108.Bister K, Jansen HW. Oncogenes in retroviruses and cells: biochemistry and molecular genetics. Adv Cancer Res. 1986;47:99–188. doi: 10.1016/s0065-230x(08)60199-2. [DOI] [PubMed] [Google Scholar]
- 109.Bister K, Duesberg PH. Structure and specific sequences of avian erythroblastosis virus RNA: evidence for multiple classes of transforming genes among avian tumor viruses. Proc Natl Acad Sci U S A. 1979;76:5023–7. doi: 10.1073/pnas.76.10.5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lai MM, Hu SS, Vogt PK. Avian erythroblastosis virus: transformation-specific sequences form a contiguous segment of 3.25 kb located in the middle of the 6-kb genome. Virology. 1979;97:366–77. doi: 10.1016/0042-6822(79)90347-7. [DOI] [PubMed] [Google Scholar]
- 111.Hihara H, Yamamoto H, Shimohira H, Arai K, Shimizu T. Avian erythroblastosis virus isolated from chick erythroblastosis induced by lymphatic leukemia virus subgroup A. J Natl Cancer Inst. 1983;70:891–7. [PubMed] [Google Scholar]
- 112.Boerner JL, Danielsen A, Maihle NJ. Ligand-independent oncogenic signaling by the epidermal growth factor receptor: v-ErbB as a paradigm. Exp Cell Res. 2003;284:111–21. doi: 10.1016/s0014-4827(02)00096-4. [DOI] [PubMed] [Google Scholar]
- 113.Vennstrom B, Fanshier L, Moscovici C, Bishop JM. Molecular cloning of the avian erythroblastosis virus genome and recovery of oncogenic virus by transfection of chicken cells. J Virol. 1980;36:575–85. doi: 10.1128/jvi.36.2.575-585.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yamamoto T, Hihara H, Nishida T, Kawai S, Toyoshima K. A new avian erythroblastosis virus, AEV-H, carries erbB gene responsible for the induction of both erythroblastosis and sarcomas. Cell. 1983;34:225–32. doi: 10.1016/0092-8674(83)90153-8. [DOI] [PubMed] [Google Scholar]
- 115.Yamamoto T, et al. The erbB gene of avian erythroblastosis virus is a member of the src gene family. Cell. 1983;35:71–8. doi: 10.1016/0092-8674(83)90209-x. [DOI] [PubMed] [Google Scholar]
- 116.Nishida T, et al. Comparison of genome structures among three different strains of avian erythroblastosis virus. Gann. 1984;75:325–33. [PubMed] [Google Scholar]
- 117.Hayman MJ, Royer-Pokora B, Graf T. Defectiveness of avian erythroblastosis virus: synthesis of a 75K gag-related protein. Virology. 1979;92:31–45. doi: 10.1016/0042-6822(79)90212-5. [DOI] [PubMed] [Google Scholar]
- 118.Rettenmier CW, Anderson SM, Riemen MW, Hanafusa H. gag-Related polypeptides encoded by replication-defective avian oncoviruses. J Virol. 1979;32:749–61. doi: 10.1128/jvi.32.3.749-761.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hayman MJ, et al. Identification and characterization of the avian erythroblastosis virus erbB gene product as a membrane glycoprotein. Cell. 1983;32:579–88. doi: 10.1016/0092-8674(83)90477-4. [DOI] [PubMed] [Google Scholar]
- 120.Privalsky ML, Sealy L, Bishop JM, McGrath JP, Levinson AD. The product of the avian erythroblastosis virus erbB locus is a glycoprotein. Cell. 1983;32:1257–67. doi: 10.1016/0092-8674(83)90307-0. [DOI] [PubMed] [Google Scholar]
- 121.Hayman MJ, Beug H. Identification of a form of the avian erythroblastosis virus erb-B gene product at the cell surface. Nature. 1984;309:460–2. doi: 10.1038/309460a0. [DOI] [PubMed] [Google Scholar]
- 122.Privalsky ML, Bishop JM. Subcellular localization of the v-erb-B protein, the product of a transforming gene of avian erythroblastosis virus. Virology. 1984;135:356–68. doi: 10.1016/0042-6822(84)90192-2. [DOI] [PubMed] [Google Scholar]
- 123.Privalsky ML, Ralston R, Bishop JM. The membrane glycoprotein encoded by the retroviral oncogene v-erb-B is structurally related to tyrosine-specific protein kinases. Proc Natl Acad Sci U S A. 1984;81:704–7. doi: 10.1073/pnas.81.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kris RM, et al. Antibodies against a synthetic peptide as a probe for the kinase activity of the avian EGF receptor and v-erbB protein. Cell. 1985;40:619–25. doi: 10.1016/0092-8674(85)90210-7. [DOI] [PubMed] [Google Scholar]
- 125.Decker SJ. Phosphorylation of the erbB gene product from an avian erythroblastosis virus-transformed chick fibroblast cell line. J Biol Chem. 1985;260:2003–6. [PubMed] [Google Scholar]
- 126.Downward J, et al. Close similarity of epidermal growth factor receptor and v-erb-B oncogene protein sequences. Nature. 1984;307:521–7. doi: 10.1038/307521a0. [DOI] [PubMed] [Google Scholar]
