Abstract
The transcriptional output at a genomic locus in eukaryotes is determined, in part, by the pattern of histone modifications that are read and interpreted by key effector proteins. The histone deacetylase activity of the evolutionarily-conserved Rpd3S/Sin3S complex is crucial for suppressing aberrant transcription from cryptic start sites within intragenic regions of actively transcribed genes. Precise targeting of the complex relies on the chromatin binding activities of the MRG15 and Pf1 subunits. Whereas the molecular target of the MRG15 chromodomain (CD) has been suggested to be H3K36me2/3, the precise molecular target of the Pf1 plant homeodomain 1 (PHD1) has remained elusive. Here we show that Pf1 PHD1 binds preferentially to the unmodified extreme N-terminus of histone H3 (H3K4me0) but not to H3K4me2/3, which are enriched in the promoter and 5′ regions of genes. Unlike previously characterized CD and PHD domains that bind to their targets with micromolar affinity, both MRG15 CD and Pf1 PHD1 bind to their targets with >100 μM affinity, offering an explanation for why both MRG15 CD and Pf1 PHD1 domains are required to target the Rpd3S/Sin3S complex to chromatin. Our results also suggest that bivalency, rather than cooperativity, is the operative mechanism by which Pf1 and MRG15 combine to engage H3 in a biologically significant manner. Finally, the studies reveal an unanticipated role of Pf1 PHD1 in engaging the MRG15 MRG domain, albeit in a Pf1 MRG-binding domain (MBD)-dependent manner, implying a key role for the MRG15 MRG-Pf1 MBD interaction in chromatin targeting of the Rpd3S/Sin3S complex.
Keywords: Transcription repression, combinatorial readout, histone code, histone interactions, NMR
Introduction
The transcriptional status of a genomic locus is determined, in part, by the pattern of post-translational modification of histones, the major protein component of chromatin.1–3 Among the various histone modifications, the roles of histone (lysine) acetylation and deacetylation are especially well-characterized and are generally correlated with transcriptional activation or repression, respectively. The enzymes that determine steady-state levels of histone acetylation exhibit broad substrate specificity, are usually members of multi-protein complexes, and rely on the non-enzymatic subunits of these complexes to target their activities. Targeting often relies on interactions between these subunits and the pre-existing patterns of histone modifications. What these chromatin signals are and how they are read and interpreted are important questions in contemporary chromatin transcription biology.
The ~0.6 MDa mammalian Rpd3S/Sin3S corepressor complex constitutes one of only five major histone deacetylase (HDAC)-containing complexes that shares some of the core subunits with the 1.2–2 MDa Rpd3L/Sin3L complex.4,5 Both complexes are evolutionarily conserved from yeast to human but perform distinct functions. Whereas the Rpd3L/Sin3L complex is broadly involved in promoter-based repression, the Rpd3S/Sin3S complex effects repression of aberrant gene transcription from cryptic start sites, besides inhibiting transcription elongation, within the intragenic regions of actively transcribed genes.5–14 Chromatin interactions involving two Rpd3S/Sin3S-specific subunits MRG15 and Pf1 (Eaf3p and Rco1p, respectively, in yeast) are critical for targeting the corepressor complex to transcribed loci.10,12 Studies in yeast showed that the Eaf3p chromodomain (CD) and the Rco1p PHD1 domains were especially important for targeting as deletions of either of these domains or heterologous replacements of the PHD domain severely diminished or abrogated the ability of the complex to associate with nucleosomes.10
At the molecular level, the chromodomain of Eaf3p, the yeast MRG15 ortholog, has been shown to recognize H3K36me2/3, 15,16 a chromatin signal enriched in the intragenic regions of actively transcribed genes.9,11,14 However, the precise molecular target for Pf1/Rco1p PHD1 is not known. Here we characterize the molecular interactions involving MRG15 CD with di- and tri-methylated H3 peptides. Additionally, we identify unmodified histone H3 as a target for Pf1 PHD1 interactions and investigate whether Pf1 PHD1 and MRG15 CD can bind cooperatively to histone H3 peptides bearing binding sites for both domains. Finally, we also tested whether the PHD1 and CD domains could interact with each other (and thereby contribute towards the cooperativity in H3 binding) in the context of much larger or full-length proteins.
Results
The MRG15 Chromodomain is a General Methyllysine Binding Module
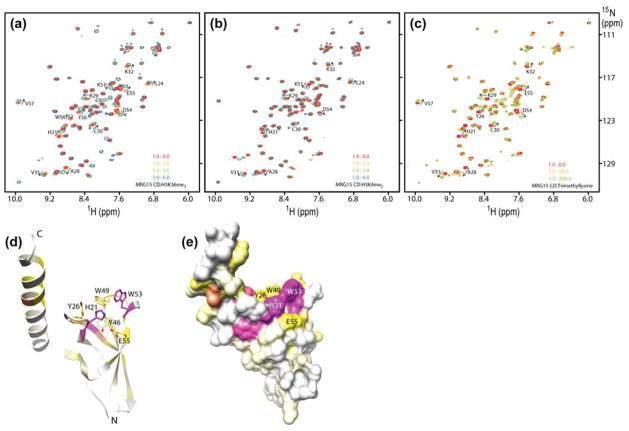
To characterize the interaction between MRG15 CD and its target H3K36me2/3, we used solution NMR spectroscopy. 15N-labeled MRG15 CD was titrated with histone H3 peptides (residues 28–44) di- or tri-methylated at Lys36 (H3K36me2/3). Small, but significant perturbations were detected for a subset of resonances in the NMR spectrum of MRG15 CD, implying a specific interaction (Fig. 1a). The positions of the resonances shifted as a function of added peptide, characteristic of a complex with fast dissociation kinetics. Least-squares fitting of chemical shift deviations as a function of added ligand for multiple resonances yielded equilibrium dissociation constants (Kds) of 0.58 ± 0.18 mM and 1.14 ± 0.39 mM for H3K36me3 and H3K36me2 peptides, respectively. Given the low affinity of these interactions, we evaluated the specificity of the interaction by titrating an H3 peptide (residues 1–12) peptide dimethylated at Lys4 (H3K4me2), a signal that is enriched in the 5′ regions of actively transcribed genes.17 The H3K4me2 peptide induced similar perturbations in the NMR spectrum of MRG15 CD as the H3K36me2/3 peptides (Fig. 1b) and bound with only about three-fold lower affinity (3.19 ± 0.83 mM) than the H3K36me2 peptide. To test whether a significant portion of the binding affinity resided within the methyllysine moiety, we titrated MRG15 CD with free trimethyllysine. The trimethyllysine induced a similar pattern of perturbations in the NMR spectrum of MRG15 CD as the aforementioned peptides and bound with 11 ± 3.3 mM affinity (Fig. 1c). Collectively, these results indicate that the MRG15 CD binds with low affinity to di- or tri-methylated peptides with little sequence preference and with most of the binding energy coming from favorable methyllysine binding interactions.
