Abstract
The biased usage of synonymous codons affects translational efficiency of genes. We studied codon usage patterns of genes that are perfectly conserved at the amino acid level among three important mosquito vector species: Aedes aegypti (vector of dengue virus), Anopheles gambiae (vector of malaria) and Culex quinquefasciatus (vector of lymphatic filariasis and West Nile Virus). Although these proteins have same amino acid sequences, non-random usage of synonymous codons is evident among the orthologous genes. The coding sequences of these genes were simulated to generate random mutation sites to be further investigated for patterns of codon bias. It was found that codon usage bias is significantly higher in genes that represented perfectly conserved proteins than genes where variation was apparent at the amino acid sequence. Our results suggest that genes coding for perfectly conserved proteins are highly biased with optimized codons and may be under stringent translational selection in these vector species.
Keywords: Vector mosquitoes, Selection, Codon bias, Translational efficiency, Codon context
1. Introduction
Usage of synonymous codons is non-random during decoding of genes to proteins. As a result, specific codons are more frequently represented than other synonymous codons during translation of genes. This is commonly referred to as codon usage bias or simply codon bias. Whereas codon bias is widespread both in prokaryotes and eukaryotes, the extent of bias is variable within and between species (Plotkin and Kudla 2011). Various factors such as expression level, gene length, composition bias (%G+C content and GC skew), recombination rates, and RNA stability are likely to influence the extent of codon bias of genes (Akashi 1997; Powel and Moriyama 1997; Moriyama and Powell 1998; Powell et al. 2003). Variations in codon optimization among genes provide differential efficiency in the translation of genes. The selection associated with this process is commonly termed as ‘translational selection’. Numerous reports have shown links between codon bias and selection for translational efficiency (reviewed in Hershberg and Petrov 2008). It has been observed that the extents of biased usage of codons are often correlated with the cognate tRNA gene copies in some species including mosquitoes (Rocha 2004, Behura et al. 2010; Behura and Severson 2011). Our recent studies have shown that genes that are related to translation, energy metabolism and carbohydrate metabolism may be under translational selection in Aedes aegypti and Anopheles gambiae mosquitoes (Behura and Severson 2011).
The Anopheles gambiae, Aedes aegypti and Culex quinquefasciatus mosquitoes are major vectors malaria, dengue and lymphatic filariasis respectively. The genome sequences of the three mosquito species have been completed (Holt et al., 2002; Nene et al. 2007 and Arensburger et al. 2010). These projects (www.vectorbase.org) have provided opportunities to better understand structure, function and evolution of mosquito genes. In spite of recent progresses in mosquito genomics, many critical aspects of gene structure and evolution are not well studied in vector mosquitoes (Severson and Behura, 2012). In this study, we analyze the codon bias patterns of genes that code for perfectly conserved proteins among the three mosquito species. This is important because it may provide seminal information on selection of perfectly conserved proteins pertaining to translational efficiency. Here, we conduct genome-wide search to identify genes that code for perfectly conserved proteins among the three species and investigate the patterns of codon usage patterns of the orthologous genes. We perform simulation of coding sequences of these genes and show that simulated gene sequences that are perfectly conserved at the protein level are associated with significantly higher codon usage bias than genes that lack such conservation. Our results suggest that genes coding for perfectly conserved proteins may be under differential translational selection compared to genes that lack such evolutionary conservation at the protein level.
2. Materials and Methods
2.1 Data
The orthologous relationships, percentage sequence identity as well as gene length of all predicted coding genes of the three mosquito species were downloaded from VectorBase (www.vectorbase.org) via data depository at Biomart. From these, genes were filtered using the criteria that they are 1:1 orthologs among the three species and that they are 100% similar at the protein level. The D. melanogaster orthologs (1:1) of these genes, if present, were also obtained from Biomart for use as out-group in phylogenetic analyses.
2.2 Codon bias analysis
The codon counts as well as the codon context analyses of these genes were determined for each species using the Anaconda Software (Moura et al. 2007). Because the amino acid sequences are exactly the same among the three species for these genes, variation in cumulative frequencies of codons directly relates to the biased usage of synonymous codons among species. The codon frequency was calculated as the proportion of the codon to the total number of codons represented by the orthologous genes. For comparison of codon contexts, frequency of each codon context was compared with the expected frequency. The expected frequency was calculated from the total counts of each amino acid pair divided by the product of codon degeneracy of the two amino acids. As the amino acid sequences are conserved, the expected frequencies of codon sequences for each amino acid pair remains unchanged among the three species. Thus, the ratio of observed to expected frequencies of codon contexts shows the biased usage of synonymous codon pairs among the orthologous genes.
The synonymous codon usage order (SCUO) was used as an index of measurement of biased usage of codons. SCUO index is determined by comparing the observed entropy of each amino acid site to the expected value, where the expected value of the entropy assumes random usage of all synonymous codons of the amino acid (Zeeberg 2002). The SCUO index is suitable for performing comparative estimates of synonymous codon usage bias within and between genomes as demonstrated by Wan et al. 2004. We used CodonO software (Angellotti et al. 2007), available at http://www.sysbiology.org/CodonO, to locally calculate synonymous codon usage order (SCUO) of each gene. The effective number of codons (ENC) and base composition of codon sequences including GC and GC3 contents were determined by using ‘codonw’ program (http://sourceforge.net/projects/codonw/).
The paralogous copies of the A. aegypti genes were obtained from VetcorBase. The expression data generated in A. aegypti during an earlier study (Behura et al. 2011) was used. The microarray expression data is publicly available at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE16563. The UTR (untranslated regions) sequences of genes were also downloaded from VectorBase. To compare the regulatory elements of UTRs, we chose 4 genes (out of the 10 genes studied) where both the 5′ and 3′ UTRs have been identified among the three species. The putative regulatory elements in the UTR sequences were predicted by the UTRScan tool (http://itbtools.ba.itb.cnr.it/utrscan).
