Abstract
Absorption of light by the visual pigment rhodopsin triggers a rapid cis-trans photoisomerization of its retinal chromophore and a series of conformational changes in both the retinal and protein. The largest structural change is an outward tilt of transmembrane helix H6 that increases the separation of the intracellular ends of H6 and H3, and opens up the G-protein binding site. In the dark state of rhodopsin, Glu247 at the intracellular end of H6 forms a salt bridge with Arg135 on H3 to tether H6 in an inactive conformation. The Arg135-Glu247 interaction is broken in the active state of the receptor, and Arg135 is then stabilized by interactions with Tyr223, Met257 and Tyr306 on helices H5, H6 and H7, respectively. To address the mechanism of H6 motion, solid-state NMR measurements are undertaken of Metarhodopsin I (Meta I), the intermediate preceding the active Metarhodopsin II (Meta II) state of the receptor. 13C NMR dipolar recoupling measurements reveal an interhelical contact of 13Cζ-Arg135 with 13Cε-Met257 in Meta I, but not with 13Cζ-Tyr223 or 13Cζ-Tyr306. These observations suggest that helix H6 has rotated in the formation of Meta I, but that structural changes involving helices H5 and H7 have not yet occurred. Together, our results provide insights into the sequence of events leading up to the outward motion of H6, a hallmark of G protein-coupled receptor activation.
Keywords: G protein-coupled receptor, visual pigment, rhodopsin, solid-state NMR spectroscopy
INTRODUCTION
The visual receptors are members of the large family of G protein-coupled receptors (GPCRs).1,2 These receptors all contain seven transmembrane helices, designated H1-H7 (Figure 1), and have the ability to activate intracellular heterotrimeric G proteins. However, the visual receptors are unique in being activated by light-induced isomerization of a covalently bound retinal chromophore rather than by binding of a diffusible ligand. Nevertheless, much of our current understanding of the structure and activation of GPCRs has emerged from structure-function studies of the visual receptors, particularly the vertebrate dim-light photoreceptor rhodopsin.
Figure 1.
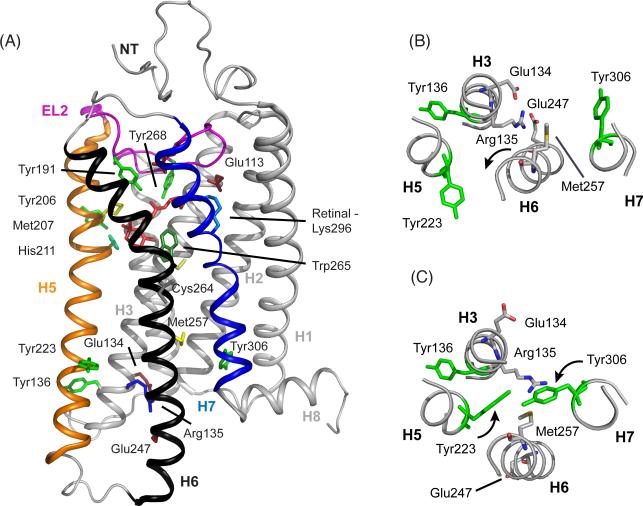
Crystal structures of rhodopsin (PDB access code = 1GZM) and Meta II (PDB access code = 3PXO) highlighting key residues associated with photoactivation.88 (A) The full receptor structure is shown. The retinal is buried between the transmembrane helices on the extracellular side of the protein with the second extracellular loop (EL2) forming a cap on the retinal binding site. Motion of helices H5, H6 and H7 during activation opens up the G-protein binding site on the intracellular side of the receptor, while helices H1–H4 form a tightly packed core. (B) Cross section of the intracellular side of rhodopsin in the region of the Arg1353.50 - Glu2476.30 salt bridge. Only selected helices are shown for clarity. (C) Structure of the intracellular side of Meta II in the region of the ionic lock. There are several differences between inactive rhodopsin (panel B) and Meta II (panel C). Rotation of H6 breaks the Arg1353.50 - Glu2476.30 contact and moves Met2576.40 into the H3-H6 interface. The intracellular end of H6 tilts outward, allowing the side chain of Arg1353.50 to extend and contact Met2576.40. Rotation of H5 and H7 place Tyr2235.58 and Tyr3067.53 into contact with Arg1353.50. Hydrogen-bonds between these tyrosines and Arg1353.50 stabilize the active state of the receptor.
It has been known since the pioneering EPR studies of Hubbell and coworkers using site-directed spin labeling that activation of rhodopsin involves the outward rotation of transmembrane helix H6.3 However, a mechanistic understanding of how retinal isomerization is coupled to H6 motion has been lacking. The first high-resolution crystal structure of rhodopsin was obtained in 2000 of the inactive, dark state of the receptor (Figure 1).4 This structure revealed the position of highly conserved residues within the large family of Class A GPCRs, but provided few clues about the mechanism of receptor activation. The crystal structure of opsin, which captures the outward rotation of H6, revealed several key elements of the activation mechanism.5,6 In the dark state of the rhodopsin, Arg1353.50 forms a salt bridge with Glu2476.30 on H6 (footnote †). In opsin, the rotation of H6 moves Glu2476.30 toward H5 where it forms a new electrostatic interaction with Lys2315.66. H6 motion also places Met2576.40 in contact with Arg1353.50 on transmembrane helix H3. However, this latter interaction appears to require the outward tilt of H6, which displaces the intracellular end of H6 by ~5 Å relative to H3.7 That is, the interface between H3 and H6 is tightly packed and the outward motion of H6 allows the side chain of Arg1353.50 to extend toward Met2576.40 and make contact (see Figure 1C). In addition, the opsin crystal structure showed that the intracellular ends of both helices H5 and H7 undergo rotation to allow two highly conserved tyrosine residues to hydrogen bond with Arg1353.50. Tyr2235.58 on H5 and Tyr3067.53 on H7 rotate toward Arg1353.50 in the active receptor and contribute to breaking of the Arg1353.50 – Glu2476.30 salt bridge.5,6 The combined effect of these structural changes is to open up a cavity on the intracellular side of the receptor that serves as the binding site for the C terminal tail of the Gα subunit of transducin, the heterotrimeric G protein that associates with light-activated rhodopsin.
Several observations argue that the motions of helices H5, H6 and H7 are part of a common activation mechanism for Class A GPCRs. First, several of the residues described above are highly conserved.1 Arg1353.50 is part of the conserved ERY motif on helix H3 (i.e. residues Glu1343.49-Arg1353.50-Tyr1363.51). Tyr2235.58 is the most highly conserved residue on H5, while Tyr3067.53 is part of a conserved NPxxY sequence on H7. Second, recent crystal structures of the β2-adrenergic receptor with bound agonists show an outward displacement of H6 and confirm that this signature structural change for rhodopsin activation is present in a ligand-activated GPCR.8,9 Nevertheless, the large changes in H6 in these crystal structures are only observed when the receptor is in complex with an antibody mimicking the G protein. The structural changes of the β2-adrenergic receptor10 and other ligand-activated GPCRs11,12 without stabilizing antibodies exhibit structural changes that are much smaller than those observed in the active state of rhodopsin. One possible explanation for the differences between rhodopsin and the ligand-activated receptors is that rhodopsin has evolved several mechanisms to reduce basal activity, including the salt-bridge between Arg1353.50 and Glu2476.30, which are essential for a visual receptor. In contrast, ligand-activated receptors typically exhibit high levels of basal activity. These receptors are inherently dynamic molecules that interconvert between transiently populated conformational states.13 As a result, if the visual receptors and ligand-activated receptors share a common activation mechanism, there are likely interactions that restrict H6 motion in rhodopsin that are released following photo-isomerization, but prior to the conformational step that generates the active receptor.
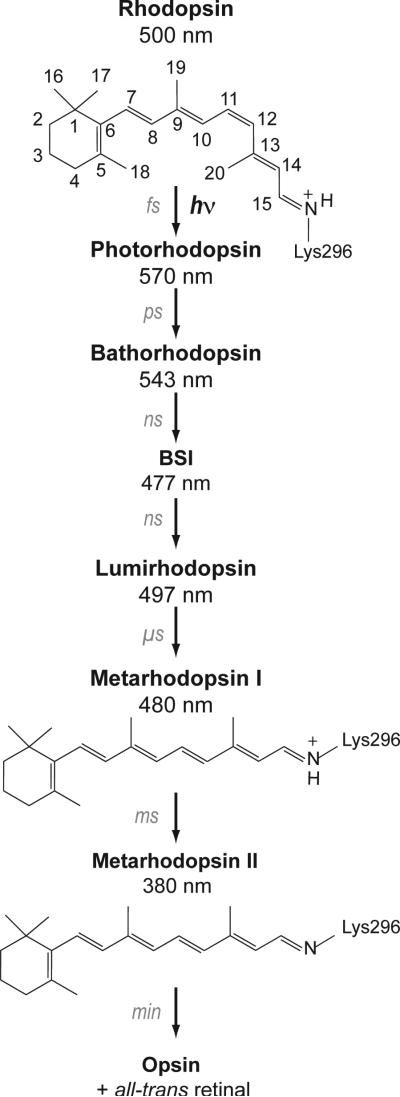
Figure 2 presents the photoreaction pathway leading from dark, inactive rhodopsin to the active Metarhodopsin II (Meta II) intermediate. In the dark-state, the retinal chromophore is bound as a protonated Schiff base to Lys2967.43 on transmembrane helix H7. Absorption of light triggers the rapid isomerization of the retinal from the 11-cis to the all-trans configuration. The early intermediates contain a conformationally distorted all-trans retinal chromophore.14 Calorimetric studies have shown that ~30 kcal/mol of the absorbed light energy is stored in Bathorhodopsin, the first relatively long-lived intermediate.15 The trapped energy is released as the retinal relaxes16-19 and the surrounding amino acids reorient in the transitions to the Blue-shifted Intermediate, Lumirhodopsin and Metarhodopsin I (Meta I). Meta I immediately precedes the activated Meta II state. The current study focuses on the orientation and interactions involving helix H6 in Meta I in order to establish whether conformational changes occur in this helix leading up to the active Meta II state.
Figure 2.
Photoreaction of rhodopsin. Structures of the 11-cis and all-trans retinal chromophores and the photoreaction intermediates of rhodopsin are shown. Absorption of light results in 11-cis to all-trans isomerization of the retinal. The retinal-protein complex subsequently relaxes thermally through a series of spectrally well-defined intermediates: Photorhodopsin, Bathorhodopsin, the Blue Shifted Intermediate (BSI), Lumirhodopsin, and Meta I. Deprotonation of the retinal Schiff base nitrogen occurs in the formation of the Meta II intermediate. Meta I exists in a pH- and temperature-dependent equilibrium with the active Meta II intermediate.89 High pH and low temperature favor Meta I. Evidence for substantial structural changes in Meta I comes from time resolved absorbance measurements showing that the transition to Lumirhodopsin is the last step in the photoreaction sequence that does not exhibit differences between lipid and detergent environments.90 In addition, Meta I is the first intermediate that cannot be fully photo-converted back to rhodopsin.91
The retinal chromophore in the Meta I intermediate has an all-trans configuration and exhibits an absorption maximum (λmax) at 480 nm. There are no high-resolution crystal structures of Meta I. However, a 5.5 Å resolution structure of Meta I obtained by electron cryo-microscopy of 2D crystals showed no significant displacements of the transmembrane helices as compared to the dark-state of rhodopsin.20 The largest change in Meta I relative to rhodopsin was in the region of Trp2656.48 on helix H6,20 which suggested a local change either in the conformation of the Trp2656.48 side chain or in the rotational orientation of the H6 helix.