Figure 1.
MRG15 chromodomain is a methyllysine binding module. Titrations of 15N-labeled MRG15 CD with (a) H3K36me3 (28–44), (b) H3K4me2 (1-12), and (c) trimethyllysine. 1H-15N correlated spectra of MRG15 CD were recorded as a function of increasing amounts of trimethyllysine or methyllysine-containing peptides in NMR buffer comprising 20 mM Tris-d11 acetate-d4 (pH 7.5), 1 mM DTT-d10 and 0.2% NaN3 at 25 °C. MRG15 CD concentrations used for the experiments in panels a-c were 0.2 mM except for panel b (0.75 mM). Chemical shift perturbations induced by H3K36me3 (28–44) mapped on to the (d) polypeptide backbone and (e) molecular surface of the MRG15 chromodomain (PDB: 2F5K).18 The perturbations are color ramped from white (corresponding to 0 ppm) to yellow (average + 1 standard deviation = 0.055 ppm) to magenta (average + 3 standard deviations = 0.115 ppm). Side chains of residues that form the canonical methyllysine binding pocket are shown in panel d.
The fast dissociation kinetics of the MRG15 CD-H3K36me3 complex precluded the observation of intermolecular 1H-1H NOEs vital for structure determination. We assigned the backbone resonances of apo-MRG15 CD and mapped the chemical shift perturbations induced by the H3K36me3 peptide on to the structure of the apo-MRG15 CD. Chemical shift mapping reveals a contiguous surface of strongly perturbed residues including His21, Tyr26, Tyr46, Trp49, Trp53, and Glu55 that line the putative methyllysine binding pocket (Fig. 1d & e); the equivalent side chains of Tyr26, Tyr46, and Trp49 in the orthologous Eaf3p form the aromatic cage that encloses the methyllysine residue, which provides confirmation that MRG15 binds in an equivalent manner.16 In the course of our studies, we found that the interactions between MRG15 CD and the methyllysine peptides were strongly sensitive to the pH used for the titrations. Although all the aforementioned studies were conducted at pH 7.5, our initial studies of the interaction between MRG15 CD and the H3K36me3 peptide conducted at pH 6 yielded little or no perturbations in the MRG15 spectrum even in the presence of 6-fold excess peptide (data not shown). The lack of interaction at acidic pH is likely caused by the positive charge on His21 and the resulting unfavorable electrostatic interaction with the positively charged methyllysine moiety.
Structural analyses have suggested reclassification of the MRG15 and Eaf3p chromodomains as a chromobarrel domain, as the domain lacks the canonical methyllysine peptide-binding cleft found in chromodomains.18,19 In chromobarrel domains, the cleft is occupied by a strand contributed by the domain itself which precludes extensive interactions with histone peptides, although the methyllysine binding pocket is intact. This might explain why the domain binds to di- or tri-methylated peptides with little sequence preference and with only slightly better affinity than to free trimethyllysine (note that the α-amino and -carboxyl groups in free trimethyllysine were not ‘capped’ by charge-neutralizing groups and thus could diminish the affinity of the interaction).
The Pf1 PHD1 Domain Binds to the N-terminus of Unmodified H3
To identify potential binding partners for Pf1 PHD1, we did pull-down assays using a panel of peptides spanning various segments of histone H3 N-terminus, which is a common target for interactions with PHD domains.20 Whereas the unmodified histone H3 peptide spanning residues 1–20 bound to Pf1 PHD1, H3 peptides spanning residues 27–46 and 18–37 failed to do so, suggesting that the extreme N-terminus harbored a binding site for the PHD domain (Fig. 2a). Since many PHD domains harbor methyllysine-binding activity,20 H3 peptides bearing K4me1/2/3, K9me3 and K36me3 modifications were tested for binding to Pf1 PHD1. Whereas trimethylation of Lys9 and Lys36 produced no effect on Pf1 PHD1-binding compared to the corresponding unmodified peptides, methylation of Lys4 significantly attenuated PHD1 binding activity. By contrast, in pulldowns conducted in parallel with Pf1 PHD2, none of the H3 peptides that were tested bound to this PHD domain (Fig. 2a). Collectively, these results suggest that whereas Pf1 PHD1 preferentially targets the unmodified N-terminus of histone H3, Pf1 PHD2 has no H3-binding activity.
Figure 2.
Pf1 PHD1 binds preferentially to unmodified histone H3. (a) SDS-PAGE analysis following a pulldown experiment conducted with GST-Pf1 PHD1 or GST-Pf1 PHD2 and an array of histone H3 peptides. The protein bands were visualized using Coomassie staining. 1H-15N correlated spectra of 15N-labeled Pf1 PHD1 resulting from titrations with increasing amounts of (b) H3 (1-12) and (c) H3K4me2 (1-12). All titrations were conducted at a concentration of 100 μM Pf1 PHD1 in NMR buffer comprising 20 mM Tris-d11 (pH 7.0), 50 mM NaCl, 10 μM ZnSO4, 15 mM DTT, and 0.2% NaN3 at 25 °C. (d) Chemical shift perturbations induced by H3 (1-12) mapped on to the molecular surface of a homology model of Pf1 PHD1 based on the structure of the CHD4 PHD2 domain (PDB: 2L75).22 The perturbations are color ramped from white (corresponding to 0 ppm) to yellow (average + 1 standard deviation = 0.168 ppm) to magenta (average + 3 standard deviations = 0.338 ppm).