2.3 Codon sequence simulation
The coding sequences of the individual orthologous genes were concatenated (5,511 bp sequence). Prior to concatenating the gene sequences, the topologies of individual gene trees were compared. Individual trees were generated by BayesPhylogenies 1.1 (Pagel and Meade 2004) using general time reversible model and the D. melanogaster as the out-group species. Total 10,000 iterations were performed. The topology information of the individual trees was assessed by BayesTree 1.3 program (http://www.evolution.reading.ac.uk/BayesTrees.html). In each gene tree we consistently observed 0.99 as the probability value of the most frequent topology. Similar topology was obtained from individual trees as well as the tree generated from the concatenated genes which agreed with the known phylogeny among mosquitoes and Drosophila (Arensburger et al., 2010).
The concatenated sequences of each of the three species were used to simulate the coding sequences. Simulation was performed using the ‘EvolveAGene 3’ program (Hall 2008). The average number of changes per site of the orthologous genes was determined by using DnaSP (Rozas et al. 2007) that was used as the average branch length in generating the genealogy of the simulated sequences. The 0.016 probability value for selection of amino acid replacement is considered as realistic intensity of such selection as suggested by Carroll et al. (2007). Moreover, the dN/dS of pair-wise comparison of all the 1:1:1 orthlogous genes identified from the three genome sequences (www.vectorbase.org) also shows a similar median value (0.0156) among the three mosquito species. Although this value will change among unrelated species, the evolved gene sequences from the simulation closely mimic orthologous genes of closely related specie where dN/dS approximates to 0.016 (Carol et al. 2007).
The sequence relatedness of the simulated sequences with that of the observed data (the concatenated codon sequences of the 10 genes of the three species) was confirmed by phylogenetic methods implemented in MEGA4 (Tamura et al. 2004). The phylogeny was inferred using the Neighbor-Joining method (Saitou and Nei 1987). The evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al. 2004) and they were measured in the units of the number of base substitutions per site. The multiple alignments were performed by Muscle program (Edgar 2010). DNA to protein translation of sequences was also performed by MEGA4. The simulated sequences were separated into two groups; 1) sequences that translated to the same amino acid sequences because of introduction of silent changes and 2) sequences that translated to amino acid sequences where site variation was produced. Then we compared the codon usage bias of sequences that were perfectly conserved at the amino acid level with codon bias of genes that were not perfectly conserved at the amino acid level.
We also performed another simulation of the coding sequences wherein the codon positions were randomized. By doing this, we simulated the genes to have different composition of A, T, G and C at 1st, 2nd and 3rd positions of codons. The codon position shuffling was performed using the R statistical package ‘seqinr’ (http://cran.r-project.org/web/packages/seqinr).
2.4. Evolutionary analysis of codon bias
We used BayesTraits Continous model (Pagel 1999) to perform regression between species phylogenies and the variation of codon bias (as variable traits of ENC and GC3 content) of the orthologous genes. The ENC and GC3 values were used as trait values against one hundred trees phylogenies generated by BayesPhylogenies. The trees were rooted at D. melanogaster gene. This particular analysis was performed with concatenated sequences of 9 genes (not the 10 genes) as one of the mosquito genes lacked orthology in D. melanogaster. The tree scaling parameter ‘lambda’ was made fixed to either 0 or 1 to test whether codon bias traits varied in correlated manner with trees. The maximum likelihood method implemented in the Continuous program was used to infer whether lambda = 0 or lambda = 1 yielded significant differences in the likelihoods. This was determined by likelihood ratio tests between the two models.
2.5 Statistical analysis
All the statistical analyses were performed using R statistical packages. The codon usage bias of perfectly conserved proteins and of proteins with sequence variation generated from simulation analyses was compared by Tukey’s test in R (http://cran.r-project.org/web/packages/DTK/index.html). The default value of 0.05 was used as the threshold significance level of the test. The Poisson logistic regression was performed using ‘glm’ function in R. The ‘boot’ package was used to perform bootstrap analysis of spearman correlation between codon bias and GC contents. The likelihood ratio tests were performed by ‘lmtest’.