In contrast to the low-resolution structure of Meta I, a number of biophysical studies have revealed conformational changes that stretch from the retinal binding site on the extracellular side of the receptor to the G-protein binding site on the intracellular surface in Meta I. On the extracellular side of rhodopsin, Fourier transform infrared (FTIR) spectroscopy shows that Glu1223.37 becomes more strongly hydrogen bonded in the transition to Meta I.21 Glu1223.37 is located on helix H3 near the retinal β-ionone ring, and hydrogen bonds to the backbone carbonyl of His2115.46 on H5. These residues are part of a hydrogen-bonding network that extends to the second extracellular loop (EL2). Coupled motion of the retinal and H5 has been implicated in the transition to the active Meta II state.22
On the intracellular side, FTIR measurements of rhodopsin containing p-azido-L-phenylalanine, an engineered non-native amino acid, show that there are strong changes in polarity at the ends of helices H5 and H6 in the formation of Meta I. These changes are consistent with a rotation of H6 and movement of the intracellular end of H5 away from H3.23 Furthermore, Meta I is the first substrate for rhodopsin kinase,24 which binds to Meta I and Meta II with equal affinity,25 and has been reported to be able to bind the G protein transducin.26,27 Together these results imply that a conformational change on the extracellular side of the receptor, induced by retinal isomerization, leads to changes on the intracellular surface of Meta I.
Solid-state NMR spectroscopy provides a complementary approach to X-ray crystallography and FTIR spectroscopy for probing the structure of Meta I and defining the structural changes that occur as light energy is channeled into rhodopsin. Previous solid-state NMR studies on Meta I were restricted to the chromophore and showed that the retinal polyene chain is in a relaxed conformation.17,18,28,29 Here, we describe 13C and 15N solid-state magic angle spinning (MAS) NMR measurements on Meta I trapped in digitonin that target structural changes in the protein centered on helix H6. Digitonin is unusual compared to other detergents as the hydrophobic end of the digitonin is composed of a rigid spirostan steroid tail rather than flexible fatty acyl chains. It has been used extensively to stabilize the Meta I intermediate30-32 since it was first shown to be effective in blocking the transition from Meta I to Meta II.33 The ability of digitonin to trap Meta I is comparable to the use of cholesterol previously used to trap Meta I in 2D crystals of rhodopsin.20 Both digitonin and cholesterol have a rigid steroidal structure that prevents the Meta I to Meta II transition. In contrast, the transition to the Meta II intermediate is facilitated by solubilization of rhododpsin in n-dodecyl-β-D-maltoside (DDM) detergent, which creates a fluid environment. Our structural measurements on Meta I can be compared with previous measurements of rhodopsin and Meta II. The comparison with rhodopsin allows us to infer what structural changes occur between the dark state and Meta I, while the comparison with Meta II allows us to infer what structural changes are associated with activation. 13C dipolar assisted rotational resonance (DARR) NMR measurements reveal an interhelical contact between 13Cζ-labeled Arg1353.50 and 13Cε-labeled Met2576.40 indicating that transmembrane helix H6 rotates in the formation of Meta I. However, the NMR data show that EL2 and H5 have not shifted into their active state conformations. Our results help to define the sequential structural changes occurring between rhodopsin and Meta I that prime the receptor for transition to the active Meta II state.
EXPERIMENTAL METHODS
Materials
13C-labeled amino acids were purchased from Cambridge Isotope Laboratories (Andover, MA). A 5% digitonin stock solution was prepared by dissolving 1 g of digitonin (Calbiochem, La Jolla, CA) in 20 mL of boiling distilled water. The mixture was heated at 95 °C for 5 min to ensure that all of the digitonin had dissolved. Following storage at 4 °C overnight, the solution was filtered through a 0.2 micron sterile bottle top filter and any insoluble material was discarded.34 The 5% stock solution was stored at -20 °C.
Expression and Purification of 13C- and15 N-Labeled Rhodopsin
A stable tetracyclineinducible HEK293S cell line containing the opsin gene was used to express stable isotope labeled rhodopsin.35 Cultured cells were grown in suspension using a bioreactor (New Brunswick Scientific). The calcium reduced36 growth medium (Dulbecco's modified Eagle's medium, Sigma, St. Louis, MO) was supplemented with specific 13C-labeled amino acids (Cambridge Isotope Laboratories, Andover, MA), 10% dialyzed, heat-inactivated fetal bovine serum,36 Pluronic F-68 (0.1%), dextran sulfate (300 mg/L), penicillin (100 units/mL) and streptomycin (100 μg/mL). On day 4 after incubation, cells were fed with glucose (2.4 g/L). Opsin gene expression was induced 5 days after inoculation by addition to the growth medium of both tetracycline (2 mg/L) and sodium butyrate (5 mM). Cells were harvested on day 7, and the cell pellets were stored at -80 °C.35
The cell pellet obtained from a 3 L bioreactor culture was resuspended in 150 mL phosphate buffered saline (PBS) containing protease inhibitors (0.4 mM PMSF and 50 μg/mL benzamidine). Unlabeled 11-cis retinal was added in two steps totaling 250 nmol per gram of cell pellet. The rhodopsin-containing cells were then pelleted and suspended in PBS (40 mL/L of culture) containing DDM (1% w/v) for 4 h at room temperature. Subsequent purification by immunoaffinity chromatography using the rho-1D4 antibody was carried out according to existing protocols30-32 that were modified to lower the detergent concentration for NMR. For detergent exchange, rhodopsin was washed with 25 column volumes of PBS containing DDM (0.02% w/v), 25 column volumes of PBS containing digitonin (0.1% w/v), and 25 column volumes of PBS containing digitonin (0.02-0.05% w/v). After washing, the column was equilibrated with 10 column volumes of 2 mM phosphate buffer (pH = 7.0) containing digitonin (0.02-0.05% w/v). Rhodopsin was eluted in 2 mM phosphate buffer (pH = 7.0) containing digitonin (0.02-0.05% w/v) and 100 μM C-terminal nonapeptide. The eluted rhodopsin fractions were pooled and concentrated to a final volume of ~400 μL using Centricon devices with a 10 kDa molecular weight cut-off (Amicon, Bedford, MA), followed by further concentration under a stream of argon gas to a volume of ~100 μL. All buffers were prepared fresh before purification.
Solid-State NMR Spectroscopy
NMR spectra of digitonin-solubilized rhodopsin were acquired at static magnetic field strengths of 8.5 T or 14.1 T on Bruker AVANCE spectrometers using either a two-channel or a three-channel 4 mm MAS probe at spinning rates of 8-12 kHz. 1D 13C spectra were collected using ramped amplitude cross polarization with a contact time of 2 ms and an acquisition time of 16 ms. Intermolecular 13C...13C correlations were obtained using the DARR pulse sequence with a mixing time of 600 ms. The 1H radiofrequency field strength during mixing was matched to the MAS speed for each sample, satisfying the n=1 matching condition. Two-pulse phase-modulated proton decoupling of 80-90 kHz was used during both evolution and acquisition.37,38 For 2D data sets, 1024 points in the f2 dimension and 64 points in the f1 dimension were acquired. The 13C solid-state MAS NMR spectra were externally referenced to the 13C resonance of neat TMS at 0.00 ppm at room temperature. Using TMS as the external reference, we calibrated the carbonyl resonance of solid glycine to 176.46 ppm. The 15N solid-state MAS NMR spectra of rhodopsin and Meta I were referenced to the 15N resonance of 5.6 M aqueous NH4Cl at 0.0 ppm at room temperature. 15N-labeled glycine was used as an external reference taking the reported value of 8.1 ppm for glycine relative to 5.6 M aqueous NH4Cl.39 This 15N chemical shift reference was used, instead of the IUPAC recommendation of liquid ammonia40, for comparison with the results of previous studies on retinal Schiff bases.36,41
NMR measurements were first made on rhodopsin in the dark. For conversion to Meta I, the NMR rotor was ejected, the cap removed and the sample illuminated for 1-2 min at 4 °C using a 400 W lamp with a 495 nm high pass filter. On the basis of UV/Vis absorption spectra and NMR spectra of Meta I, the conversion from rhodopsin is > 90%. After conversion, we estimate that there is a loss of < 10% of the Meta I intermediate to Meta II and opsin before the sample is cooled to 190 K for NMR measurements. The residual rhodopsin and Meta II components in the Meta I sample do not interfere with the assignments of resonances originating from Meta I. For conversion to Meta II, samples in DDM were illuminated in the NMR rotor at room temperature. We estimate that the conversion from rhodopsin to Meta II is > 90% and the loss of Meta II to opsin is < 5% before the sample is cooled to 190 K. For both Meta I and Meta II, following conversion the NMR rotor is recapped, reinserted into a pre-cooled NMR probe and frozen within 5-10 min using cold N2 gas. NMR spectra of dark rhodopsin, Meta I and Meta II were obtained at a sample temperature of 190 K. The Meta I and II intermediates are completely stable at this temperature.
Restrained Molecular Dynamic Simulations of Meta I
The NMR constraints collected here were used to perform restrained molecular dynamics (MD) simulations with NAMD 2.843 and employed the CHARMM27r parameters for proteins and lipids.44-52 Retinal parameters were obtained from Saam and coworkers.53 The simulation system started with the high-resolution crystal structure of Lumirhodopsin54 embedded in a DOPC:DOPE (1 : 3) CHARMM-GUI55 bilayer by the replacement method with the cytoplasmic and extracellular (or intradiscal) regions solvated by water and ionized using VMD.56 There were a total of 97,459 atoms: 348 protein residues, 22 crystallographic waters, 9250 TIP3P water molecules, 36 Na+ ions, 39 Cl- ions, and 240 lipids (60 DOPC and 180 DOPE). Simulations were run at a temperature of 303 K and a pressure of 1 bar in the NPAT ensemble, in which the pressure, temperature, and area of the membrane were kept constant, using Langevin dynamics and the particle mesh Ewald method for full system periodic electrostatics. We arrive at the model of Meta I by equilibrating the membrane-protein system and then applying the restraints obtained previously by NMR,22,57,58 and described in Results (e.g. Met257-Arg135, 5 Å with k = 10 kcal/mol Å2). To allow the lipids to equilibrate, the simulations were first run for a total of 5 ns with decreasing constraints on all heavy atoms, protein backbone and retinal from 20 to 0.1. A time step of 1 fs was used for the first two steps of equilibration and then switched to 2 fs. To allow the protein to equilibrate, the simulations were run for an additional 6 ns before restraints were applied. Restraints were applied with the extra bonds feature in NAMD and with k values between 10 – 20 kcal/mol Å2, and run for 2 ns.