To gain deeper insights into the H3 binding activity of Pf1 PHD1, we characterized the interaction using solution NMR spectroscopy. 15N-labeled Pf1 PHD1 was titrated with unmodified H3 peptides spanning residues 1-12 (Fig. 2b). A subset of Pf1 PHD1 resonances shifted to new positions as a function of the added ligand, characteristic of a specific interaction with fast dissociation kinetics. A longer unmodified H3 peptide spanning residues 1-42 perturbed the same subset of resonances and induced perturbations to comparable degrees as the H3 (1-12) peptide (Supp. Fig. S1). Non-linear least squares fitting of the chemical shift deviations as a function of added ligand for multiple resonances yielded comparable Kds of 208 ± 14 μM for H3 (1-12) and 122 ± 10 μM for H3 (1-42). By contrast, an H3K4me2 (1-12) peptide, even at 20-fold excess, produced significantly diminished perturbations in the Pf1 PHD1 spectrum, perturbed only a small subset of resonances, and bound to the PHD domain with at least 15-fold lower affinity (Fig. 2c). Collectively, these results confirm a low-affinity interaction between Pf1 PHD1 and H3 and map the minimal PHD-interaction segment of H3 to the first 12 residues at the N-terminus.
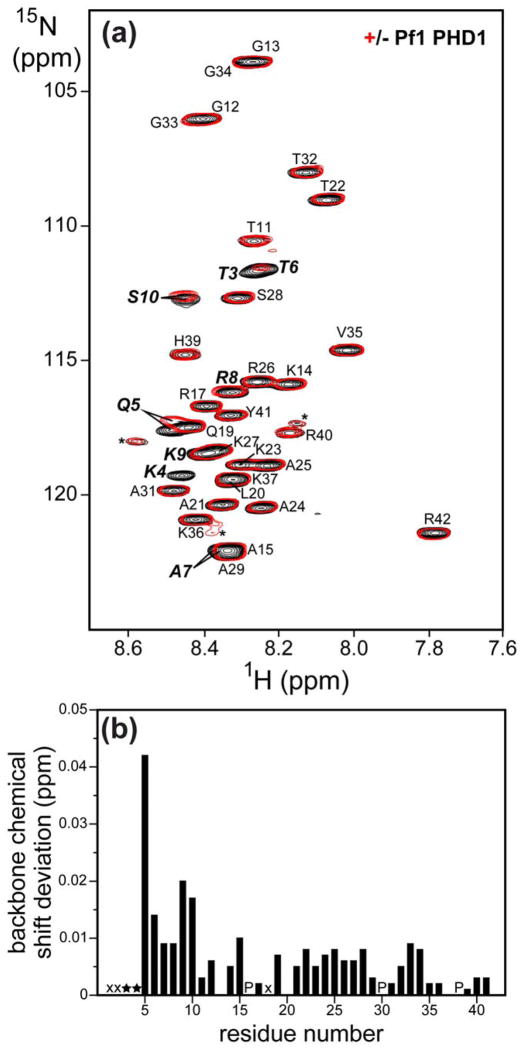
To map the segment of histone H3 involved in interactions with the Pf1 PHD1 domain directly and more precisely, we assigned the backbone resonances of H3 (1-42), and compared the 1H-15N correlated spectra of 15N-labeled H3 (1-42) in the absence and presence of unlabeled PHD1 (Fig. 3a). A small subset of resonances in the H3 spectrum exhibited perturbations in the presence of PHD1. The amide resonances of Thr3 and Lys4 were broadened beyond detection upon PHD1 addition while that of Thr6 was significantly broadened (Fig. 3b). Importantly, the most significant chemical shift changes did not extend beyond the first ten residues. These results suggest that the extreme N-terminus of H3 comprising the first ten residues is the segment targeted by the Pf1 PHD1 domain.
Figure 3.
The extreme N-terminus of histone H3 engages Pf1 PHD1. (a) 1H-15N correlated spectra of 15N-labeled H3 (1-42) recorded in the absence (black) and presence (red) of 2 equivalents of unlabeled Pf1 PHD1. The spectra were recorded at a concentration of 80 μM 15N-H3 in NMR buffer comprising 20 mM Tris-d11 (pH 6.0), 50 mM NaCl, 10 μM ZnSO4, 15 mM DTT, and 0.2% NaN3 at 25 °C. Sequence-specific assignments are annotated. Minor peaks corresponding to resonances from minor conformers, presumed to result from cis-trans proline isomerization, are denoted by asterisks. (b) Backbone amide chemical shift deviations in the H3 peptide induced by Pf1 PHD1 graphed as a function of residue number. H3 resonances that disappeared upon PHD1 addition are denoted by stars while amide proton resonances in fast exchange with solvent are denoted by ‘x’s; prolines are denoted by ‘P’s.