Results
3.1. Differential codon bias of orthologous genes coding for perfectly conserved proteins
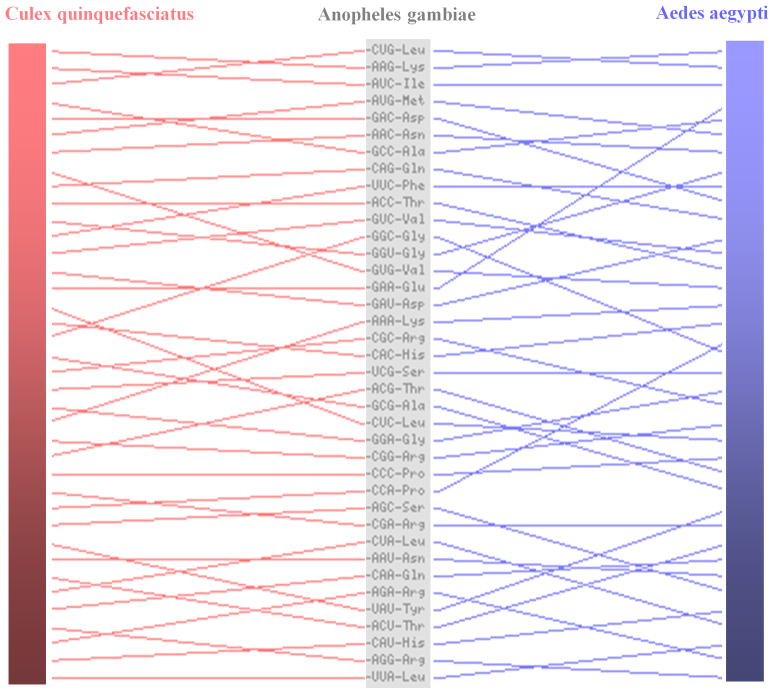
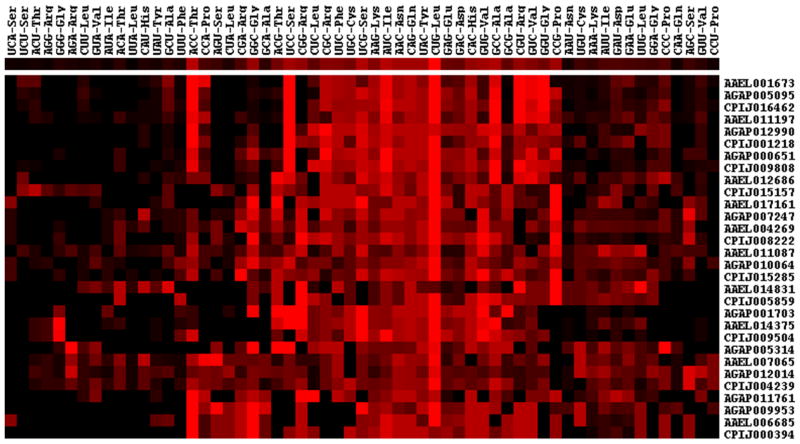
By analyzing a total of 47,475 predicted coding genes (AgamP3 + AaegL1 + CpipJ1 official gene sets) of the three mosquito species, only 30 genes were identified in 10 ortholog (1:1:1) trios that have same amino acid sequences among three species (Table 1). Multiple alignments of the codon sequences of these ortholog trios show variation in codons in spite of perfect conservation at the amino acid sequence level (Figure 1). Because the translated amino acid sequences of these genes are same, this result indicates that the synonymous codons are used with varying biases among the three species. For example, CUG is the most frequent codon (rank-1) for coding Leu residues of these genes in A. gambiae but it is the second and third most used codon (rank-2 and rank-3) in A. aegypti and C. quinquefasciatus genes, respectively (Figure 2). The variation is not due to difference in the amount of leucine amino acids in these proteins (these proteins are perfectly conserved) but due to usage bias of synonymous codons among these mosquitoes. Hierarchical clustering based on rank order correlation of codons frequencies further shows that nearly 50% of codons are coordinately used in a biased manner in these genes (Figure 3). The codon sequences of the genes further show a characteristic feature of GC percentage among the 1st, 2nd and 3rd positions of codons in these genes. The 3rd position of codons has higher GC percent than the 1st and 2nd positions in each gene (Supplementary Figure 1). Because synonymous codons are mostly variable at the 3rd position, the elevated GC content is possibly associated with biased usages of synonymous codons. The codon context frequencies are also variable among the orthologous genes (Supplementary Figure 2), indicating that the conserved proteins are biased in the usage of synonymous codon pairs. This is evident from differential usage of synonymous codon pairs for each neighboring amino acid pair of the conserved proteins among the three species (Supplementary Table 1).
Table 1.
List of genes analyzed in the study. They are 1:1:1 orthologous genes that are predicted to code for perfectly conserved proteins among the three mosquitoes.
| Gene Name | Ortholog Trios (Agam/Aaeg/Cqui) |
|---|---|
| Actin | AGAP005095/ AAEL001673/ CPIJ016462 |
| LSM protein (RNA-binding protein) | AGAP005314/ AAEL017161/ CPIJ015157 |
| EF hand (calcium-binding protein) | AGAP007247/ AAEL004269/ CPIJ008222 |
| RPS23 | AGAP012990/ AAEL012686/ CPIJ001218 |
| Adaptor protein 2 | AGAP001703/ AAEL014375/ CPIJ009504 |
| Actin | AGAP000651/ AAEL011197/ CPIJ009808 |
| RNA polymerase (rpb10) | AGAP011761/ AAEL014831/ CPIJ005859 |
| ARF (ADP ribosylation factor) | AGAP012014/ AAEL007065/ CPIJ004239 |
| G-protein | AGAP009953/ AAEL006685/ CPIJ000394 |
| RNA polymerase (rpb5) | AGAP010064/ AAEL011087/ CPIJ015285 |
Figure 1.
Multiple alignments of amino acid sequences (top) and codon sequences (bottom) of orthologous genes.
Figure 2.
Rank order displacement map of codon usage frequency (cumulative) of the orthologous genes among the three mosquitoes. The rank order of codon usages is shown in decreasing order from top to bottom for A. gambiae and then compared with A. aegypti and C. quinquefasciatus genes (the lines indicate the relative rank order positions).
Figure 3.
Hierarchical clustering pattern of codon usages among 30 genes coding perfectly conserved proteins. The codons are listed in rows and genes are in columns. The clustering is based on rank order of relative synonymous codon usage (RSCU) of each codon in each gene. Red indicates preferred usage of a codon in the gene and black indicates rare usage of codon in the corresponding gene.
3.2. Base composition and codon bias
We performed sequence simulation of the orthologous genes by reshuffling codon positions (1st, 2nd and 3rd positions of codons) of the orthologous genes. The purpose of this simulation was to determine the effect of base compositions on the biased usage the synonymous codons. The results of bootstrap correlation analysis show significant correlation (Spearman) between GC content at the third position and the effective number of codons (ENC, a non-directional measure of codon bias) among the simulated sequences (Rho = 0.88, p < 0.01 and 95% C.I. ~ 0.8 – 1.0).