RESULTS
Trapping of the Metarhodopsin I Intermediate
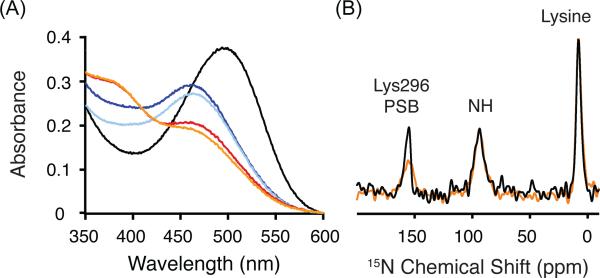
The Meta I intermediate is characterized by a visible absorption maximum at 480 nm. Figure 3A presents absorption spectra showing the conversion of rhodopsin (λmax = 500 nm, black line) to Meta I (λmax = 480 nm, blue line) following illumination by light with wavelengths longer than 495 nm. The stability of Meta I in digitonin is temperature dependent. At 4 °C, rhodopsin is fully converted to the Meta I intermediate and is stable for over 30 min (light blue line). When rhodopsin in digitonin is illuminated at 20 °C, a mixture of Meta I and Meta II (λmax = 380 nm) is formed (red line). This mixture is stable for over 30 min (orange line). The temperature dependence of the transition to Meta II has previously been characterized.30,59,60 The Meta II substate that is obtained at 20 °C in digitonin likely corresponds to Meta IIa on the basis of EPR measurements showing that digitonin effectively blocks the large increase in the mobility of a spin label at residue 2506.33 near the intracellular end of H6.60 The outward tilt of H6 is associated with the Meta IIa to Meta IIb transition.60
Figure 3.
Trapping of the Meta I intermediate in digitonin. (A) UV-Vis spectra provide a means to follow the conversion of rhodopsin (λmax = 500 nm, black line) to Meta I (λmax = 480 nm, blue line) after illumination by light (> 495 nm) at 4 °C. Meta I remains stable in digitonin for over 30 min at 4 °C (light blue line). Conversion at 20 °C leads to a mixture of Meta I and Meta II (red line), which does not change appreciably over 30 min (orange line). (B) One dimensional 15N spectra of rhodopsin (black) and Meta I (orange) labeled at 15Nζ-lysine are shown that were obtained using 1H-15N cross polarization. The 15Nζ-Lys2967.43 chemical shift is observed as a distinct narrow peak at 156.8 ppm in rhodopsin (black). In Meta I, the resonance shifts slightly and broadens. The 15Nζ resonances for the other 15Nζ-labeled lysines in rhodopsin are observed as a broad peak ~8.0 ppm.
A second characteristic feature of the Meta I state is that the retinal chromophore has an all-trans configuration and is attached to Lys2967.43 as a protonated Schiff base. To address the protonation state of the retinal Schiff base linkage, Figure 3B presents the 15N MAS NMR spectra of rhodopsin (black) and Meta I (orange) containing 15Nζ-labeled lysine. The 15Nζ-Lys2967.43 resonance in rhodopsin (black) is observed at 156.8 ppm. In Meta I, the 15Nζ-Lys2967.43 resonance broadens and shifts slightly to 155.7 ppm. The similar 15N chemical shifts between rhodopsin and Meta I demonstrate that the Schiff base nitrogen is protonated. Deprotonation of the Schiff base has a dramatic effect on the 15Nζ-Lys2967.43 chemical shift, which is observed at 282.2 ppm in Meta II.57
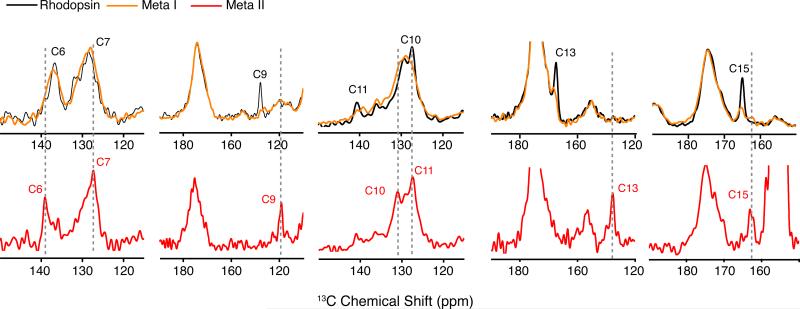
We also measured the 13C chemical shifts of Meta I regenerated with several selectively 13C-labeled retinals (Figure 4). There is a good correspondence of the NMR frequencies with other studies on Meta I.17,18,28 For example, the 13C10, 13C11 retinal resonances in Meta I at 130.6 ppm and 139.2 ppm, respectively, are in agreement with previous measurements of Meta I trapped in lipids.17 Although both saturated lipids and digitonin result in observed broadening of the 13C resonances of the polyene chain,28 we can show in Figure 4 that in digitonin there is good conversion from rhodopsin (> 90%) with less than 10% decay to Meta II or opsin. The ability to photo-convert rhodopsin to Meta I may be associated with the reduction in light scattering from detergent micelles, as compared to lipid multilayers.
Figure 4.
One-dimensional 13C MAS NMR spectra of rhodopsin and Meta I. Rhodopsin was regenerated with 11-cis retinal selectively 13C labeled at different carbons along the polyene chain and the β-ionone ring. Overlap of the 13C MAS NMR spectra of rhodopsin (black) and Meta I (orange) shows that most of the retinal resonances broaden considerably in Meta I as compared to the sharp narrow resonances observed in rhodopsin and Meta II (red). There is nearly complete conversion of rhodopsin to Meta I. For example, the 13C9 retinal resonance in rhodopsin falls in an uncrowded region of the spectrum. The residual intensity of the 13C9 resonance in the Meta I spectrum is <10% of its original intensity in rhodopsin. Also, there is <10% conversion of rhodopsin to Meta II. For example, the spectra of Meta I containing 13C13-labeled retinal show a complete absence of a resonance associated with Meta II.
Structural Changes in the Region of EL2
The second extracellular loop (EL2) of rhodopsin is part of the retinal-binding pocket.4 NMR measurements on Meta II have revealed structural changes in EL222 and in the vicinity of the retinal chromophore.61 We compare our previous results on rhodopsin and Meta II with NMR measurements of the Meta I intermediate in the region of EL2.
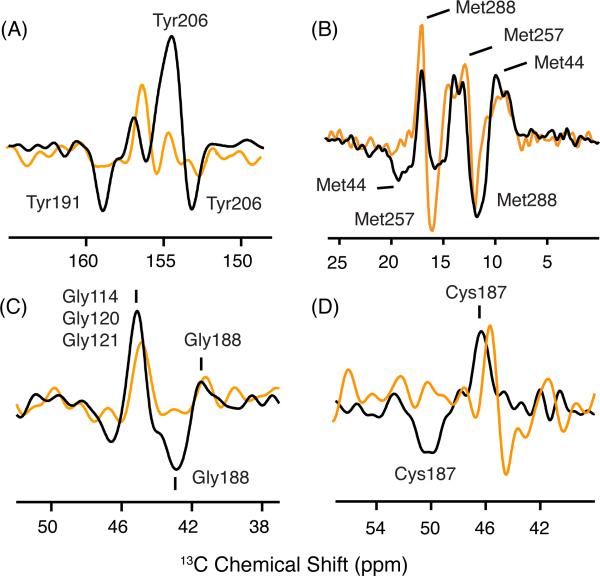
Characteristic changes in 13C chemical shifts have been observed previously in NMR difference spectra between rhodopsin and Meta II for tyrosine, methionine, glycine and cysteine on EL2 and EL3.22,61 Figure 5A shows the difference spectra for 13Cζ-tyrosine between rhodopsin and Meta I (orange) and between rhodopsin and Meta II (black). Positive peaks correspond to rhodopsin and negative peaks correspond to Meta I or Meta II. The 13Cζ-tyrosine chemical shift is sensitive to hydrogen bonding of the side chain Cζ-OH group.62 There are two distinct resonances that appear upon the formation of Meta II.22 The downfield resonance at 158.9 ppm corresponds to Tyr191 on EL2 (i.e. Tyr191EL2), which becomes more strongly hydrogen bonded in Meta II. The upfield resonance at 153.1 ppm corresponds to Tyr2065.41, which becomes more weakly hydrogen bonded in Meta II. These two resonances are not observed in Meta I indicating that EL2 and H5 have not shifted into the conformation characteristic of the active state.
Figure 5.
13C MAS difference spectra of rhodopsin, Meta I and Meta II. Difference spectra are shown for rhodopsin - Meta I (orange) and rhodopsin - Meta II (black) using rhodopsin isotopically labeled with 13Cζ-tyrosine, 13Cε-methionine, 13Cα-glycine and 13Cβ-cysteine. Positive peaks correspond to rhodopsin and negative peaks correspond to Meta I or Meta II. (A) The tyrosine 13Cζ difference spectrum between rhodopsin and Meta II exhibits two positive and two negative resonances. The two Meta II resonances at 158.9 and 153.1 ppm have been assigned to Tyr191EL2 and Tyr2065.41, respectively.22 These resonances are not observed in the rhodopsin - Meta I difference spectrum. (B) The methionine 13Cε difference spectrum between rhodopsin and Meta II exhibits three distinct positive and two negative resonances. The resonances associated with Met44 and Met288 have been assigned in both rhodopsin and Meta II.22 The Met257 resonance does not change between rhodopsin and Meta II. A strong negative cross peak is observed at 15.8 ppm and assigned to Met257 in Meta I. (C) The glycine 13Cα difference spectrum exhibits two positive and two negative resonances. The upfield resonances in rhodopsin at 41.5 ppm and in Meta II at 42.9 ppm have been assigned to Gly188EL2.22 The appearance of a negative resonance at the position of Gly188EL2 is not observed in the rhodopsin - Meta I difference spectrum. (D) In the cysteine 13Cβ difference spectrum between rhodopsin and Meta II, the chemical shift of Cys187EL2 changes from 46.8 ppm to 50.1 ppm.22 The disulfide bridge between Cys187EL2 and Cys1103.25 tethers EL2 to the helical bundle.
Figure 5B presents difference spectra for 13Cε-methionine between rhodopsin and Meta I (orange) and between rhodopsin and Meta II (black). In the rhodopsin - Meta II difference spectrum, we have previously assigned the positive resonance at 17.2 ppm (rhodopsin) and the negative resonance at 12.8 ppm (Meta II) to Met2887.35 based on its disappearance in the M288L mutant.22 Met2887.35 is located on H7 and is at the interface between EL2 and the extracellular end of H7. The rhodopsin - Meta I difference spectrum indicates that Met288 in Meta I has moved into an environment characteristic of the active state, suggesting local structural changes in H6, H7 or EL2.