To identify the surface of Pf1 PHD1 involved in interactions with H3, we assigned the backbone resonances of the PHD domain. Chemical shift deviations induced by the H3 (1-12) and the H3 (1-42) peptides revealed highly similar trends (Supp. Fig. S2), providing further confirmation that residues 1-12 harbored all the determinants necessary for the interaction with the PHD domain. The chemical shift changes induced by H3 (1-12) were mapped on to a homology model of the PHD domain. The Pf1 PHD1 homology model was built using DeepView21 and utilized the structure of the CHD4 PHD2 domain as the template, since the two domains share 50% sequence identity and 67% sequence similarity over a 58-residue segment.22 The mapping analysis reveals a contiguous surface of strongly perturbed residues including Gly66, Leu68, His73, Leu92, Gly95, and Trp97 that comprise the canonical binding pocket for histone H3 peptides that bind through their extreme N-terminus (Fig. 2d).22–24 Indeed, the pattern of chemical shift deviations noted for the PHD domain closely resembled the one reported for CHD4 PHD1, another H3K4me0 binder (Supp. Fig. S2).22 Collectively, these results suggest that the Pf1 PHD1 domain interacts with H3 in a similar manner as a subset of PHD domains that bind unmodified H3.22–25
The MRG15 CD and Pf1 PHD1 Domains Bind H3 Independently
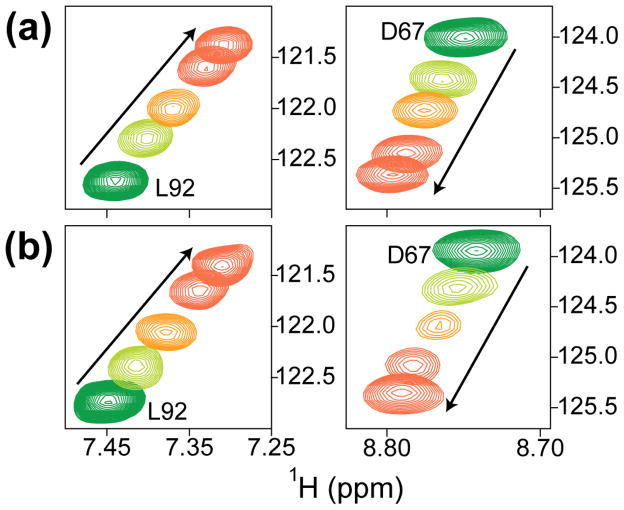
We asked whether the MRG15 CD and Pf1 PHD1 domains could cooperate in binding to H3. Since cooperative mechanisms feature direct interactions between proteins or domains that bind to a common target, we first recorded the NMR spectrum of a 1:1 mixture of 15N-labeled MRG15 CD and 15N-labeled Pf1 PHD1. Both MRG15 CD and Pf1 PHD1 resonances were largely unperturbed in the resulting spectrum relative to those recorded for the individual domains (Supp. Fig. S3), implying the lack of a direct interaction between these domains, at least in the absence of their respective histone targets. We then asked whether the two domains could interact with each other when titrated with an H3K36Cme3 (1-42) peptide (H3 Lys36 was mutated to cysteine and modified chemically to yield a trimethylated product at this position26). The chemical shift perturbations induced by H3K36Cme3 (1-42) were more profound for the Pf1 PHD1 resonances compared to those of MRG15 CD; however, these changes strongly resembled those produced by the unmodified H3 (1-42) peptide in titrations conducted solely for Pf1 PHD1 (Supp. Fig. S1 and Fig. 4a & b). Importantly, there was no significant enhancement in the affinity of the Pf1 PHD1-H3 interaction deduced from these titrations (Kd = 225 ± 11 μM in the presence of 15N-MRG15 CD vs. 122 ± 10 μM in its absence), implying that MRG15 CD and Pf1 PHD1 bound to their H3 targets independently and non-cooperatively.
Figure 4.
Pf1 PHD1 binds histone H3 with similar affinities both in the absence and presence of MRG15 CD. Expanded plots of 1H-15N correlated spectra showing perturbations induced by H3K36Cme3 (1-42) of two Pf1 PHD1 correlations in (a) the absence and (b) the presence of an equimolar amount of 15N-MRG15 CD. Spectra recorded at molar ratios of H3K36Cme3 (1-42) peptide:protein of 0:1, 1:1, 2:1, and 5:1 (green, light green, orange & red) are shown. Titrations were conducted in NMR buffer comprising 20 mM Tris (pH 7.5), 50 mM NaCl, 10 μM ZnSO4, 15 mM DTT, and 0.2% NaN3 at 25 °C. All spectra were recorded under identical solution conditions and were processed and displayed with the same parameters.
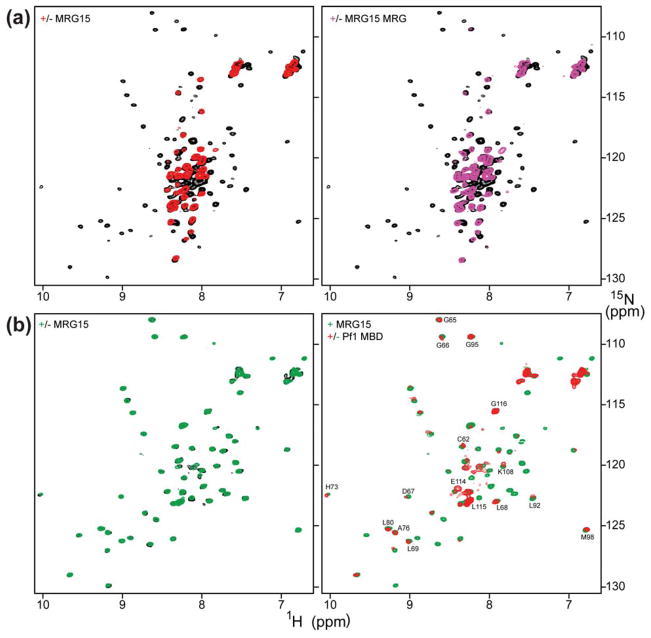
Since the MRG15 and Pf1 proteins have been shown to interact with high-affinity through their MRG and MRG-binding domains (MBD), respectively,27 we sought to test whether CD and PHD1 constructs harboring these interacting segments in the respective proteins could bind H3 with high affinity. Towards this end, we generated samples of 15N-labeled Pf1 PHD1/MBD (residues 48–241) and full-length MRG15 (residues 1–323) and recorded NMR spectra of Pf1 in the absence and presence of MRG15. As expected, the Pf1 spectrum was characterized by dispersed amide proton corresponding to the folded PHD1 domain as well as many intense and poorly-dispersed resonances corresponding to intrinsically unstructured segments following this domain (Fig. 5a). Addition of an equimolar amount of full-length MRG15, rather unexpectedly, led to the disappearance of most of the resonances belonging to the PHD1 domain, although a subset of the resonances – almost exclusively from the unstructured segments – were unaffected. This implied that the PHD1 domain interacted with some segment of MRG15, although, it precluded further NMR analysis of PHD1 interactions with the histone H3 peptide. Interestingly, the MRG15 CD appeared to be not involved in PHD1 interactions because the amide proton resonances characteristic of this domain could be readily detected in the 1D 1H NMR spectrum of the Pf1 PHD1/MBD-MRG15 complex (Supp. Fig. S4). Collectively, these results suggest that the CD and PHD1 domains do not interact with each other even in the context of full-length MRG15 or extended segments of the Pf1 protein.
Figure 5.
Pf1 PHD1 targets the MRG15 MRG domain. (a) 1H-15N correlated spectra of 15N-PHD1/MBD in the absence (black) and presence (red) of an equimolar amount of full-length MRG15 (left panel) and MRG15 MRG (right panel). (b) 1H-15N correlated spectra of 15N-PHD1 in the absence (black) and presence (green) of an equimolar amount of full-length MRG15 and in the presence of equimolar amounts of full-length MRG15 and Pf1 MBD (red). Pf1 PHD1 resonances that are relatively unperturbed upon MRG15 binding are identified. Titrations were conducted in NMR buffer comprising 20 mM Tris (pH 7.5), 50 mM NaCl, 10 μM ZnSO4, 15 mM DTT, and 0.2% NaN3 at 25 °C. All spectra were recorded under identical solution conditions and were processed and displayed with the same parameters.