The ENC being a non-directional measure of codon bias, it is suitable for comparing differential A/T versus G/C content of synonymous positions to determine the extent of mutational bias and codon bias among genes (Vicario et al. 2007). To determine if ENC of genes coding perfectly conserved proteins among the three mosquito species is affected by differential G/C- and A/T- composition of the third positions of codons, we performed generalized linear model fitting of each base at the third position (A3/ G3/ C3/ T3) as predictor variable with ENC as the dependent variable by Poisson logistic regression (Table 2). Results show that different bases at the 3rd positions have differential effect on ENC of the orthologous genes among the three mosquito species. However, it shows that G and/or C at 3rd positions have negative effect on ENC indicating that increase in the G/C content at 3rd position will reduce ENC with differential magnitude (as indicated by the co-efficient of regression). Off note, decrease of ENC indicates increase of codon bias (ENC values ranges from 61 to 20 where ENC = 61 suggests no bias of codon usage and ENC = 20 means maximum bias of codon usage). Thus, the regression analysis clearly suggests that GC content at the silent positions positively influences biased usage of synonymous codons of the genes among the three species albeit with differential magnitude.
Table 2.
Logistic regression estimates of effect of base composition at silent positions on codon bias (effective number of codons) of genes.
| Estimate | std. error | z-value | Pr(>|z|) | |
|---|---|---|---|---|
| Aedes aegypti | ||||
| T3s | 0.021064 | 0.003002 | 0.702 | 0.483 |
| C3s | −0.499102 | 0.027579 | −18.097 | <2e-16 |
| A3s | 0.395321 | 0.025629 | 15.424 | <2e-16 |
| G3s | 0.006531 | 0.000433 | 0.291 | 0.771 |
| Culex quinquefasciatus | ||||
| T3s | 0.03675 | 0.00295 | 1.244 | 0.214 |
| C3s | −0.58019 | 0.02687 | −21.591 | <2e-16 |
| A3s | 0.53595 | 0.02752 | 19.473 | <2e-16 |
| G3s | −0.25906 | 0.0224 | −11.566 | <2e-16 |
| Anopheles gambiae | ||||
| T3s | 0.26042 | 0.03502 | 7.437 | 1.03e-13 |
| C3s | −0.4251 | 0.03404 | −12.488 | <2e-16 |
| A3s | 0.61248 | 0.03096 | 19.784 | <2e-16 |
| G3s | −0.31529 | 0.02577 | −12.234 | <2e-16 |
Our previous study on global codon bias pattern between A. aegypti and A. gambiae suggested that A. gambiae genes are associated with relatively higher codon bias than A. aegypti (Behura et al. 2011). The correlation (Spearman ‘Rho’) between global GC composition of the coding sequences and ENC is 0.72 in A. gambiae compared to only 0.55 and 0.6 of C. quinquefasciatus and A. aegypti genome respectively. To test that higher codon bias may be related to higher global G/C bias, we performed likelihood ratio test of two nested models where the general hypothesis relied that all the four bases at the 3rd position affected ENC (ENC ~ A3s + T3s + G3s + C3s) whereas the special model hypothesized that only A and T affected the codon bias (ENC ~ A3s + T3s). The results of these model testing showed that the log-likelihoods of the second assumption (A/T affecting ENC) are significantly higher (p < 2.2e−16, df = 2) than that of the first assumption in each of the three species. However, the Chi-square values of log-likelihood ratios (assuming asymptotic distribution) are more than two-fold higher in the Culicinae species (A. aegypti and C. quinquefasciatus) compared to the Anophelinae. This suggests that effect of A/T changes in the third positions of codons is relatively more prominent in the Culicinae than in Anophelinae. This may shift the balance between codon bias versus mutation bias more towards codon bias in A. gambiae than in A. aegypti and in C. quinquefasciatus.
3.3. Elevated codon usage bias of genes coding for perfectly conserved proteins
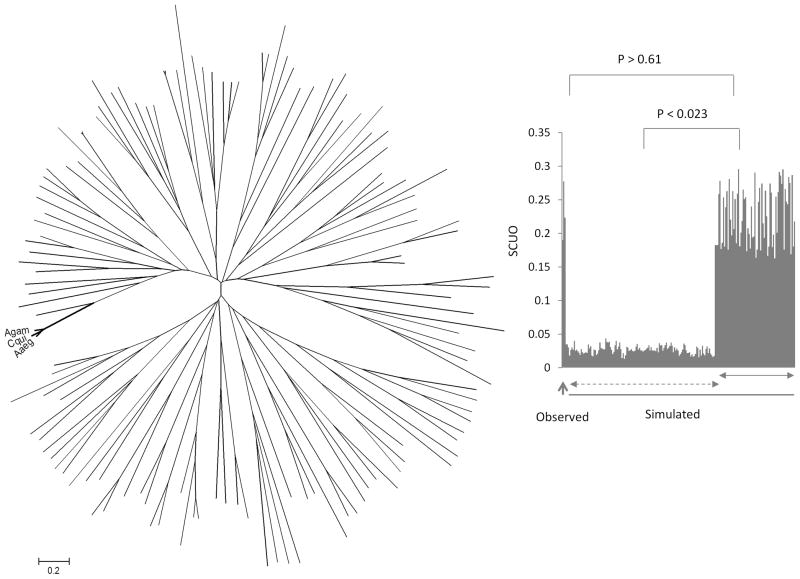
The simulation results of coding sequences show that genes that are perfectly conserved at amino acid level are significantly different in the synonymous codon usage orders (SCUO) from that of genes where protein products are not perfectly conserved. The Tukey’s test showed statistically significant (p < 0.023) codon bias between the two gene groups rejecting null hypothesis that both groups have similar codon usage (Figure 4). The perfectly conserved proteins were found to have higher codon usage bias (> 10 fold) than proteins lacking such property. There is, however, no significant difference in codon usage bias of perfectly conserved proteins generated from the simulation data and the observed data (p = 0.61). Thus, these results suggest that perfectly conserved proteins are relatively more biased in codon usage than genes that lack such property.