The chemical shift for Met2576.40 in Meta II was assigned at 14.7 ppm based on a cross-peak with Arg1353.50.63 A corresponding negative resonance is not observed in the difference spectrum because the resonance does not change between rhodopsin and Meta II. In contrast, a strong negative cross peak is observed at 15.8 ppm in Meta I (discussed below).
In the rhodopsin-Meta II difference spectrum, we have also assigned the positive resonance at 10.5 ppm (rhodopsin) and negative resonance at 19.2 ppm (Meta II) to Met441.39.61 Met441.39 is located near the retinal Schiff base. The positive resonance at 10.5 ppm is observed in the rhodopsin - Meta I difference spectrum. However, the negative resonance at 19.2 ppm is not observed in Meta I. These results suggest that the environment of Met441.39 near the protonated Schiff base in Meta I is different from rhodopsin, but does not yet correspond to the active Meta II state.
Figure 5C presents the rhodopsin - Meta II difference spectrum (black) for 13Cα-glycine. We have assigned the positive resonance at 41.5 ppm and the negative resonance at 42.9 ppm to Gly188EL2 based on its disappearance in the G188A mutant.64 The appearance of the positive peak at ~42 ppm in the rhodopsin – Meta I spectrum (orange) indicates that Gly188EL2 located in EL2 is influenced by the conversion to Meta I. However, the absence of the negative peak at 42.9 ppm argues that the EL2 has not yet adopted the final conformation observed in the Meta II state.22 The large positive peak at 45.5 ppm in Meta II is associated with several glycines on H3 (Gly1143.29, Gly1203.35 and Gly1213.36).22,61 The observation of a positive peak at this position in the rhodopsin-Meta I difference spectrum suggests a possible structural change in H3 in the transition to Meta I. Gly1143.29 on H3 is adjacent to Glu1133.28 and one helical turn from Cys1103.25. Changes in the interactions of the protonated Schiff base with its counterion are likely to induce structural changes in the helix backbone in this region. For example, the largest changes in the crystal structure of Lumirhodopsin, relative to rhodopsin, were observed in two helical turns of H3 centered on Gly1203.35 and Gly1213.36.54
Figure 5D shows the changes in the 13Cβ resonances of the highly conserved Cys1103.25-Cys187EL2 disulfide group. Cys1103.25 is at the extracellular end of H3 and Cys187EL2 is part of the β4 strand of EL2. The Cys187EL2 13Cβ resonance shifts from 46.8 ppm to 50.1 ppm upon conversion of wild-type rhodopsin to Meta II trapped in DDM.22 In digitonin, the dark state 13Cβ resonance of Cys187EL2 is at 45.5 ppm and shifts to 44.4 ppm in Meta I. The change in chemical shift of Cys187EL2 suggests a conformation or environment distinct from either the dark state or Meta II.
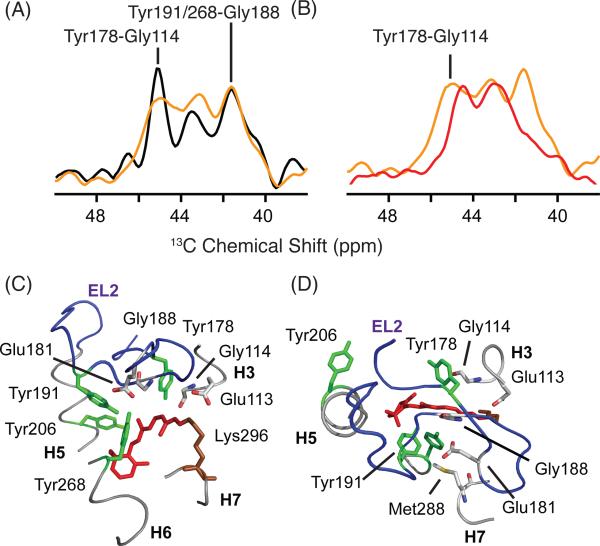
Two-dimensional (2D) MAS NMR dipolar recoupling experiments provide support for the conclusion that EL2 has not yet shifted into its active state conformation. Figure 6A presents slices through the 13Cζ-tyrosine diagonal resonance from 2D DARR NMR spectra of rhodopsin (black) and Meta I (orange). In rhodopsin, at least four (of five possible) 13Cζ Tyr - 13Cα Gly cross-peaks are detected. The cross-peaks at 42.0 ppm and 45.5 ppm are assigned to contacts between Tyr2686.48 and Gly188EL2, and between Tyr178EL2 and Gly1143.29, respectively.22 In Meta I, these contacts are observed. The most substantial change between rhodopsin and Meta I is broadening of the Tyr178EL2 - Gly1143.29 cross-peak. In Meta II (Figure 6B, red), the Gly188EL2 – Tyr2686.51 and the Gly188EL2 - Tyr191EL2 cross-peaks are absent. The loss of these cross-peaks is attributed to a reorganization of the EL2 hydrogen-bonding network upon activation, respectively.22
Figure 6.
Two-dimensional DARR NMR spectra of 13Cζ-tyrosine, 13Cα-glycine-labeled rhodopsin. Slices through the diagonal resonances of 13Cζ-tyrosine showing through space 13C...13C contacts with 13Cα-glycine are shown for rhodopsin (black) and Meta I (orange) in panel (A) and for Meta I (orange) and Meta II (red) in panel (B). Cross peaks between 13Cζ- tyrosine and 13Cα-glycine provide a way to monitor changes on the extracellular side of the rhodopsin. In the rhodopsin crystal structure (1U19), there are six 13Cζ-tyrosine - 13Cα-glycine contacts located in the extracellular region of rhodopsin involving 5 tyrosines and 5 glycines: Tyr10NT-Gly3NT, 3.9 Å; Tyr10NT -Gly280EL3, 4.4 Å; Tyr29NT -Gly101EL1, 4.0 Å; Tyr178EL2 -Gly1143.29, 4.5 Å; Tyr191EL2 -Gly188EL2, 5.2 Å; Tyr2686.51 -Gly188 EL2, 5.3 Å. The slices shown in (A) reveal that the Tyr-Gly contacts are similar between rhodopsin and Meta I. (C, D) Structure of the extracellular side of rhodopsin (PDB access code = 1GZM) viewed from the side (C) and from the extracellular surface. The structure highlights the position of EL2 over the retinal binding site and the positions of several of the amino acids discussed.
Structural Changes in Transmembrane Helices H5 and H6
The difference spectra of 13Cζ-tyrosine and 13Cε-methionine in Figures 5A and B also provide insights into the structural changes occurring in the transmembrane helices. Tyr2065.41 on H5 does not adopt its active state conformation in Meta I (Figure 5A). This tyrosine is part of a hydrogen-bonding network with His2115.46 (H5), Glu1223.37 (H3), Trp1263.41 (H3) and Ala1664.55 (H4). The conversion to Meta II is associated with a rearrangement of this network such that the side chain of His211 hydrogen bonds directly with the side chain of Glu1223.37.65,66 This rearrangement appears to be driven by direct contact of the retinal β-ionone ring with H5.22,6113C dipolar couplings reveal close contacts between Met2075.42 and the retinal C6 and C7 carbons upon conversion to Meta II.22,61
The rhodopsin - Meta I difference spectrum of 13Cε-methionine (Figure 5B) reveals a large negative resonance at 15.8 ppm. This frequency is close to that of Met2576.40 in Meta II.63 The tentative assignment of the 15.8 ppm resonance in Meta I to 13Cε-Met2576.40 raises the possibility that H6 has rotated in Meta I to place the Met2576.40 side chain in an environment similar to that in Meta II.
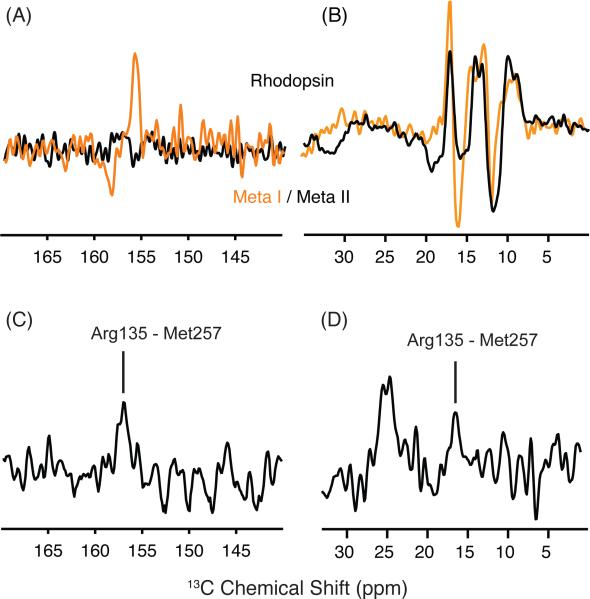
Figure 7A presents difference spectra between rhodopsin and Meta I (orange) and between rhodopsin - Meta II (black) for rhodopsin containing 13Cζ-labeled arginine. As above, the positive peaks correspond to rhodopsin and negative peaks correspond to Meta I or Meta II. The rhodopsin - Meta II difference spectrum in the region of the 13Cζ arginine chemical shift does not reveal any substantial changes indicating the chemical shift of Arg1353.50 is similar in rhodopsin and Meta II.
Figure 7.
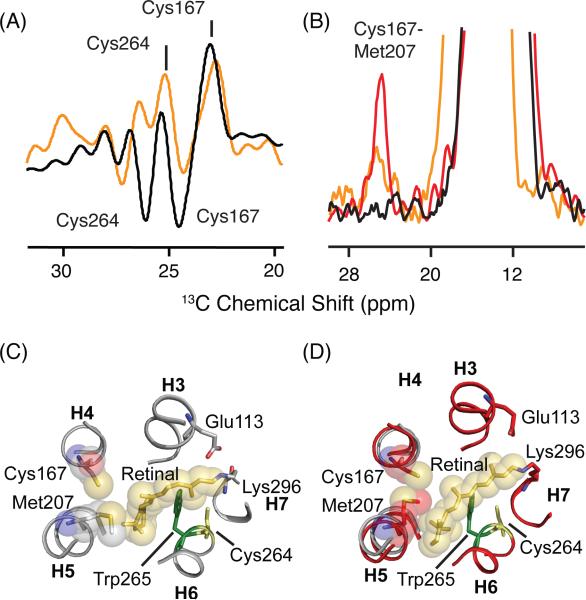
13Cζ-arginine and 13Cε-methionine changes in the transition to Meta I. (A) Rhodopsin - Meta I (orange) and rhodopsin - Meta II (black) difference spectra are shown in the region of 13Cζ -arginine. The 13Cζ -arginine chemical shifts do not appear to differ between rhodopsin and Meta II, as indicated by the flat baseline in region of the difference spectrum. In contrast, the rhodopsin - Meta I difference spectrum reveals a single 13Cζ -arginine has changed in the transition to Meta I. (B) Rhodopsin - Meta I (orange) and rhodopsin - Meta II (black) difference spectra are shown in the region of 13Cε-methionine. A strong negative peak at 15.7 ppm corresponds to a new 13Cε-methionine resonance in Meta I. (C,D) Slices extracted from a 2D DARR NMR spectrum reveal a cross peak between 13Cζ -Arg1353.50 and 13Cε-Met2576.40 in Meta I on both sides of the diagonal in 2D spectrum.