The Pf1 PHD1 Domain Targets the MRG15 MRG Domain
To map the precise segment of MRG15 involved in Pf1 PHD1 interactions, we recorded an NMR spectrum of a sample of an equimolar mixture of 15N-Pf1 PHD1/MBD with unlabeled MRG15 MRG. Once again, the Pf1 spectrum is characterized by the disappearance of resonances corresponding to the PHD1 domain (Fig. 5a), implying that the PHD1 domain targeted the MRG domain. We then asked whether the MRG-binding activity of Pf1 PHD1 was intrinsic to the domain or required the presence of Pf1 MBD. The NMR spectrum of a 1:1 mixture of 15N-Pf1 PHD1 (residues 48–116) and full-length MRG15 exhibited little or no perturbations compared to the apo-PHD1 (Fig. 5b). However, the addition of one equivalent of Pf1 MBD (in trans), to this sample yielded significant perturbations characterized by the disappearance of a majority of backbone amide correlations with only those belonging to residues forming the putative H3-binding pocket and at the extreme C-terminus of the domain either partially or mostly unaffected, respectively (Fig. 5b). Titration of this mixture with the H3K36Cme3 (1–42) peptide severely but selectively broadened the PHD1 resonances belonging to the putative H3-binding pocket (Supp. Fig. S5). Interestingly, no ‘shifting’ of resonances, characteristic of short-lived complexes, was detected in these titrations, potentially suggesting a higher-affinity interaction for H3K36Cme3 (1–42). Collectively, these results suggest that Pf1 PHD1 binds to a composite surface presented by the MRG15 MRG and Pf1 MBD domains and that the histone H3 binding surface of Pf1 PHD1 is distinct from the one used for engaging the MRG domain.
Discussion
The rich diversity of post-translational histone modifications is matched only by the diversity of the molecular mechanisms employed to read and interpret these modifications.20,28 A variety of chromatin binding effector proteins containing more than one chromatin binding module have been shown to deploy so-called ‘paired modules’ to engage two discrete histone segments, each carrying a separate signal within the same polypeptide or in different polypeptides within the same or adjacent nucleosomes. This type of combinatorial readout incorporating elements of Boolean logic provides a facile, yet powerful, mechanism for tuning transcriptional response.
The activities of the Pf1 PHD1 and MRG15 CD domains represent a variation on this theme, as these domains are located in two distinct subunits of the Rpd3S/Sin3S complex, but combine to read specific signals on histone H3 found in the intragenic regions of actively transcribed genes. Our studies clarify the precise form of these signals – H3 lacking in Lys4 methylation (i.e. H3K4me0) and H3K36me2/3 – that are recognized by the PHD1 and CD domains, respectively.10,12 The involvement of the H3K4me0 signal in this process can be readily rationalized, since the degree and the level of H3 Lys4 methylation are thought to decrease from the 5′-end to the 3′-end of transcribed genes in anti-correlation with the H3K4me0 signal.17 The enrichment profile of H3K4me0 thus mirrors that of the H3K36me2/3 signals.9,11,14
We note that a previous study suggested the involvement of the H3K4me3 signal in Rpd3S/Sin3S recruitment.12 However, these studies were conducted following overexpression of Sin3B, MRG15, and Pf1 proteins in 293T cells and partial purification of the associated complex. It is plausible that additional endogenous proteins such as RBP2, which possesses strong H3K4me3-binding activity and also associates with the Rpd3S/Sin3S complex,29–32 could have co-purified with this complex and mediated the association with H3K4me3-modified nucleosomes. In the studies described herein, bacterially-expressed and purified Pf1 PHD1 recombinant proteins were used that conclusively show that this domain binds preferentially to H3K4me0 over H3K4me3. Indeed, yeast Rpd3S/Sin3S complexes harboring intact Rco1p PHD1 exhibited significant binding activity for unmodified nucleosomes.10 Additionally, at the sequence level, Pf1 PHD1 appears to belong to a class of PHD domains that bind H3K4me0.23 Finally, neither trimethyllysine nor acetyllysine interacted with Pf1 PHD1 in NMR titrations even at molar concentrations (G.S.K. and I.R., unpublished data), excluding the possibility of the PHD domain targeting either of these better characterized histone modifications. We note that Pf1 PHD1 has been shown to associate with phospholipids, but phospholipid binding involves a polybasic segment that is distinct from the H3 binding site.33 How, and indeed if, these disparate activities of the PHD domain are linked is presently unclear.
Even though the Pf1 protein has two PHD domains and could potentially function as a paired chromatin binding module, unlike PHD1, the PHD2 domain lacks histone H3 binding activity. This is consistent with the observation that, at the sequence level, PHD2 does not belong to either of the two sub-classes of PHD1 domains known to bind unmethylated or methylated H3 Lys4.23 This is also confirmed by the lack of any interaction with various small molecule mimics of histone modifications including trimethyllysine, acetyllysine, dimethylarginine, phosphoserine, and phosphothreonine in NMR titrations of Pf1 PHD2 (G.S.K. and I.R., unpublished data). This suggests that the PHD2 domain might have a non-traditional function, perhaps as a protein-protein interaction domain. Since Pf1 also serves as a molecular scaffold linking MRG15 with Sin3 and the rest of the Rpd3S/Sin3S complex,12,34,35 it is plausible that the PHD2 domain has a function in Sin3 binding. In support of this notion, deletion of PHD2 abrogated the Sin3 binding activity of Pf1.12
Although the measured affinities for various PHD domains that bind H3K4me0 range from low micromolar to 30 μM,22,24,36 the Pf1 PHD1 appears to bind H3K4me0 much more weakly (~100–200 μM). We are presently unable to definitively explain the basis for the lower affinity of the interaction, considering most of the affinity determinants noted for the other PHD-H3K4me0 complexes appear to be preserved in Pf1 PHD1. However, we note that the Pf1 PHD1 domain features a two-amino acid insertion that is not found in the other H3K4me0 binders in one of the loops adjacent to the binding site that might serve to diminish the affinity of the interaction. The insertion as well as the other portions of the PHD domain are very well conserved in Pf1 orthologs. Given the high degree of sequence conservation between the Pf1 PHD1 and the Rco1p PHD1 domains (43% sequence identity and 59% sequence similarity over a 51-residue segment), especially in the H3 binding site, we anticipate the PHD domain of the yeast protein, which is biologically well characterized, to share many of the features of the mammalian ortholog.