Figure 4.
Phylogenetic (Neighbor-joining) tree analysis of observed and simulated coding sequences of the genes. The observed genes are indicated by Agam, Cqui and Aaeg representing the three mosquitoes. All other sequences represent simulated coding genes generated by the EvolveAgene 3 program. The distance scale is shown at the bottom. On the right: Comparison of codon bias (SUCO values on the Y-axis) of observed (shown by an upward arrow) and the simulated sequences. The sequences shown by a solid horizontal arrow are the simulated coding sequences that represented amino acid sequences without any variation. The simulated sequences shown by a dashed horizontal arrow coded for proteins where variation in the amino acid sequences were found. The significance values of differences in codon bias are indicated above the graph.
Studies have shown that genes that are highly biased in codon usages are expressed at higher level than genes with low codon bias in A. aegypti and A. gambiae mosquitoes (Behura and Severson 2011). Thus, based on our simulation results described above, we hypothesized that genes coding for perfectly conserved protein should have elevated expression than genes that lack such property. To test this hypothesis, we identified paralogous copies of the A. aegypti genes studied in this analysis (Table 1) and compared the expression levels of genes that code for perfectly conserved proteins in the other two mosquito species and paralogous genes that lack such property (Supplementary Table 2). This was investigated using the genome-wide microarray expression data generated for A. aegypti in an earlier study (Behura et al. 2011). We found that A. aegypti genes coding for conserved proteins across the three mosquito species have consistently higher expression than the paralogous genes whose protein products are not perfectly conserved across the three genomes. In some cases, we found nearly 2-fold higher expression of gene (i.e. AAEL004269) coding perfectly conserved protein (calcium-binding protein) than its paralogous copy (i. e. AAEL009327) whose translational product is not perfectly conserved among the three mosquitoes.
We also investigated whether the genes whose protein products are perfectly conserved among the species may contain some common UTR (untranslated regions) regulatory elements that may dictate elevated expression of these genes. In spite of the fact that the official genes of these mosquitoes are not comprehensively annotated for the UTR sequences, we were able to identify 4 genes (out of the 10 genes studied) where both the 5′ and 3′ UTRs have been identified among the three species. We used these UTR sequences to bioinformatically identify putative regulatory UTR elements among the orthologous genes (Supplementary Table 3). We found several such elements when searched against a public database of UTR elements using UTRScan tool (http://itbtools.ba.itb.cnr.it/utrscan). However, these elements are not common among the orthologous genes suggesting that regulation by UTR elements is unlikely to affect translational selection of these specific genes across the three species.
3.4. Phylogenetic relationship of codon bias
We asked whether the observed bias of codon usages and its association with GC3 content is an evolutionary effect of phylogeny of the mosquito species. To determine that we used Drosophila melanogaster as an outgroup species to generate Bayesian phylogenies of the orthologous genes (concatenated). A total number of 100 trees (rooted at D. melanogaster) were analyzed by maximum likelihood method implemented in continuous regression model (Pagel 1999) to test dependence/non-dependence of traits on phylogenies. The log likelihoods of the individual trees were estimated at two values (0 and 1) of the tree-scaling parameter ‘lambda’ (Supplementary Table 4). The parameter lambda reveals whether the phylogeny correctly predicts the patterns of covariance among species on a given trait. When the observed trait evolved independent of species phylogeny, this parameter takes the value 0. On the other hand, when traits are evolving according to the tree topology, lambda takes the value of 1. The data shown in Supplementary Table 4 clearly shows that the likelihood estimates are consistently higher among the trees when lambda = 0 compared to lambda = 1. We performed likelihood ratio test between the two model estimates but found no statistically significant difference in the log-likelihoods in any tree. This suggested that codon bias and GC3 content may not be solely affected by species phylogeny. Whether there is a bias in this intrinsic balance in codon bias dependency with species phylogeny, two intermediate kappa values close to 0 and 1 (0.1 and 0.9 respectively) were used to further estimate the log-likelihoods. The intermediate values of lambda are expected to overestimate the variance of traits with species phylogeny (Pagel 1999). Thus, we expected that the likelihood at lambda = 0.1 to be higher than the corresponding estimates at lambda = 0. Also, likelihoods at lambda = 0.9 should be more than the estimates at lambda = 1. The data in Supplementary Table 4 shows that the estimates are elevated when lambda shifts from 0 to 0.1 but such expectation is not observed when lambda shifts from 1 to 0.9. This indicates that covariance of codon bias is biased more towards non-dependency than dependency with the species phylogeny of these mosquitoes.
Discussion
Our current investigation addresses an important selection process (translational selection) that may regulate the coding efficiency of perfectly conserved proteins among closely related species. An earlier study by us showed that higher codon bias was associated with higher expression of genes in mosquitoes suggesting possible role of codon usage bias in the selection of translational efficiency of mosquito genes (Behura and Severson 2011). Elevated codon usage bias is known to have association with translational selection based on tRNA analysis (Rocha 2004). How perfectly conserved proteins are selected for translational efficiency is also an important question. In this report, we show that genes coding for perfectly conserved proteins are highly biased in codon usage patterns. Whether translational selection is a cause or consequence of codon usage bias of such genes has not been explicitly tested. However, we have shown indirect evidences based on expression data of A. aegypti that elevated codon bias is associated with higher expression level of genes. The paralogous genes whose protein sequences are not perfectly conserved among the three mosquito species are consistently expressed at lower level than genes coding for perfectly conserved proteins. Our sequence simulation results suggest that the latter category of genes (coding perfectly conserved proteins) is associated with elevated codon usage bias than genes that lack this property. This indicates that optimized codons may result in increase in translational efficiency of these genes. In mosquitoes, tRNA copies show significant correlation with codon usage bias that suggests a role of codon bias in modulating translational efficiency of mosquito genes (Behura and Severson 2011). However, it is possible that translational selection may be achieved by interplay of other factors along as well. For example, persistent negative correlation between codon bias and intron contents has been observed suggesting a possible evolutionary link between the two factors in eukaryotic species (Vinogradov 2001).