- 127.Ullrich A, et al. Human epidermal growth factor receptor cDNA sequence and aberrant expression of the amplified gene in A431 epidermoid carcinoma cells. Nature. 1984;309:418–25. doi: 10.1038/309418a0. [DOI] [PubMed] [Google Scholar]
- 128.Hatanpaa KJ, Burma S, Zhao D, Habib AA. Epidermal growth factor receptor in glioma: signal transduction, neuropathology, imaging, and radioresistance. Neoplasia. 2010;12:675–84. doi: 10.1593/neo.10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mellinghoff IK, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–24. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 130.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 131.Paez JG, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 132.Pao W, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yamamoto H, Toyooka S, Mitsudomi T. Impact of EGFR mutation analysis in non-small cell lung cancer. Lung Cancer. 2009;63:315–21. doi: 10.1016/j.lungcan.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 134.Plowman GD, et al. Ligand-specific activation of HER4/p180erbB4, a fourth member of the epidermal growth factor receptor family. Proc Natl Acad Sci U S A. 1993;90:1746–50. doi: 10.1073/pnas.90.5.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Plowman GD, et al. Molecular cloning and expression of an additional epidermal growth factor receptor-related gene. Proc Natl Acad Sci U S A. 1990;87:4905–9. doi: 10.1073/pnas.87.13.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schechter AL, et al. The neu gene: an erbB-homologous gene distinct from and unlinked to the gene encoding the EGF receptor. Science. 1985;229:976–8. doi: 10.1126/science.2992090. [DOI] [PubMed] [Google Scholar]
- 137.Bargmann CI, Hung MC, Weinberg RA. Multiple independent activations of the neu oncogene by a point mutation altering the transmembrane domain of p185. Cell. 1986;45:649–57. doi: 10.1016/0092-8674(86)90779-8. [DOI] [PubMed] [Google Scholar]
- 138.Slamon DJ, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–82. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 139.Cobleigh MA, et al. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol. 1999;17:2639–48. doi: 10.1200/JCO.1999.17.9.2639. [DOI] [PubMed] [Google Scholar]
- 140.Amin DN, Campbell MR, Moasser MM. The role of HER3, the unpretentious member of the HER family, in cancer biology and cancer therapeutics. Semin Cell Dev Biol. 2010;21:944–50. doi: 10.1016/j.semcdb.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Muraoka-Cook RS, Feng SM, Strunk KE, Earp HS., 3rd ErbB4/HER4: role in mammary gland development, differentiation and growth inhibition. J Mammary Gland Biol Neoplasia. 2008;13:235–46. doi: 10.1007/s10911-008-9080-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kung HJ, Boerkoel C, Carter TH. Retroviral mutagenesis of cellular oncogenes: a review with insights into the mechanisms of insertional activation. Curr Top Microbiol Immunol. 1991;171:1–25. doi: 10.1007/978-3-642-76524-7_1. [DOI] [PubMed] [Google Scholar]
- 143.Maki Y, Bos TJ, Davis C, Starbuck M, Vogt PK. Avian sarcoma virus 17 carries the jun oncogene. Proc Natl Acad Sci U S A. 1987;84:2848–52. doi: 10.1073/pnas.84.9.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li J, Vogt PK. The retroviral oncogene qin belongs to the transcription factor family that includes the homeotic gene fork head. Proc Natl Acad Sci U S A. 1993;90:4490–4. doi: 10.1073/pnas.90.10.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chang HW, et al. Transformation of chicken cells by the gene encoding the catalytic subunit of PI 3-kinase. Science. 1997;276:1848–50. doi: 10.1126/science.276.5320.1848. [DOI] [PubMed] [Google Scholar]
- 146.Curran T, Teich NM. Candidate product of the FBJ murine osteosarcoma virus oncogene: characterization of a 55,000-dalton phosphoprotein. J Virol. 1982;42:114–22. doi: 10.1128/jvi.42.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Vogt PK. The story of Jun. Arch Biochem Biophys. 1995;316:1–4. doi: 10.1006/abbi.1995.1001. [DOI] [PubMed] [Google Scholar]
- 148.Vogt PK. Jun, the oncoprotein. Oncogene. 2001;20:2365–77. doi: 10.1038/sj.onc.1204443. [DOI] [PubMed] [Google Scholar]