The mammalian MRG15 CD also shares many of the features of its yeast counterpart, Eaf3p, including its ability to interact with low affinity with H3K36me2/3.15,16 The ability of MRG15 CD to interact with free trimethyllysine and with H3K4me2 with only slightly lower affinity than H3K36me2/3 implies that sequence context does not play a significant role and that the domain should be regarded as a general trimethyllysine module. The presence of MRG15/Eaf3p in Tip60/NuA4 histone acetyltransferase (HAT) complexes associated with transcriptional activation and the targeting of these complexes to the promoter regions enriched in H3K4me3 suggest that the MRG15/Eaf3p CDs may also play a role in H3K4me3 binding.
The low affinity of the individual Pf1 PHD1-H3K4me0 and MRG15 CD-H3K36me2/3 interactions is surprising, but notwithstanding this unexpected feature, both sets of interactions have been shown to be biologically significant in yeast.10 Interestingly, this combinatorial readout mechanism does not appear to rely on direct, cooperative interactions between the Pf1 PHD1 and MRG15 CD domains. We recently characterized the interaction between MRG15 and Pf1 involving the MRG and MBD domains and found it to be of high affinity (10–20 nM),27 consistent with their roles as constitutively associated subunits of the Rpd3S/Sin3S complex. Our findings suggest a model where multivalency rather than cooperativity is the operative mechanism for chromatin targeting of the Rpd3S/Sin3S complex by the Pf1 PHD1 and MRG15 CD domains (Fig. 6), as has been described recently for another HDAC-containing corepressor complex.37 The bivalent nature of the interaction likely serves to enhance the residence time of the Rpd3S/Sin3S complex for histone deacetylation. By combining two low affinity interactions, the Rpd3S/Sin3S complex is thus poised for rapid dissociation from histones, particularly when the H3K36me2/3 signals are erased.
Figure 6.
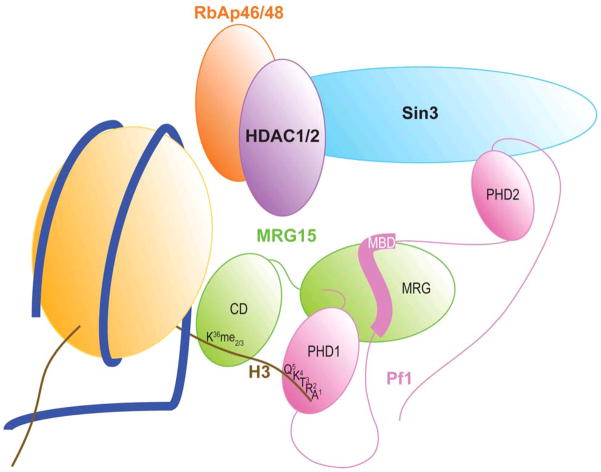
A molecular model for bivalent recognition of histone H3 by the Pf1 and MRG15 subunits of the Rpd3S/Sin3S complex. The Pf1 PHD1 and MRG15 CD domains engage different segments of H3 in a non-cooperative manner but the two relatively weak interactions likely translate into a more efficient interaction due to the nanomolar affinity interaction between Pf1 MBD and MRG15 MRG that is bolstered by another interaction with the Pf1 PHD1 domain that collectively tether the two chromatin-binding domains in cis in the Rpd3S/Sin3S complex. Note that although the cartoon shows Pf1 and MRG15 contacting the same histone H3 polypeptide, it is conceivable that they contact different H3s in the same or adjacent nucleosomes.
The surprising discovery of a Pf1 MBD-dependent interaction between MRG15 MRG and Pf1 PHD1 adds another layer of complexity to the mechanism of chromatin targeting by these proteins. Although the chromatin binding domains of MRG15 and Pf1 are separated from the MRG and MBD domains by rather long linker segments (~65–80 residues), the interaction between Pf1 PHD1 and MRG15 MRG is likely to lead to a dramatic reduction in this flexibility, thereby perhaps tuning Pf1 and MRG15 to engage appropriately spaced H3 signals within the same polypeptide or in different polypeptides within the same or adjacent nucleosomes, as was described recently for the NURD corepressor complex.37 The unanticipated role of Pf1 PHD1 in Rpd3S/Sin3S complex assembly by likely further stabilizing the MRG15 MRG-Pf1 MBD interaction confirms the suspicion that PHD domains are functionally diverse than they appear.
Materials and Methods
Production of MRG15, MRG15 CD, Pf1 PHD1, Pf1 PHD1/MBD and histone H3 peptides
The coding sequences for full-length human MRG15 and MRG15 CD (residues 1–90) was amplified using PCR, inserted into the pMCSG7 vector,38 and the cloned gene segments confirmed via DNA sequencing. E. coli BL21(DE3) cells containing the expression vector for MRG15 CD were grown at 37 °C in LB broth. Upon reaching an OD600nm of 0.6, the temperature was shifted to 20 °C, protein expression was induced with 1 mM isopropyl-beta-D-thiogalactopyranoside (IPTG), and cells were harvested 16 h later. The cell pellets were suspended in a 50 mM Tris-HCl buffer (pH 8) containing 0.15 M sodium chloride, 8 M urea, and 5 mM Tris-(2-carboxy-ethyl) phosphine (TCEP) hydrochloride (Buffer A); 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 μM leupeptin, 1 mM pepstatin, and 0.1% Triton X-100 were added, lysed using sonication, and centrifuged. The supernatant was incubated with Talon resin (EMD Biosciences) for 15 min, washed extensively with Buffer A lacking urea, and eluted using the wash buffer containing 250 mM imidazole. The eluted proteins were incubated with tobacco etch virus (TEV) protease for 4 hours at 22 °C, followed by overnight incubation at 4 °C. This mixture was centrifuged and the MRG15 CD protein was further purified via reversed-phase HPLC using a C18 column and a linear gradient of 0.1% trifluoroacetic acid (TFA) and 0.1% TFA in 80% acetonitrile. Protein fractions were pooled and lyophilized. Samples that were labeled with 15N and/or 13C were expressed and purified in the same fashion except the cells were grown in a M9 minimal media containing 15N-ammonium sulfate and/or 13C-D-glucose (Cambridge Isotopes). The identity of the protein and the extent of isotope incorporation were confirmed by electrospray ionization-mass spectrometry (ESI-MS).