In contrast to the absence of changes in the rhodopsin – Meta II difference spectrum, the rhodopsin - Meta I difference spectrum reveals a change in chemical shift of an arginine 13Cζ resonance from 155.7 ppm in rhodopsin to 157.3 ppm in Meta I. The linewidth of this resonance is consistent with it corresponding to a single arginine residue. To confirm that this arginine is Arg1353.50, we obtained 2D DARR NMR spectra of rhodopsin labeled with both 13Cζ-arginine and 13Cε-methionine. Figures 7C and 7D present slices from the 2D DARR NMR spectrum of Meta I in the regions of the 13Cζ-arginine and 13Cε-methionine cross-peaks, respectively. Figure 7C reveals a 13Cζ-Arg1353.50 - 13Cε-Met2576.40 cross-peak at 156.9 ppm in the slice taken through the 13Cε-methionine diagonal resonance, while Figure 7D reveals a 13Cε-methionine - 13Cζ-arginine cross-peak at 16.2 ppm in the slice taken through the 13Cζ-arginine diagonal resonance. The intensities of the 13Cε-methionine - 13Cζ-arginine cross-peaks in Meta I relative to the diagonal resonances are roughly the same intensity as in Meta II63 where the internuclear distance is 4.5 - 5 Å. We attribute the appearance of a 13Cζ-Arg1353.50 - 13Cε-Met2576.40 cross-peak to rotation of H6 (see Discussion).
In both active opsin and Meta II, the rotation of H6 that places Met2576.40 in contact with Arg1353.50 is accompanied by the rotation of H5 and H7.63 This rotation has implications for two salt-bridges formed by Arg1353.50 in the dark-state of rhodopsin. The first salt bridge between Arg1353.50 and Glu2476.30, often referred to as the ionic lock, must be broken upon rotation of H6. The second salt bridge between Arg1353.50 and Glu1343.49 is disrupted with the rotation of Tyr2235.58 on H5 and Tyr3067.53 on H7. Tyr2235.58 and Tyr3067.53 form hydrogen bonds with the arginine guanidinium side chain to stabilize Arg1353.50 in a protonated state.63
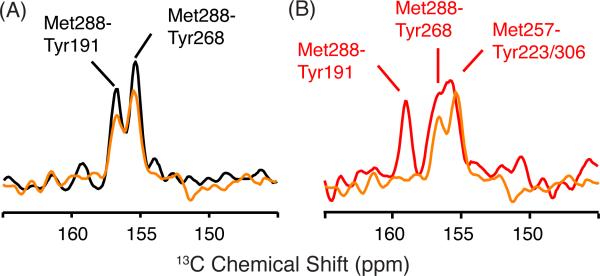
To test whether there are parallel changes in Meta I, 2D DARR NMR spectra were obtained of rhodopsin, Meta I and Meta II using rhodopsin labeled with 13Cε-methionine and 13Cζ-tyrosine. Figure 8 presents slices through the 13Cε-methionine diagonal resonance corresponding to Met2887.35 in the 13Cζ-tyrosine region of rhodopsin (Figure 8A; black) at 17.2 ppm and Meta I (Figure 8A,B; orange) at 12.8 ppm. In Figure 8A, we observe two cross-peaks that we assign to the two tyrosines closest to Met2887.35 (i.e. Tyr2686.48 at 3.9 Å and Tyr191EL2 at 5.2 Å).63 Upon conversion to Meta I, these cross-peaks do not change position or intensity. However, in Meta II the Met2887.35 - Tyr191EL2 and Met2887.35-Tyr2686.48 cross-peaks shift downfield, and two overlapping cross-peaks assigned to Met2576.40 - Tyr2235.58 and Met2576.40 - Tyr3067.53 appear at 155.7 ppm.63
Figure 8.
Two-dimensional 2D DARR NMR spectra of 13Cζ tyrosine, 13Cε methionine-labeled rhodopsin. (A) Slices through the 13Cζ tyrosine diagonal resonance from 2D DARR NMR spectra of rhodopsin (black) and Meta I (orange) labeled with 13Cζ-tyrosine and 13Cε-methionine highlight the region of Tyr-Met cross-peaks. In rhodopsin, we observe cross-peaks between Met2887.35 -Tyr191EL2 and Met2887.35 -Tyr2686.51. Upon conversion to Meta I, the intensity of the cross-peaks does not change appreciably. (B) Slices through the 13Cζ-tyrosine diagonal resonance from 2D DARR NMR spectra of rhodopsin (black) and Meta II (red) are shown using the same labeling strategy as in (A).
We interpret the results in Figures 7 and 8 in terms of rotation of H6 in Meta I without the corresponding rotation of Tyr2235.58 on H5 and Tyr3067.53 on H7. This interpretation is consistent with several observations. First, while we observe an Arg1353.50 - Met2576.40 cross- peak in both Meta I and Meta II, we only observe the Met2576.40 - Tyr2235.58 and Met2576.40 - Tyr3067.53 cross-peaks in Meta II. Second, we observe a change in a 13Cζ-arginine resonance in Meta I, but not in Meta II. We interpret this as due to breaking of the Arg1353.50 – Glu2476.30 interaction upon formation of Meta I, and re-establishing strong hydrogen bonding interactions of Arg1353.50 with Tyr2235.58 and Tyr3067.53 in Meta II. Third, the rhodopsin-Meta I difference spectrum for 13Cε-methionine of wild-type rhodopsin in Figure 7A is remarkably similar to the rhodopsin – Meta II difference spectrum for 13Cε-methionine of the Y223F mutant.63 In the Y223F mutant, there is a rapid decay of the Meta II state to opsin, which suggests that in wild-type Meta II the Tyr2235.58 - Arg1353.50 interaction is holding H5 in an active orientation, whereas in Meta II of the Y223F mutant, H5 has rotated back to an inactive orientation. FTIR measurements of the Y223F mutant show a shift in the pH-dependent equilibrium between Meta I and Meta II back toward Meta I.63
Although we do not observe rotation of Tyr2235.58 on H5 in Meta I, 13C-difference and DARR NMR spectra of Meta I and Meta II labeled with 13Cβ-cysteine show there is a change in the H4-H5 interface. In the 1D 13C difference spectra of 13Cβ-cysteine-labeled rhodopsin, the Cβ carbons exhibit considerable changes in both Meta I and in Meta II (Figure 9A). We can assign the positive peaks at 23.2 ppm and 25.8 ppm to Cys1674.56 and Cys2646.47, respectively. These assignments were made based on the interhelical cross-peaks to His2115.46 and Tyr3017.48 respectively.22 In Meta II, these resonances shift to 24.4 ppm and 26.2 ppm, respectively.
Figure 9.
Chemical shift changes in 13Cβ-cysteine in the transition from rhodopsin to Meta I. (A) Rhodopsin - Meta I (orange) and rhodopsin - Meta II (black) difference spectra in the region of reduced 13Cβ-cysteine resonances. (B) Slices taken through 2D 13C DARR NMR spectra of rhodopsin (black), Meta I (orange) and Meta II (red) exhibit a cross-peak between Met2075.42 and Cys1674.56. The intensity of the Cys1674.56 - Met2075.42 cross-peak in Meta I is intermediate between rhodopsin and Meta II, suggesting that there is partial rotation of H5. (C) Structure of rhodopsin (PDB access code = 1GZM) in the region of the retinal binding site. The structure highlights the positions of Cys1674.56 and Met2075.42. (D) Structure of Meta II (PDB access code = 3PXO) showing displacement of H5 and closer interaction of Cys1674.56 and Met2075.42.
Cys1674.56 can serve as a probe for the motion of H5 relative to H4. In the formation of Meta II, a strong cross-peak is observed between 13Cε-Met2075.42 on H5 and 13Cβ-Cys1674.56 on H4 (Figure 9B).61 The side chain of Met2075.42 rotates from its original position towards H4 to give space for the β-ionone ring of the retinal to pack against H5 in Meta II. In contrast, only a weak cross-peak is observed between Met2075.42 and Cys1674.56 in Meta I (Figure 9B). These data show that although the ligand-binding pocket in the region of H5 has responded to retinal isomerization, the motion is not as large as that observed in Meta II.
DISCUSSION
In this study, solid-state 13C and 15N NMR measurements of the Meta I intermediate are used to address the structural changes that precede receptor activation in rhodopsin. We find that the largest conformational change in Meta I is rotation of transmembrane helix H6. The chemical shift changes associated with EL2 and the rotations of Tyr2235.58 on H5 and Tyr3067.53 on H7, which are observed in Meta II, have not occurred in Meta I. We discuss these observations in connection with the sequence of events that occur in the transition from rhodopsin to Meta I and from Meta I to the active Meta II conformation.
EL2 has not Adopted an Active Conformation in Meta I
In rhodopsin, EL2 folds into two β-strands (β3 and β4) that form a plug over the retinal-binding site and prevent access of hydroxylamine to the protonated Schiff base linkage. The β-strands are constrained in the dark-state by a conserved disulfide bond between Cys1103.25 (H3) and Cys187EL2 (β4) and a network of hydrogen bonding interactions. NMR chemical shift measurements of Meta II indicate a rearrangement of hydrogen bonding interactions between EL2 and amino acids on transmembrane helices H5 and H6.22 These changes in EL2, relative to rhodopsin, have suggested that EL2 adopts an active conformation that is coupled to the motion of transmembrane helices H5, H6 and H7.22
Several observations in the current study suggest that EL2 has not adopted an active conformation in Meta I. First, the characteristic Meta II chemical shifts of Cys187EL2, Gly188EL2 and Tyr191EL2 on the β4 strand of EL2 are not observed in Meta I. Second, a contact between Gly188EL2 on EL2 and Tyr2686.48 at the extracellular end of H6, which is present in rhodopsin, is still observed in Meta I, but absent in Meta II.22 Third, the Met2887.35-Tyr2686.48 and Met2887.35-Tyr191EL2 cross-peaks have similar intensities in rhodopsin and Meta I indicating that EL2 has not markedly changed its position relative to the extracellular ends of H6 and H7. The only indication of substantial change in the vicinity of EL2 is the chemical shift change of Met288, which may reflect changes in the position of H6 and H7, as discussed below, rather than changes in the position of EL2.