Our study further shows that usage bias of codon pair sequences (codon context) may have an influential role on translational efficiency of these genes. Codon context is an important feature of codon selection as the neighboring codons correspond to the A-site (aminoacylation site) and P-sites (peptide synthesis site) of the ribosome during translation (Buckingham 1994; Irwin et al. 1995; Moura et al. 2007, Tats et al. 2008).
The use of SCUO as a measure of non-randomness in the usage of synonymous codons is appropriate for this study. SCUO represents an accurate estimation of codon usage bias of genes and also a useful index for comparison of codon bias within and between genomes (Wan et al. 2004, Angellotti et al. 2007). The A. aegypti genes showed relatively lower codon usage bias than A. gambiae and C. quinquefasciatus orthologs. The evolution of codon usage bias is most likely independent of the three mosquito species. The evolutionary divergence time is estimated at 145–160 million years between Anophelinae and Culicinae and 52–54 million years between Culex and Aedes genera (Arensburger et al. 2010). However, phylogenetic inertia of codon bias is often described in some species including fruit flies (Heger et al. 2007).
Our data shows significant differences in the synonymous codon usage bias of genes that translate to perfectly conserved amino acid sequences from genes that code for proteins with variation in the amino acid sequence. The coding sequences that represented perfectly conserved proteins contain most of the optimized codons identified from whole genome analysis (Behura and Severson 2011) unlike the coding sequences where sequence variation was observed at the amino acid level (data not shown). The elevated usage of optimized codons by genes coding for perfectly conserved proteins further suggests translational efficiency of such genes. Whether translational selection of perfectly conserved proteins has any functional significance in these vector species is yet to be discovered. However, accumulating evidences suggest that de-optimization of codon sequences without altering the protein sequences may have significant effect on organisms such as viruses (Mueller et al., 2006; Coleman et al., 2008). Our recent work on gene expression of A. aegypti to dengue virus infection also suggested that several intrinsic features such as intron, gene positions and sequence evolution along with codon usage bias have significant effect on transcriptional responsiveness of the mosquito to infection (Behura and Severson 2012). It was found from this study that codon bias has the most profound effect on gene expression to dengue infection compared to any other sequence features.
The role of base substitution on biased usage of synonymous codons is explained differentially by neutralist and selectionist theories (Hey 1999). While neutralist theory explains base substitution of coding sequences on the basis of directional mutation pressure and assumes neutral effect on such changes, the selectionist theory claims role of selective forces that shape the base substitutions leading to codon bias. Our results from simulated sequences (codon position reshuffling) show that A/T and G/C biased synonymous positions are significantly different and that G/C significantly affects the extent of codon usage bias of the genes. However, our logistic regression analysis clearly shows that A/T and G/C composition have significantly different effect on the effective number of codons of the genes. According to the selection-mutation-drift theory, codon bias of genes is evolved as a result of balance between the forces of selection and mutation within finite populations (Bulmer 1991). In this model, balance among mutation pressure, genetic drift, and weak selection acts in favor of translationally superior codons. Thus, we think that the interplay between mutational process and selective forces modulate the evolution of synonymous codons among the three mosquitoes. In many cases, codon bias is primarily affected by mutational processes and only secondarily by selective forces acting on coding sequences (Chen et al. 2004, Rao et al. 2011). In general, mutations from a rare codon to a frequent codon are beneficial for the organism, whereas mutations in the opposite direction are likely to be deleterious. Assuming this mechanism to be true in the mosquito species, it is likely that codon optimization of perfectly conserved proteins is beneficial for these mosquitoes. Although the phenotypic benefit of this selection is not known in mosquitoes, literature evidence suggests that biased usage of codons may have fitness benefit on the organism (such as alcohol tolerance of fruit flies) (Carilini 2004). Although, further studies are required to determine the functional role of codon selection in mosquitoes, the results of the current study highlights the evolutionary significance of genetic variation of perfectly conserved proteins in closely related species.
Supplementary Material
Highlights.
Genes coding for perfectly conserved proteins among three mosquito species are studied.
These genes have higher codon bias than genes coding for variable protein sequences.