The MRG domain of MRG15 (residues 155–323) and the MRG-binding domain of Pf1 (residues 200–241) were expressed and purified as described previously.39 Full-length MRG15 was expressed and purified as the recombinant MRG15 MRG polypeptide except that the protein was further purified using a HiTrap-SP cation-exchange column (GE Healthcare) to remove residual DNA contamination. The protein was loaded on to the column pre-equilibrated with 50 mM Tris (pH 7.9) buffer, washed extensively, and eluted with the same buffer containing 500 mM NaCl.
Peptides corresponding to unmodified histone H3 (residues 1-12), H3K4me2 (1-12), H3K36me2 (28-44), and H3K36me3 (28-44) were chemically synthesized using automated procedures (University of Utah DNA/Peptide Synthesis Facility) and purified to homogeneity by reversed-phase HPLC; peptide identities were confirmed by ESI-MS. The H3 (1-12) and H3K4me2 (1-12) peptides harbor a non-native, C-terminal tyrosine residue.
The coding sequences for human Pf1 PHD1 domain (residues 48–116) and Pf1 PHD1/MBD (residues 48–241) were amplified by PCR, sub-cloned into the pMCSG10 vector, and the sequence verified by DNA sequencing. Protein expression for both constructs followed the same protocol as for the MRG15 CD. To limit protein degradation, the protein was purified by selectively lysing the outer membrane and removing the periplasmic proteins via osmotic shock treatment followed by centrifugation. The pellets were resuspended in 50 mM Tris buffer (pH 8) containing 0.2 M NaCl, 5 mM dithiothreitol (DTT), 20 μM ZnSO4, 2 mM PMSF, 2 μM leupeptin, and 2 mM pepstatin, lysed, centrifuged and the supernatant incubated with glutathione sepharose 4B resin (GE Healthcare) for 30 min at 4 °C. The resin was washed extensively with 50 mM Tris buffer (pH 8) containing 0.2 M NaCl, 5 mM DTT, 20 μM ZnSO4 and incubated with TEV protease at 22 °C overnight. The flow-through containing the cleaved protein was collected and purified to homogeneity via reversed-phase HPLC as described for the MRG15 CD protein. Uniformly 15N- and/or 13C-labeled proteins were produced using the same procedure except that the cells were grown in M9 minimal medium containing 15N-ammonium sulfate and/or 13C-D-glucose, respectively, except the post-induction temperature used was 25 °C for higher yield. The identity of the protein and the extent of isotope incorporation were established by ESI-MS prior to NMR studies.
The coding sequence for human histone H3 (1-42) was amplified by PCR, introduced into the pET-30 Xa vector (Novagen), and the sequence confirmed by DNA sequencing. The protein was expressed and purified as described for MRG15 CD; however, instead of using the TEV protease, the N-terminal His6-tag was removed via treatment with Factor Xa and the resulting peptide purified via reversed-phase HPLC. The H3 K36C (1-42) mutant was generated via site-directed mutagenesis using the QuikChange protocol (Stratagene). The mutant protein was expressed and purified in the same manner as the wild-type protein. To generate the H3 K36Cme3 (1-42) peptide, the purified protein was treated with 2-bromoethyl-trimethylammonium bromide (Sigma-Aldrich) at 50 °C for 5 h.26 The desired N-trimethylated aminoethylcysteine product was isolated by passing through a PD-10 column. Uniformly 15N- and 13C-labeled H3 (1-42) peptides were produced using the same procedures, except cells were grown in M9 minimal medium containing 15N-ammonium sulfate and/or 13C-D-glucose, respectively. ESI-MS was used to verify peptide identities and isotope enrichment levels.
NMR samples and titrations
NMR samples were prepared by dissolving lyophilized MRG15 CD and histone H3 peptides in 20 mM Tris-acetate buffer (pH 7.5) containing 1 mM DTT-d10 and 0.2% NaN3. Pf1 PHD1 and Pf1 PHD1/MBD were refolded by dissolving the dry, lyophilized powder in 50 mM Tris buffer (pH 7) containing 6 M guanidine hydrochloride (Gdn-HCl) and 50 mM DTT and incubated at 42 °C for 5 h. After the addition of 1 mM ZnSO4, the sample was passed through a Superdex 75 gel filtration column (GE Healthcare) pre-equilibrated with 50 mM Tris buffer (pH 7.3) containing 0.2 M NaCl, 20 mM DTT and 15 μM ZnSO4. Fractions containing the purified protein were pooled, concentrated and exchanged into NMR buffer containing 20 mM Tris-d11 (pH 7.3), 50 mM NaCl, 10 μM ZnSO4, 15 mM DTT, and 0.2% NaN3; the pH was adjusted to 7.0 or 7.5 prior to NMR titrations.
Protein concentrations were measured spectrophotometrically.40 MRG15 CD concentrations for NMR titrations with various ligands ranged between 0.195 mM and 0.75 mM, whereas those for Pf1 PHD1, Pf1 PHD1/MBD and H3 (1-42) ranged between 0.05 mM and 0.15 mM. The pH of the solution after each addition during the titrations was carefully adjusted to the starting value. The ligands mimicking various histone modifications including acetyllysine, trimethyllysine, dimethylarginine, phosphoserine, and phosphothreonine were purchased from Sigma-Aldrich and used without further purification.