The NMR measurements indicating that EL2 has not changed conformation are consistent with recent studies on the accessibility of the retinal Schiff base to hydroxylamine.67 In Meta II, hydroxylamine is able to hydrolyze the Schiff base linkage between the retinal chromophore and the side chain of Lys2967.43. However, in both rhodopsin and Meta I, the protonated Schiff base is not accessible to hydroxylamine.67 (This conclusion disagrees with an earlier study68 where hydroxylamine was shown to react at 275 K with a metarhodopsin intermediate, presumably Meta I, in a pH independent fashion. Later experiments on Meta I trapped at 240 K were not able to confirm this result,67 which opened up the possibility that in the earlier experiments the reactive metarhodopsin intermediate was Meta III, which is known to react with hydroxylamine.59) As a result, the increased accessibility of the Schiff base to hydroxylamine in Meta II is likely due to change in the packing interactions of EL2 with the retinal.22 In this regard, Meta I with EL2 tightly packed against the retinal may serve as a model for Class A GPCRs where the main function of EL2 seems to be as an agonist diffusion barrier as in the angiotensin II type 1 receptor.69
Although EL2 does not appear to have reached the structure adopted in Meta II, there are marked changes in the NMR resonances associated with the retinal chromophore. In rhodopsin and Meta II, narrow, distinct NMR resonances are observed, implying that there are well-defined conformations associated with the active and inactive states of rhodopsin. In contrast, we observe substantial broadening of the 15N and 13C resonances of the Schiff base and retinal in Meta I. The broadening of the 13C resonances was previously interpreted in terms of heterogeneity in the conformation of the polyene chain of the retinal chromophore.28 Alternatively, broadening of the 15Nζ-Lys2967.43 and 13C-retinal resonances may reflect the loss a single well-defined hydrogen bonding contact with Glu1133.28 and water in the binding pocket. As such, the broadening may be associated with the formation of a complex counterion involving Glu1133.28, Glu181EL2 and water.
The concept of a complex counterion has previously been discussed. Yan et al.32 proposed that there is a switch in the Schiff base counterion from Glu1133.28 to Glu181EL2 in the formation of Meta I. In the dark state of rhodopsin, Glu1133.28 is part of a hydrogen-bonding network stretching to Glu181EL2 via the backbone carbonyl of Cys187EL2 and a structural water molecule.70 Yan et al. 32 have observed a dramatic shift in the pKa of the protonated Schiff base upon mutation of Glu181EL2 in EL2 and have suggested that Glu181EL2 is the predominant counterion in the Meta I state. Lüdeke et al.71 have found that Glu181EL2 is unprotonated in both rhodopsin and Meta I, and have argued that together Glu1133.28 and Glu181EL2 form a complex counterion.
Rotation of H6 Occurs in the Formation of Meta I
A hallmark of rhodopsin activation is the outward displacement of helix H6 from the helical bundle.3,7,72 Cross-linking of the intracellular ends of H3 and H6 blocks this motion and prevents receptor activation.3,73 Hubbell and coworkers have demonstrated that the outward motion of H6 occurs following the internal proton transfer from the retinal protonated Schiff base to Glu1133.28.72
In the cryo-EM structure of Meta I, Schertler and coworkers found that there was increased electron density on the side of H6 facing the β-ionone ring at the level of Trp2656.48.20 This density is consistent with the displacement of Trp2656.48 as seen in the structure of active opsin.6 However, there was no change observed in the position of the intracellular end of H6, as in opsin, indicating that H6 does not tilt outward in Meta I. One way to reconcile the cryo-EM structure of Meta I showing a displacement of Trp2656.48 with the NMR results showing a contact between Met2576.40 and Arg1353.50 is to propose that H6 rotates in Meta I without an outward displacement.
The displacement of the Trp2656.48 side chain may also be reflected in recent deuterium NMR studies of Brown and coworkers.74 In rhodopsin, the Trp2656.48 side chain is packed against the C18 and C20 methyl groups. These methyl groups exhibit much more restricted rotational motion than the C19 methyl group in the 6-s-trans, 12-s-trans, 11-cis retinal chromophore in rhodopsin because of steric clashes within the retinal molecule (e.g. the C18 methyl group clashes with the C8H proton in a twisted 6-s-cis geometry and the C20 methyl group clashes with the C10H proton in a twisted 12-s-trans geometry). However, differences in the activation energy (Ea) for rotational motion of these methyl groups between rhodopsin and Meta I and between Meta I and Meta II may have contributions from steric clashes with the protein.74 The Ea of the C19 methyl group increases in the transition from rhodopsin to Meta I and from Meta I to Meta II, and is in fact greater than the Ea for C20 methyl group in Meta II. These observations are consistent with 13C...13C distance measurements showing that the Trp2656.48 side chain moves from a position in contact with the C20 methyl group to a position in contact with the C19 methyl group upon receptor activation.58,75
The proposed rotation of H6 in Meta I is in agreement with recent azido labeling studies on rhodopsin where p-azido-L-phenylalanine is incorporated into positions at the intracellular end of H6.23 These studies show that there are large changes in the vibrations of the azido label at position 2506.33 between Lumirhodopsin and Meta I. However, no further changes in the azido vibrations are observed in the formation of Meta II suggesting that structural changes in this region of the receptor are complete in Meta I. These results on azido-labeled rhodopsin were interpreted in terms of a rotation of H6 to break the Arg1353.50-Glu2476.30 salt bridge.
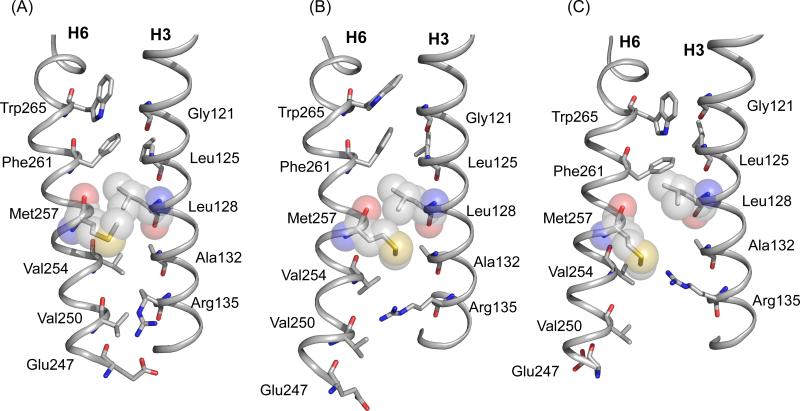
Figure 10 shows the residues in the interface between H3 and H6. The distance between the labeled 13Cε-Met2576.40 - 13Cζ-Arg1353.50 carbons is ~11 Å in dark rhodopsin (dashed line in Figure 10A). The conversion to Meta I brings the labeled 13C sites to within ~6 Å. Rotation of H6 alone does not reduce the distance between these sites, arguing that the side chains of Arg1353.50 and Met2576.40 reorient in Meta I. MD simulations guided by NMR restraints suggest that the side chain of Met2576.40 ratchets past Leu1283.43 (Figure 10B). The rotation of H6 changes the packing interactions in the H3 and H6 interface in several ways that facilitate side chain motion. The proposed rotation of H6 moves Val2546.37 away from Arg1353.50, which otherwise blocks the motion of the arginine side chain toward Met2576.40, and also moves Met2576.40 toward Ala1323.47. The small side chain of Ala1323.47 provides space for bending of the Met2576.40 side chain toward Arg1353.50. As a result the H3-H6 ionic lock between Arg1353.50 and Glu2476.30 is broken while the Lys2315.66 and Glu2476.30 interaction seen in the active form of opsin is not formed in Meta I despite a movement of these residues towards each other. The loss of the Glu2476.30 salt bridge in Meta I can explain the observed increased hydration specific for this intermediate.76
Figure 10.
Structural changes in H3 and H6 upon rhodopsin activation. Packing interactions are shown between H3 and H6 in the crystal structure of rhodopsin (PDB access code = 1GZM) (A), Meta I (B) and in the crystal structure of Meta II (PDB access code = 3PXO) (C). The Meta I structure is based on MD simulations guided by NMR constraints. NMR measurements between Arg1353.50 and Met2576.40, in combination with azido labeling studies23 and EPR3 measurements of Meta I and Meta II, are consistent with rotation of H6. Trp2656.48 in the conserved CWxP motif on H6 is locked in place in dark rhodopsin by the 11-cis retinal chromophore. Retinal isomerization releases the packing constraints on the Trp2656.48 indole ring. Gly1213.36 on H3 is strictly conserved in the visual receptors.1 The small side chain facilitates packing of the Trp2656.48 side chain in dark rhodopsin, but does not hinder H6 rotation. Leu1283.43 and Met2576.40 are closely packed in dark rhodopsin. Leu1283.43 is highly conserved (78%) in GPCRs and is part of a tightly packed transmembrane core.1 The rotation of H6 is facilitated by the small side chain at position 1323.48. This position is highly conserved in the GPCRs as either an alanine (36%) or a serine (50%).63 The Arg1353.50 side chain interacts with Glu2476.30 in the dark and is prevented from moving toward the Met2576.40 by Val2546.37. Val2546.37 is conserved (63%) as a β-branched amino acid across the Class A GPCRs. Rotation of H6 breaks the Arg1353.50- Glu2476.30 interaction and removes the steric interaction with Val2546.37. The side chain of Glu2476.30 is in an intermediate position between Arg1353.50 and Lys2315.66; a Glu2476.30 - Lys2315.66 salt-bridge forms in Meta II.
H5 has not Adopted an Active Conformation in Meta I
One of the triggers for activation of rhodopsin is the steric interaction between the β-ionone ring of the retinal and transmembrane helix H5 in the region of Met2075.42 and Phe2125.47. Contact of the β-ionone ring with H5 leads to rearrangement of hydrogen bonds between H3 and H5 on both the extracellular and intracellular sides of the receptor. On the extracellular end of H5, Tyr2065.41 and His2115.46 form hydrogen bonds with Trp1263.41 and Glu1223.37 on H3 in the dark-state of rhodopsin. These hydrogen bonds are disrupted upon activation. On the intracellular end of H5, Tyr2235.58 is oriented toward the surrounding lipid membrane in the dark-state of rhodopsin, but rotates upon activation into the helical bundle to hydrogen bond with Arg1353.50 of the conserved ERY sequence. The question is whether these changes occur in concert with rotation of H6 in Meta I or with the outward displacement of H6 in Meta II.
Ye et al.23 observed only small changes in the azido vibrations between rhodopsin and Meta I when the azido label was incorporated at position 2275.62 on H5, whereas much larger changes were observed upon formation of Meta II. These results differ from those using the azido probes on H6 (discussed above) and were interpreted in terms of a small movement of H5 in response to rotation of H6. In agreement with these studies, we observe a small increase in intensity of the Cys1674.56-Met2075.42 cross-peak in the formation of Meta I, but a much larger increase in the formation of Meta II (Figure 9B). We interpret the change of the Cys1674.56 - Met2075.42 contact in terms of displacement of H5 relative to H4 and rearrangement of the position of the Met2075.42 side chain (Figures 9C and D), but conclude that the H5 helix has not completely shifted into its active state orientation.