Base composition has significant role in translational selection of these genes
Codon bias of these genes is unlikely to be related to species evolution
The neutralist and selectionist theories explain the observed codon bias pattern
Acknowledgments
This work was possible because of the undergraduate mentoring program (the “Building Bridges Mentoring Program”) at the University of Notre Dame. The research was supported in part from a grant from NIH-NIAID (RO1-AI059342) to DWS and an undergraduate scholarship from the ‘Gates Millennium Scholars Program’ to OR. The authors are thankful to two anonymous reviewers whose suggestion has helped improve the paper.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akashi H. Codon bias evolution in Drosophila. Population genetics of mutation-selection drift. Gene. 1997;205:269–278. doi: 10.1016/s0378-1119(97)00400-9. [DOI] [PubMed] [Google Scholar]
- Angellotti MC, Bhuiyan SB, Chen G, Wan XF. CodonO: codon usage bias analysis within and across genomes. Nucleic Acids Res. 2007;35:W132–6. doi: 10.1093/nar/gkm392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arensburger P, Megy K, Waterhouse RM, Abrudan J, Amedeo P, Antelo B, Bartholomay L, Bidwell S, Caler E, Camara F, Campbell CL, Campbell KS, Casola C, Castro MT, Chandramouliswaran I, Chapman SB, Christley S, Costas J, Eisenstadt E, Feschotte C, Fraser-Liggett C, Guigo R, Haas B, Hammond M, Hansson BS, Hemingway J, Hill SR, Howarth C, Ignell R, Kennedy RC, Kodira CD, Lobo NF, Mao C, Mayhew G, Michel K, Mori A, Liu N, Naveira H, Nene V, Nguyen N, Pearson MD, Pritham EJ, Puiu D, Qi Y, Ranson H, Ribeiro JM, Roberston HM, Severson DW, Shumway M, Stanke M, Strausberg RL, Sun C, Sutton G, Tu ZJ, Tubio JM, Unger MF, Vanlandingham DL, Vilella AJ, White O, White JR, Wondji CS, Wortman J, Zdobnov EM, Birren B, Christensen BM, Collins FH, Cornel A, Dimopoulos G, Hannick LI, Higgs S, Lanzaro GC, Lawson D, Lee NH, Muskavitch MA, Raikhel AS, Atkinson PW. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science. 2010;330:86–88. doi: 10.1126/science.1191864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behura SK, Severson DW. Coadaptation of isoacceptor tRNA genes and codon usage bias for translation efficiency in Aedes aegypti and Anopheles gambiae. Insect Mol Biol. 2011;20:177–187. doi: 10.1111/j.1365-2583.2010.01055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behura SK, Severson DW. Intrinsic features of Aedes aegypti genes affect transcriptional responsiveness of mosquito genes to dengue virus infection. Infect Genet Evol. 2012 doi: 10.1016/j.meegid.2012.04.027. http://dx.doi.org/10.1016/j.meegid.2012.04.027. [DOI] [PMC free article] [PubMed]
- Behura SK, Stanke M, Desjardins CA, Werren JH, Severson DW. Comparative analysis of nuclear tRNA genes of Nasonia vitripennis and other arthropods, and relationships to codon usage bias. Insect Mol Biol. 2010;19(Suppl 1):49–58. doi: 10.1111/j.1365-2583.2009.00933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham RH. Codon context and protein synthesis: enhancements of the genetic code. Biochimie. 1994;76:351–354. doi: 10.1016/0300-9084(94)90108-2. [DOI] [PubMed] [Google Scholar]
- Bulmer M. The selection-mutation-drift theory of synonymous codon usage. Genetics. 1991;129:897–907. doi: 10.1093/genetics/129.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlini DB. Experimental reduction of codon bias in the Drosophila alcohol dehydrogenase gene results in decreased ethanol tolerance of adult flies. J Evol Biol. 2004;17:779–785. doi: 10.1111/j.1420-9101.2004.00725.x. [DOI] [PubMed] [Google Scholar]
- Carroll H, Beckstead W, O’Connor T, Ebbert M, Clement M, Snell Q, McClellan D. DNA reference alignment benchmarks based on tertiary structure of encoded proteins. Bioinformatics. 2007;23:2648–2649. doi: 10.1093/bioinformatics/btm389. [DOI] [PubMed] [Google Scholar]
- Chen SL, Lee W, Hottes AK, Shapiro L, McAdams HH. Codon usage between genomes is constrained by genome-wide mutational processes. Proc Natl Acad Sci U S A. 2004;101:3480–3485. doi: 10.1073/pnas.0307827100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JR, Papamichail D, Skiena S, Futcher B, Wimmer E, Mueller S. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;320:1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Quality measures for protein alignment benchmarks. Nucl Acids Res. 2010;38:2145–2153. doi: 10.1093/nar/gkp1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BG. Simulating DNA coding sequence evolution with EvolveAGene 3. Mol Biol Evol. 2008;25:688–695. doi: 10.1093/molbev/msn008. [DOI] [PubMed] [Google Scholar]
- Heger A, Ponting CP. Variable strength of translational selection among 12 Drosophila species. Genetics. 2007;177:1337–1348. doi: 10.1534/genetics.107.070466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R, Petrov DA. Selection on codon bias. Annu Rev Genet. 2008;42:287–299. doi: 10.1146/annurev.genet.42.110807.091442. [DOI] [PubMed] [Google Scholar]
- Hey J. The neutralist, the fly and the selectionist. Trends Ecol Evol. 1999;14(1):35–38. doi: 10.1016/s0169-5347(98)01497-9. [DOI] [PubMed] [Google Scholar]