- 149.Li Z, Tuteja G, Schug J, Kaestner KH. Foxa1 and Foxa2 are essential for sexual dimorphism in liver cancer. Cell. 2012;148:72–83. doi: 10.1016/j.cell.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nature reviews. Cancer. 2007;7:847–59. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 151.Schaffhausen BS, Roberts TM. Lessons from polyoma middle T antigen on signaling and transformation: A DNA tumor virus contribution to the war on cancer. Virology. 2009;384:304–16. doi: 10.1016/j.virol.2008.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Whitman M, Kaplan DR, Schaffhausen B, Cantley L, Roberts TM. Association of phosphatidylinositol kinase activity with polyoma middle-T competent for transformation. Nature. 1985;315:239–42. doi: 10.1038/315239a0. [DOI] [PubMed] [Google Scholar]
- 153.Cantley LC, et al. Oncogenes and phosphatidylinositol turnover. Annals of the New York Academy of Sciences. 1986;488:481–90. doi: 10.1111/j.1749-6632.1986.tb46580.x. [DOI] [PubMed] [Google Scholar]
- 154.Kaplan DR, et al. Common elements in growth factor stimulation and oncogenic transformation: 85 kd phosphoprotein and phosphatidylinositol kinase activity. Cell. 1987;50:1021–9. doi: 10.1016/0092-8674(87)90168-1. [DOI] [PubMed] [Google Scholar]
- 155.Courtneidge SA, Heber A. An 81 kd protein complexed with middle T antigen and pp60c-src: a possible phosphatidylinositol kinase. Cell. 1987;50:1031–7. doi: 10.1016/0092-8674(87)90169-3. [DOI] [PubMed] [Google Scholar]
- 156.Whitman M, Downes CP, Keeler M, Keller T, Cantley L. Type I phosphatidylinositol kinase makes a novel inositol phospholipid, phosphatidylinositol-3-phosphate. Nature. 1988;332:644–6. doi: 10.1038/332644a0. [DOI] [PubMed] [Google Scholar]
- 157.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–19. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 159.Aoki M, et al. The catalytic subunit of phosphoinositide 3-kinase: requirements for oncogenicity. J Biol Chem. 2000;275:6267–75. doi: 10.1074/jbc.275.9.6267. [DOI] [PubMed] [Google Scholar]
- 160.Sun M, Hart JR, Hillmann P, Gymnopoulos M, Vogt PK. Addition of N-terminal peptide sequences activates the oncogenic and signaling potentials of the catalytic subunit p110alpha of phosphoinositide-3-kinase. Cell Cycle. 2011;10:3731–9. doi: 10.4161/cc.10.21.17920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chaussade C, Cho K, Mawson C, Rewcastle GW, Shepherd PR. Functional differences between two classes of oncogenic mutation in the PIK3CA gene. Biochem Biophys Res Commun. 2009;381:577–81. doi: 10.1016/j.bbrc.2009.02.081. [DOI] [PubMed] [Google Scholar]
- 162.Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci U S A. 2008;105:2652–7. doi: 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Miled N, et al. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317:239–42. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- 164.Shekar SC, et al. Mechanism of constitutive phosphoinositide 3-kinase activation by oncogenic mutants of the p85 regulatory subunit. J Biol Chem. 2005;280:27850–5. doi: 10.1074/jbc.M506005200. [DOI] [PubMed] [Google Scholar]
- 165.Georgescu MM. PTEN Tumor Suppressor Network in PI3K-Akt Pathway Control. Genes Cancer. 2010;1:1170–7. doi: 10.1177/1947601911407325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. J Clin Oncol. 2010;28:1075–83. doi: 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 168.Samuels Y, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 169.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–8. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 170.Li J, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–7. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 171.Feldman ME, Shokat KM. New inhibitors of the PI3K-Akt-mTOR pathway: insights into mTOR signaling from a new generation of Tor Kinase Domain Inhibitors (TORKinibs). Curr Top Microbiol Immunol. 2010;347:241–62. doi: 10.1007/82_2010_64. [DOI] [PubMed] [Google Scholar]
- 172.Sherr CJ. Principles of tumor suppression. Cell. 2004;116:235–46. doi: 10.1016/s0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- 173.Lovat F, Valeri N, Croce CM. MicroRNAs in the pathogenesis of cancer. Semin Oncol. 2011;38:724–33. doi: 10.1053/j.seminoncol.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 174.Morris KV, Vogt PK. Long antisense non-coding RNAs and their role in transcription and oncogenesis. Cell Cycle. 2010;9:2544–7. doi: 10.4161/cc.9.13.12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Druker BJ. Translation of the Philadelphia chromosome into therapy for CML. Blood. 2008;112:4808–17. doi: 10.1182/blood-2008-07-077958. [DOI] [PubMed] [Google Scholar]