Support for the conclusion that H5 has not adopted an active conformation in Meta I comes from both NMR and FTIR spectroscopy. The NMR chemical shift of Tyr2065.41 provides a probe of the hydrogen-bonding network centered on His2115.46, where a distinctive upfield chemical shift of Tyr2065.41 is observed in the rhodopsin-Meta II difference spectrum. We do not observe this chemical shift change in the rhodopsin - Meta I difference spectrum (Figure 5A). In addition, we do not observe the contact between Tyr2235.58 and Met2576.40 in Meta I that is associated with the rotation of Tyr2235.58 toward Arg1353.50 observed in Meta II (see Figure 1C). In the deuterium NMR studies cited above,74 the Ea for rotation of the C18 methyl group increases in Meta I and then decreases in Meta II. These changes were interpreted in terms of a steric clash of the β-ionone ring with H5 in Meta I, and then displacement of H5 (to lessen the steric clash with the β-ionone ring) in Meta II.
In FTIR studies on Meta I and Meta II, the vibrational frequency at 1734 cm-1 associated with the Glu1223.37 carboxyl group has provided an excellent probe of the hydrogen bonding interactions between H3 and H5. Siebert and coworkers found that the 1734 cm-1 vibration shifts to 1701 cm-1 in the transition from rhodopsin to Meta I indicating that Glu1223.37 becomes more strongly hydrogen bonded.21,77 However, in the transition to Meta II, the Glu1223.37 vibration shifts to 1745 cm-1, a signature of weaker hydrogen bonding. These results agree with both the NMR and azido-labeling studies described above showing that the position or orientation of H5 changes between rhodopsin and Meta I, and then undergoes further changes between Meta I and Meta II.
Sequence of Events in the Formation of the Active Meta II State
One of the challenges for understanding how light activates the visual receptor rhodopsin has been to delineate the chain of molecular events leading to the fully active Meta II conformation. The pioneering work of Hubbell and coworkers revealed that the defining motion is the outward tilting of H6.3 However, the crystal structures of dark rhodopsin and its early photointermediates have provided few clues as to the sequence of the events that drive this motion. The crystal structures of Bathorhodopsin78, Lumirhodopsin54 and Meta I20 showed only subtle changes from the structure of the dark-state of rhodopsin. The picture that has emerged over the past decade is that the structural changes in the receptor leading up to Meta II only involve slight rearrangements of the side chains in the binding site to accommodate the all-trans chromophore. In contrast, the more recent structures of active opsin5,6 and Meta II66,79 capture substantial structural changes on the intracellular side of the receptor.
The two hallmarks of Meta II formation are the proton transfer from the protonated Schiff base to Glu1133.28 and the outward motion of H6. Both steps are set up in Meta I. The observed Schiff base 15N chemical shift in Meta I, along with vibrational spectroscopy studies,19,32,71 favor a slightly stronger protonated Schiff base – counterion interaction. The interaction may involve both Glu1133.28 and Glu181EL2 as part of a complex counterion. As a consequence of a rearrangement in the electrostatic interactions near the protonated Schiff base, Glu1133.28 may move into a more hydrophobic environment in Meta I and become the driving force for Schiff base deprotonation. Deprotonation of the Schiff base is the first of two protonation switches that must be triggered for rhodopsin activation.80
The NMR evidence for rotation of H6 in the Meta I intermediate supports the conclusions drawn from previous azido labeling measurements.23,81 Both studies suggest that H6 rotation disrupts the intracellular salt-bridge between Arg1353.50 and Glu2476.30. In contrast, the electrostatic interaction between Arg1353.50 and Glu1343.49, both highly conserved residues, remains intact in Meta I and is arguably more important for stabilizing the inactive state of the receptor. Protonation of Glu1343.49 in Meta II has been described as the second switch required for rhodopsin activation.80 The NMR studies presented above indicate that the inward rotation of Tyr2235.58 on H5 and Tyr3067.53 on H7 have not occurred in Meta I and consequently must be associated with the outward motion of H6 upon activation.
Finally, the observed structural transitions in the formation of Meta I provide insights into the conservation of residues in the ligand-activated GPCRs and common elements of receptor activation. The ionic lock between Arg1353.50 and Glu2476.30 is not highly conserved. Even in those receptor subfamilies where these complementary charged residues are conserved, crystal structures often do not reveal a direct interaction.82-85 For example, Schertler and coworkers have recently reported crystal structures of two inactive forms of the β1-adreneric receptor, one with an ionic lock between Arg3.50-Glu6.30 and the other without.86 The two forms were found with several inverse agonists bound. The lack of a stabilizing electrostatic interaction between H3 and H6 in the diffusible ligand-activated receptors may be associated with their generally high levels of basal activity relative to the dark state of rhodopsin. In contrast, Arg1353.50 and Glu1343.49 are part of the highly conserved D/ERY sequence on H3, and their interaction appears to have a much more dramatic effect in modulating receptor activity. The observation of a direct interaction in Meta I between Arg1353.50 and Met2576.40, a non-conserved, but highly important residue in the visual receptors, suggests that in the ligand-activated receptors there are subfamily specific interactions that regulate receptor activation.
CONCLUSIONS
The work presented here describes solid-state NMR studies on the Meta I intermediate in the photoreaction of the visual receptor rhodopsin. Meta I is the intermediate immediately preceding Meta II, the active state of the receptor. By comparing our current results with previous NMR measurements on Meta II, we are able to define the structural changes that result in receptor activation. The major conclusions of the manuscript are that transmembrane helix H6 has rotated in the formation of Meta I to break the intracellular ionic lock between Arg1353.50 and Glu2476.30, but that structural changes involving helices H5 and H7 have not yet occurred. The motion of H6, along with several additional smaller changes in structure, appears to prime the receptor for the transition to an active conformation.
ACKNOWLEDGEMENTS
This work was supported by NIH-NSF instrumentation grants (S10 RR13889 and DBI-9977553), and a grant from the NIH to S.O.S (GM-41412), and Kimmelman Center for Biomolecular Structure and Assembly to MS. We gratefully acknowledge the W.M. Keck Foundation for support of the NMR facilities in the Center of Structural Biology at Stony Brook. MS holds the Katzir-Makineni Professorial Chair in Chemistry.
Footnotes
The amino acid numbering used in this manuscript incorporates the residue number from the amino acid sequence of the specific receptor being discussed (e.g., Lys296) and a residue number from a generic numbering system developed by Ballesteros and Weinstein87 (e.g. Lys2967.43) that gives the position of an amino acid relative to the most conserved amino acid (designated 50) on a specific helix. In this example, Lys2967.43 on H7 is seven residues toward the N terminus from the most conserved residue on H7, Pro3037.50. Because sequence alignments are poor for the extracellular and intracellular loops, as well as for the N and C termini, a generic superscript (e.g., EL2) is used to designate the position of a non-transmembrane residue.
REFERENCES
- 1.Smith SO. Annu. Rev. Biophys. 2010;39:309–328. doi: 10.1146/annurev-biophys-101209-104901. [DOI] [PubMed] [Google Scholar]
- 2.Palczewski K. Annu. Rev. Biochem. 2006;75:743–767. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 4.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 5.Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, Hofmann KP, Ernst OP. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 6.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 7.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rasmussen SGF, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, DeVree BT, Rosenbaum DM, Thian FS, Kobilka TS, Schnapp A, Konetzki I, Sunahara RK, Gellman SH, Pautsch A, Steyaert J, Weis WI, Kobilka BK. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rasmussen SGF, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah STA, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenbaum DM, Zhang C, Lyons JA, Holl R, Aragao D, Arlow DH, Rasmussen SGF, Choi HJ, DeVree BT, Sunahara RK, Chae PS, Gellman SH, Dror RO, Shaw DE, Weis WI, Caffrey M, Gmeiner P, Kobilka BK. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu F, Wu HX, Katritch V, Han GW, Jacobson KA, Gao ZG, Cherezov V, Stevens RC. Science. 2011;332:322–327. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warne T, Moukhametzianov R, Baker JG, Nehmé R, Edwards PC, Leslie AGW, Schertler GFX, Tate CG. Nature. 2011;469:241–244. doi: 10.1038/nature09746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deupi X, Kobilka BK. Physiology. 2010;25:293–303. doi: 10.1152/physiol.00002.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eyring G, Curry B, Broek A, Lugtenburg J, Mathies R. Biochemistry. 1982;21:384–393. doi: 10.1021/bi00531a028. [DOI] [PubMed] [Google Scholar]
- 15.Cooper A. Nature. 1979;282:531–533. doi: 10.1038/282531a0. [DOI] [PubMed] [Google Scholar]
- 16.Doukas AG, Aton B, Callender RH, Ebrey TG. Biochemistry. 1978;17:2430–2435. doi: 10.1021/bi00605a028. [DOI] [PubMed] [Google Scholar]
- 17.Verdegem PJE, Bovee-Geurts PHM, De Grip WJ, Lugtenburg J, de Groot HJM. Biochemistry. 1999;38:11316–11324. doi: 10.1021/bi983014e. [DOI] [PubMed] [Google Scholar]
- 18.Feng X, Verdegem PJE, Eden M, Sandström D, Lee YK, Bovee-Geurts PHM, De Grip WJ, Lugtenburg J, de Groot HJM, Levitt MH. J. Biomol. NMR. 2000;16:1–8. doi: 10.1023/a:1008377231625. [DOI] [PubMed] [Google Scholar]
- 19.Pan DH, Mathies RA. Biochemistry. 2001;40:7929–7936. doi: 10.1021/bi010670x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruprecht JJ, Mielke T, Vogel R, Villa C, Schertler GFX. EMBO J. 2004;23:3609–3620. doi: 10.1038/sj.emboj.7600374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beck M, Sakmar TP, Siebert F. Biochemistry. 1998;37:7630–7639. doi: 10.1021/bi9801560. [DOI] [PubMed] [Google Scholar]