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JM, Wides R, Salzberg SL, Loftus B, Yandell M, Majoros WH, Rusch DB, Lai Z, Kraft CL, Abril JF, Anthouard V, Arensburger P, Atkinson PW, Baden H, de Berardinis V, Baldwin D, Benes V, Biedler J, Blass C, Bolanos R, Boscus D, Barnstead M, Cai S, Center A, Chaturverdi K, Christophides GK, Chrystal MA, Clamp M, Cravchik A, Curwen V, Dana A, Delcher A, Dew I, Evans CA, Flanigan M, Grundschober-Freimoser A, Friedli L, Gu Z, Guan P, Guigo R, Hillenmeyer ME, Hladun SL, Hogan JR, Hong YS, Hoover J, Jaillon O, Ke Z, Kodira C, Kokoza E, Koutsos A, Letunic I, Levitsky A, Liang Y, Lin JJ, Lobo NF, Lopez JR, Malek JA, McIntosh TC, Meister S, Miller J, Mobarry C, Mongin E, Murphy SD, O’Brochta DA, Pfannkoch C, Qi R, Regier MA, Remington K, Shao H, Sharakhova MV, Sitter CD, Shetty J, Smith TJ, Strong R, Sun J, Thomasova D, Ton LQ, Topalis P, Tu Z, Unger MF, Walenz B, Wang A, Wang J, Wang M, Wang X, Woodford KJ, Wortman JR, Wu M, Yao A, Zdobnov EM, Zhang H, Zhao Q, Zhao S, Zhu SC, Zhimulev I, Coluzzi M, della Torre A, Roth CW, Louis C, Kalush F, Mural RJ, Myers EW, Adams MD, Smith HO, Broder S, Gardner MJ, Fraser CM, Birney E, Bork P, Brey PT, Venter JC, Weissenbach J, Kafatos FC, Collins FH, Hoffman SL. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–49. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Irwin B, Heck JD, Hatfield GW. Codon pair utilization biases influence translational elongation step times. J Biol Chem. 1995;270:22801–22806. doi: 10.1074/jbc.270.39.22801. [DOI] [PubMed] [Google Scholar]
- Moriyama EN, Powell JR. Gene length and codon usage bias in Drosophila melanogaster, Saccharomyces cerevisiae and Escherichia coli. Nucl Acids Res. 1998;26:3188–3193. doi: 10.1093/nar/26.13.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura G, Pinheiro M, Arrais J, Gomes AC, Carreto L, Freitas A, Oliveira JL, Santos MA. Large scale comparative codon-pair context analysis unveils general rules that fine-tune evolution of mRNA primary structure. PLoS One. 2007;2:e847. doi: 10.1371/journal.pone.0000847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S, Papamichail D, Coleman JR, Skiena S, Wimmer E. Reduction of the rate of poliovirus protein synthesis through large-scale codon deoptimization causes attenuation of viral virulence by lowering specific infectivity. J Virol. 2006;80:9687–9696. doi: 10.1128/JVI.00738-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi Z, Megy K, Grabherr M, Ren Q, Zdobnov EM, Lobo NF, Campbell KS, Brown SE, Bonaldo MF, Zhu J, Sinkins SP, Hogenkamp DG, Amedeo P, Arensburger P, Atkinson PW, Bidwell S, Biedler J, Birney E, Bruggner RV, Costas J, Coy MR, Crabtree J, Crawford M, Debruyn B, Decaprio D, Eiglmeier K, Eisenstadt E, El-Dorry H, Gelbart WM, Gomes SL, Hammond M, Hannick LI, Hogan JR, Holmes MH, Jaffe D, Johnston JS, Kennedy RC, Koo H, Kravitz S, Kriventseva EV, Kulp D, Labutti K, Lee E, Li S, Lovin DD, Mao C, Mauceli E, Menck CF, Miller JR, Montgomery P, Mori A, Nascimento AL, Naveira HF, Nusbaum C, O’leary S, Orvis J, Pertea M, Quesneville H, Reidenbach KR, Rogers YH, Roth CW, Schneider JR, Schatz M, Shumway M, Stanke M, Stinson EO, Tubio JM, Vanzee JP, Verjovski-Almeida S, Werner D, White O, Wyder S, Zeng Q, Zhao Q, Zhao Y, Hill CA, Raikhel AS, Soares MB, Knudson DL, Lee NH, Galagan J, Salzberg SL, Paulsen IT, Dimopoulos G, Collins FH, Birren B, Fraser-Liggett CM, Severson DW. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- Pagel M, Meade A. A phylogenetic mixture model for detecting pattern-heterogeneity in gene sequence or character-state data. Syst Biol. 2004;53:571–581. doi: 10.1080/10635150490468675. [DOI] [PubMed] [Google Scholar]
- Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR, Moriyama EN. Evolution of codon usage bias in Drosophila. Proc Natl Acad Sci U S A. 1997;94:7784–7790. doi: 10.1073/pnas.94.15.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR, Sezzi E, Moriyama EN, Gleason JM, Caccone A. Analysis of a shift in codon usage in Drosophila. J Mol Evol. 2003;57(Suppl 1):S214–25. doi: 10.1007/s00239-003-0030-3. [DOI] [PubMed] [Google Scholar]
- Rao Y, Wu G, Wang Z, Chai X, Nie Q, Zhang X. Mutation bias is the driving force of codon usage in the Gallus gallus genome. DNA Res. 2011;18:499–512. doi: 10.1093/dnares/dsr035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP. Codon usage bias from tRNA’s point of view: redundancy, specialization, and efficient decoding for translation optimization. Genome Res. 2004;14:2279–2286. doi: 10.1101/gr.2896904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19: 2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Severson DW, Behura SK. Mosquito genomics: Progress and challenges. Ann Rev Entomol. 2012;57:143–66. doi: 10.1146/annurev-ento-120710-100651. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tats A, Tenson T, Remm M. Preferred and avoided codon pairs in three domains of life. BMC Genomics. 2008;9:463. doi: 10.1186/1471-2164-9-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicario S, Moriyama EN, Powell JR. Codon usage in twelve species of Drosophila. BMC Evol Biol. 2007;7:226. doi: 10.1186/1471-2148-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov AE. Intron length and codon usage. J Mol Evol. 2001;52:2–5. doi: 10.1007/s002390010128. [DOI] [PubMed] [Google Scholar]
- Wan XF, Xu D, Kleinhofs A, Zhou J. Quantitative relationship between synonymous codon usage bias and GC composition across unicellular genomes. BMC Evol Biol. 2004;4:19. doi: 10.1186/1471-2148-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeberg B. Shannon information theoretic computation of synonymous codon usage biases in coding regions of human and mouse genomes. Genome Res. 2002;12:944–955. doi: 10.1101/gr.213402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.