- 176.Corcoran RB, Settleman J, Engelman JA. Potential therapeutic strategies to overcome acquired resistance to BRAF or MEK inhibitors in BRAF mutant cancers. Oncotarget. 2011;2:336–46. doi: 10.18632/oncotarget.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Engelman JA, Janne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2008;14:2895–9. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- 178.Hayward WS, Neel BG, Astrin SM. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981;290:475–80. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- 179.Kool J, Berns A. High-throughput insertional mutagenesis screens in mice to identify oncogenic networks. Nature reviews. Cancer. 2009;9:389–99. doi: 10.1038/nrc2647. [DOI] [PubMed] [Google Scholar]
- 180.Berk AJ. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene. 2005;24:7673–85. doi: 10.1038/sj.onc.1209040. [DOI] [PubMed] [Google Scholar]
- 181.Munger K, et al. Mechanisms of human papillomavirus-induced oncogenesis. J Virol. 2004;78:11451–60. doi: 10.1128/JVI.78.21.11451-11460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: unraveling the biology. Trends in biochemical sciences. 2004;29:409–17. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 183.Sullivan CS, Pipas JM. T antigens of simian virus 40: molecular chaperones for viral replication and tumorigenesis. Microbiol Mol Biol Rev. 2002;66:179–202. doi: 10.1128/MMBR.66.2.179-202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Massague J. G1 cell-cycle control and cancer. Nature. 2004;432:298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- 185.Damania B. Oncogenic gamma-herpesviruses: comparison of viral proteins involved in tumorigenesis. Nat Rev Microbiol. 2004;2:656–68. doi: 10.1038/nrmicro958. [DOI] [PubMed] [Google Scholar]
- 186.Hecht JL, Aster JC. Molecular biology of Burkitt's lymphoma. J Clin Oncol. 2000;18:3707–21. doi: 10.1200/JCO.2000.18.21.3707. [DOI] [PubMed] [Google Scholar]
- 187.Smith SM, Anastasi J, Cohen KS, Godley LA. The impact of MYC expression in lymphoma biology: beyond Burkitt lymphoma. Blood Cells Mol Dis. 2010;45:317–23. doi: 10.1016/j.bcmd.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 188.Klapproth K, Wirth T. Advances in the understanding of MYC-induced lymphomagenesis. Br J Haematol. 2010;149:484–97. doi: 10.1111/j.1365-2141.2010.08159.x. [DOI] [PubMed] [Google Scholar]
- 189.Corvi R, Savelyeva L, Schwab M. Patterns of oncogene activation in human neuroblastoma cells. J Neurooncol. 1997;31:25–31. doi: 10.1023/a:1005721027709. [DOI] [PubMed] [Google Scholar]
- 190.Taylor MD, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123:465–72. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Swartling FJ. Myc proteins in brain tumor development and maintenance. Ups J Med Sci. 2012 doi: 10.3109/03009734.2012.658975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Scheurlen WG, et al. Molecular analysis of childhood primitive neuroectodermal tumors defines markers associated with poor outcome. J Clin Oncol. 1998;16:2478–85. doi: 10.1200/JCO.1998.16.7.2478. [DOI] [PubMed] [Google Scholar]
- 193.Huang PH, Xu AM, White FM. Oncogenic EGFR signaling networks in glioma. Sci Signal. 2009;2:re6. doi: 10.1126/scisignal.287re6. [DOI] [PubMed] [Google Scholar]
- 194.Zavoral M, Minarikova P, Zavada F, Salek C, Minarik M. Molecular biology of pancreatic cancer. World J Gastroenterol. 2011;17:2897–908. doi: 10.3748/wjg.v17.i24.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Morris J.P.t., Wang SC, Hebrok M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nature reviews. Cancer. 2010;10:683–95. doi: 10.1038/nrc2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Perez-Mancera PA, Tuveson DA. Physiological analysis of oncogenic K-ras. Methods Enzymol. 2006;407:676–90. doi: 10.1016/S0076-6879(05)07053-9. [DOI] [PubMed] [Google Scholar]
- 197.Poulikakos PI, Rosen N. Mutant BRAF melanomas--dependence and resistance. Cancer Cell. 2011;19:11–5. doi: 10.1016/j.ccr.2011.01.008. [DOI] [PubMed] [Google Scholar]