- 22.Ahuja S, Hornak V, Yan ECY, Syrett N, Goncalves JA, Hirshfeld A, Ziliox M, Sakmar TP, Sheves M, Reeves PJ, Smith SO, Eilers M. Nat. Struct. Mol. Biol. 2009;16:168–175. doi: 10.1038/nsmb.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye SX, Zaitseva E, Caltabiano G, Schertler GFX, Sakmar TP, Deupi X, Vogel R. Nature. 2010;464:1386–1389. doi: 10.1038/nature08948. [DOI] [PubMed] [Google Scholar]
- 24.Paulsen R, Bentrop J. Nature. 1983;302:417–419. doi: 10.1038/302417a0. [DOI] [PubMed] [Google Scholar]
- 25.Pulvermüller A, Palczewski K, Hofmann KP. Biochemistry. 1993;32:14082–14088. doi: 10.1021/bi00214a002. [DOI] [PubMed] [Google Scholar]
- 26.Tachibanaki S, Imai H, Mizukami T, Okada T, Imamoto Y, Matsuda T, Fukada Y, Terakita A, Shichida Y. Biochemistry. 1997;36:14173–14180. doi: 10.1021/bi970932o. [DOI] [PubMed] [Google Scholar]
- 27.Tachibanaki S, Imai H, Terakita A, Shichida Y. FEBS Lett. 1998;425:126–130. doi: 10.1016/s0014-5793(98)00216-6. [DOI] [PubMed] [Google Scholar]
- 28.Spooner PJR, Sharples JM, Goodall SC, Seedorf H, Verhoeven MA, Lugtenburg J, Bovee-Geurts PHM, DeGrip WJ, Watts A. Biochemistry. 2003;42:13371–13378. doi: 10.1021/bi0354029. [DOI] [PubMed] [Google Scholar]
- 29.Salgado GFJ, Struts AV, Tanaka K, Krane S, Nakanishi K, Brown MF. J. Am. Chem. Soc. 2006;128:11067–11071. doi: 10.1021/ja058738+. [DOI] [PubMed] [Google Scholar]
- 30.Franke RR, Sakmar TP, Graham RM, Khorana HG. J. Biol. Chem. 1992;267:14767–14774. [PubMed] [Google Scholar]
- 31.Resek JF, Farahbakhsh ZT, Hubbell WL, Khorana HG. Biochemistry. 1993;32:12025–12032. doi: 10.1021/bi00096a012. [DOI] [PubMed] [Google Scholar]
- 32.Yan ECY, Kazmi MA, Ganim Z, Hou JM, Pan DH, Chang BSW, Sakmar TP, Mathies RA. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9262–9267. doi: 10.1073/pnas.1531970100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matthews RG, Hubbard R, Brown PK, Wald G. J. Gen. Physiol. 1963;47:215–240. doi: 10.1085/jgp.47.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kiefer H, Krieger J, Olszewski JD, Von Heijne G, Prestwich GD, Breer H. Biochemistry. 1996;35:16077–16084. doi: 10.1021/bi9612069. [DOI] [PubMed] [Google Scholar]
- 35.Reeves PJ, Kim JM, Khorana HG. Proc. Natl. Acad. Sci. U. S. A. 2002;99:13413–13418. doi: 10.1073/pnas.212519199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eilers M, Reeves PJ, Ying WW, Khorana HG, Smith SO. Proc. Natl. Acad. Sci. U. S. A. 1999;96:487–492. doi: 10.1073/pnas.96.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. J. Chem. Phys. 1995;103:6951–6958. [Google Scholar]
- 38.Fung BM, Khitrin AK, Ermolaev K. J. Magn. Reson. 2000;142:97–101. doi: 10.1006/jmre.1999.1896. [DOI] [PubMed] [Google Scholar]
- 39.Wang JX, Balazs YS, Thompson LK. Biochemistry. 1997;36:1699–1703. doi: 10.1021/bi962578k. [DOI] [PubMed] [Google Scholar]
- 40.Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wüthrich K. J. Biomol. NMR. 1998;12:1–23. doi: 10.1023/a:1008290618449. [DOI] [PubMed] [Google Scholar]
- 41.Harbison GS, Herzfeld J, Griffin RG. Biochemistry. 1983;22:1–4. doi: 10.1021/bi00270a600. [DOI] [PubMed] [Google Scholar]
- 42.Getmanova E, Patel AB, Klein-Seetharaman J, Loewen MC, Reeves PJ, Friedman N, Sheves M, Smith SO, Khorana HG. Biochemistry. 2004;43:1126–1133. doi: 10.1021/bi030120u. [DOI] [PubMed] [Google Scholar]
- 43.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.MacKerell AD, Feig M, Brooks CL. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 45.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 46.Yin DX, Mackerell AD. J. Comput. Chem. 1998;19:334–348. [Google Scholar]
- 47.Feller SE, Gawrisch K, MacKerell AD. J. Am. Chem. Soc. 2002;124:318–326. doi: 10.1021/ja0118340. [DOI] [PubMed] [Google Scholar]
- 48.Feller SE, MacKerell AD. J. Phys. Chem. B. 2000;104:7510–7515. [Google Scholar]
- 49.Feller SE, Yin DX, Pastor RW, MacKerell AD. Biophys. J. 1997;73:2269–2279. doi: 10.1016/S0006-3495(97)78259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.MacKerell AD. J. Chim. Phys. Phys.-Chim. Biol. 1997;94:1436–1447. [Google Scholar]
- 51.Beglov D, Roux B. J. Chem. Phys. 1994;100:9050–9063. [Google Scholar]
- 52.Klauda JB, Brooks BR, MacKerell AD, Venable RM, Pastor RW. J. Phys. Chem. B. 2005;109:5300–5311. doi: 10.1021/jp0468096. [DOI] [PubMed] [Google Scholar]
- 53.Saam J, Tajkhorshid E, Hayashi S, Schulten K. Biophys. J. 2002;83:3097–3112. doi: 10.1016/S0006-3495(02)75314-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakamichi H, Okada T. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12729–12734. doi: 10.1073/pnas.0601765103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jo S, Lim JB, Klauda JB, Im W. Biophys. J. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Humphrey W, Dalke A, Schulten K. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 57.Ahuja S, Eilers M, Hirshfeld A, Yan ECY, Ziliox M, Sakmar TP, Sheves M, Smith SO. J. Am. Chem. Soc. 2009;131:15160–15169. doi: 10.1021/ja9034768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hornak V, Ahuja S, Eilers M, Goncalves JA, Sheves M, Reeves PJ, Smith SO. J. Mol. Biol. 2010;396:510–527. doi: 10.1016/j.jmb.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vogel R, Siebert F, Mathias G, Tavan P, Fan GB, Sheves M. Biochemistry. 2003;42:9863–9874. doi: 10.1021/bi034684+. [DOI] [PubMed] [Google Scholar]
- 60.Kusnetzow AK, Altenbach C, Hubbell WL. Biochemistry. 2006;45:5538–5550. doi: 10.1021/bi060101v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ahuja S, Crocker E, Eilers M, Hornak V, Hirshfeld A, Ziliox M, Syrett N, Reeves PJ, Khorana HG, Sheves M, Smith SO. J. Biol. Chem. 2009;284:10190–10201. doi: 10.1074/jbc.M805725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Herzfeld J, Das Gupta SK, Farrar MR, Harbison GS, McDermott AE, Pelletier SL, Raleigh DP, Smith SO, Winkel C, Lugtenburg J, Griffin RG. Biochemistry. 1990;29:5567–5574. doi: 10.1021/bi00475a022. [DOI] [PubMed] [Google Scholar]
- 63.Goncalves JA, South K, Ahuja S, Zaitseva E, Opefi CA, Eilers M, Vogel R, Reeves PJ, Smith SO. Proc. Natl. Acad. Sci. U. S. A. 2010;107:19861–19866. doi: 10.1073/pnas.1009405107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goncalves JA, Ahuja S, Erfani S, Eilers M, Smith SO. Prog. Nucl. Magn. Reson. Spectrosc. 2010;57:159–180. doi: 10.1016/j.pnmrs.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Patel AB, Crocker E, Reeves PJ, Getmanova EV, Eilers M, Khorana HG, Smith SO. J. Mol. Biol. 2005;347:803–812. doi: 10.1016/j.jmb.2005.01.069. [DOI] [PubMed] [Google Scholar]
- 66.Choe HW, Kim YJ, Park JH, Morizumi T, Pai EF, Krauss N, Hofmann KP, Scheerer P, Ernst OP. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 67.Katayama K, Furutani Y, Kandori H. J. Phys. Chem. B. 2010;114:9039–9046. doi: 10.1021/jp102288c. [DOI] [PubMed] [Google Scholar]
- 68.Ratner VL, Bagirov IG, Fesenko EE. Vision Res. 1981;21:251–253. doi: 10.1016/0042-6989(81)90118-8. [DOI] [PubMed] [Google Scholar]
- 69.Unal H, Jagannathan R, Bhat MB, Karnik SS. J. Biol. Chem. 2010;285:16341–16350. doi: 10.1074/jbc.M109.094870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Okada T, Fujiyoshi Y, Silow M, Navarro J, Landau EM, Shichida Y. Proc. Natl. Acad. Sci. U. S. A. 2002;99:5982–5987. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lüdeke S, Beck R, Yan ECY, Sakmar TP, Siebert F, Vogel R. J. Mol. Biol. 2005;353:345–356. doi: 10.1016/j.jmb.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 72.Knierim B, Hofmann KP, Ernst OP, Hubbell WL. Proc. Natl. Acad. Sci. U. S. A. 2007;104:20290–20295. doi: 10.1073/pnas.0710393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sheikh SP, Zvyaga TA, Lichtarge O, Sakmar TP, Bourne HR. Nature. 1996;383:347–350. doi: 10.1038/383347a0. [DOI] [PubMed] [Google Scholar]
- 74.Struts AV, Salgado GFJ, Martínez-Mayorga K, Brown MF. Nat. Struct. Mol. Biol. 2011;18:392–394. doi: 10.1038/nsmb.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Crocker E, Eilers M, Ahuja S, Hornak V, Hirshfeld A, Sheves M, Smith SO. J. Mol. Biol. 2006;357:163–172. doi: 10.1016/j.jmb.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 76.Grossfield A, Pitman MC, Feller SE, Soubias O, Gawrisch K. J. Mol. Biol. 2008;381:478–486. doi: 10.1016/j.jmb.2008.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fahmy K, Jäger F, Beck M, Zvyaga TA, Sakmar TP, Siebert F. Proc. Natl. Acad. Sci. U. S. A. 1993;90:10206–10210. doi: 10.1073/pnas.90.21.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nakamichi H, Okada T. Angew. Chem.-Int. Edit. 2006;45:4270–4273. doi: 10.1002/anie.200600595. [DOI] [PubMed] [Google Scholar]
- 79.Standfuss J, Edwards PC, D'Antona A, Fransen M, Xie G, Oprian DD, Schertler GF. Nature. 2011;471:656–660. doi: 10.1038/nature09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mahalingam M, Martinez-Mayorga K, Brown MF, Vogel R. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17795–17800. doi: 10.1073/pnas.0804541105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ye SX, Huber T, Vogel R, Sakmar TP. Nat. Chem. Biol. 2009;5:397–399. doi: 10.1038/nchembio.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AG, Tate CG, Schertler GFX. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EYT, Lane JR, Ijzerman AP, Stevens RC. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chien EYT, Liu W, Zhao QA, Katritch V, Han GW, Hanson MA, Shi L, Newman AH, Javitch JA, Cherezov V, Stevens RC. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moukhametzianov R, Warne T, Edwards PC, Serrano-Vega MJ, Leslie AG, Tate CG, Schertler GFX. Proc. Natl. Acad. Sci. U. S. A. 2011;108:8228–8232. doi: 10.1073/pnas.1100185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ballesteros JA, Weinstein H. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 88.Ahuja S, Smith SO. Trends Pharmacol. Sci. 2009;30:494–502. doi: 10.1016/j.tips.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 89.Parkes JH, Liebman PA. Biochemistry. 1984;23:5054–5061. doi: 10.1021/bi00316a035. [DOI] [PubMed] [Google Scholar]
- 90.Epps J, Lewis JW, Szundi I, Kliger DS. Photochem. Photobiol. 2006;82:1436–1441. doi: 10.1562/2006-02-01-RA-792. [DOI] [PubMed] [Google Scholar]
- 91.Furutani Y, Kandori H, Shichida Y. Biochemistry. 2003;42:8494–8500. doi: 10.1021/bi034438y. [DOI] [PubMed] [Google Scholar]