NMR spectroscopy
All NMR spectra were recorded at 25 °C on a 600 MHz Varian Inova spectrometer equipped with a pulsed-field gradient triple-resonance cold probe. NMR data processing and analysis were performed using NMRPipe41 or Felix 98.0 (Accelrys) and Sparky,42 respectively. Backbone 1H, 15N, and 13C resonance assignments for MRG15 CD, Pf1 PHD1, and histone H3 (1-42) were made by analyzing 3D CBCA(CO)NH, HNCACAB, HNCO and HN(CA)CO spectra;43 sequence-specific assignments were made, in part, using the AutoAssign software44 but were manually checked for accuracy. Backbone amide chemical shift deviations were calculated using the formula: Δδav = √(0.5 ((δHN,bound − δHN,free)2 + 0.04 (δN,bound − δN,free)2)). Non-linear regression assuming a single binding site was performed using MicroCal Origin software employing the fitting function y(x) = b(((x+1+a) − √((x+1+a)2 – 4x))/2), where y(x) is the observed deviation at molar ratio x, and a and b are the fitted parameters related to the dissociation constant and the chemical shift deviation at x = ∞, respectively.45
Pulldown Assays
Pulldown assays with biotinylated histone peptides were performed as described in Wysocka et al.46 Briefly, recombinant Pf1 PHD1 (residues 53–103) and Pf1 PHD2 (residues 268–319) fused to GST were expressed in E. coli and purified as described above for GST-Pf1 PHD1 (residues 48–116) except the proteins were eluted from the resin using glutathione. After incubating the respective GST-fusion proteins with the avidin-bound peptide beads, the beads were washed extensively in 50 mM Tris, pH 7.5, 150 mM NaCl, 0.05% NP-40, 0.3 mg/ml BSA and 1 mM dithiothreitol (DTT). Proteins bound to the avidin beads were then separated on a SDS-PAGE gel and visualized by Coomassie staining.
Supplementary Material
Highlights.
MRG15 CD binds H3K36me2/3 and H3K4me2 with low affinity and specificity
4Pf1 PHD1 binds H3K4me0 specifically but with low affinity; Pf1 PHD2 does not bind H3
Pf1 PHD1 also binds unexpectedly to MRG15 MRG in a Pf1 MBD-dependent manner
MRG15 CD and Pf1 PHD1 bind to their H3 targets via bivalent interactions
Acknowledgments
This work was supported by a grant from the NIH to I.R. (R01 GM064715). G.G.W. thanks C. David Allis for support. We thank Petar Jelinic for providing the GST-Pf1-PHD constructs. W.C. and A.P. were supported by Undergraduate Research Grant awards from Northwestern. We thank Jessica Wahi and Mthabisi Moyo for contributions towards the development of this work; Jessica was supported by a summer minority supplement from the NIH. Access to instrumentation in and institutional support for the WCAS Biological NMR Center are gratefully acknowledged.
Abbreviations
- CD
chromodomain
- PHD
plant homeodomain
- HDAC
histone deacetylase
- MRG
mortality factor on chromosome 4 related gene
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gardner KE, Allis CD, Strahl BD. OPERating ON Chromatin, a Colorful Language where Context Matters. Journal of Molecular Biology. 2011;409:36–46. doi: 10.1016/j.jmb.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Research. 2011;21:381–95. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayakawa T, Nakayama J. Physiological roles of class I HDAC complex and histone demethylase. J Biomed Biotechnol. 2011;2011:129383. doi: 10.1155/2011/129383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grzenda A, Lomberk G, Zhang JS, Urrutia R. Sin3: master scaffold and transcriptional corepressor. Biochimica et Biophysica Acta. 2009;1789:443–50. doi: 10.1016/j.bbagrm.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silverstein RA, Ekwall K. Sin3: a flexible regulator of global gene expression and genome stability. Curr Genet. 2005;47:1–17. doi: 10.1007/s00294-004-0541-5. [DOI] [PubMed] [Google Scholar]
- 7.McDonel P, Costello I, Hendrich B. Keeping things quiet: roles of NuRD and Sin3 co-repressor complexes during mammalian development. International Journal of Biochemistry and Cell Biology. 2009;41:108–16. doi: 10.1016/j.biocel.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cunliffe VT. Eloquent silence: developmental functions of Class I histone deacetylases. Current Opinion in Genetics and Development. 2008;18:404–10. doi: 10.1016/j.gde.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–92. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 10.Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–4. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- 11.Li B, Jackson J, Simon MD, Fleharty B, Gogol M, Seidel C, Workman JL, Shilatifard A. Histone H3 lysine 36 dimethylation (H3K36me2) is sufficient to recruit the Rpd3s histone deacetylase complex and to repress spurious transcription. Journal of Biological Chemistry. 2009;284:7970–6. doi: 10.1074/jbc.M808220200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jelinic P, Pellegrino J, David G. A novel mammalian complex containing Sin3B mitigates histone acetylation and RNA polymerase II progression within transcribed loci. Molecular and Cellular Biology. 2011;31:54–62. doi: 10.1128/MCB.00840-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicolas E, Yamada T, Cam HP, Fitzgerald PC, Kobayashi R, Grewal SI. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat Struct Mol Biol. 2007;14:372–80. doi: 10.1038/nsmb1239. [DOI] [PubMed] [Google Scholar]
- 14.Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell. 2005;20:971–8. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 15.Sun B, Hong J, Zhang P, Dong X, Shen X, Lin D, Ding J. Molecular basis of the interaction of Saccharomyces cerevisiae Eaf3 chromo domain with methylated H3K36. Journal of Biological Chemistry. 2008;283:36504–12. doi: 10.1074/jbc.M806564200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu C, Cui G, Botuyan MV, Mer G. Structural basis for the recognition of methylated histone H3K36 by the Eaf3 subunit of histone deacetylase complex Rpd3S. Structure. 2008;16:1740–50. doi: 10.1016/j.str.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buratowski S, Kim T. The role of cotranscriptional histone methylations. Cold Spring Harbor Symposia on Quantitative Biology. 2010;75:95–102. doi: 10.1101/sqb.2010.75.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang P, Du J, Sun B, Dong X, Xu G, Zhou J, Huang Q, Liu Q, Hao Q, Ding J. Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Research. 2006;34:6621–8. doi: 10.1093/nar/gkl989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yap KL, Zhou MM. Structure and mechanisms of lysine methylation recognition by the chromodomain in gene transcription. Biochemistry. 2011;50:1966–80. doi: 10.1021/bi101885m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Musselman CA, Kutateladze TG. Handpicking epigenetic marks with PHD fingers. Nucleic Acids Research. 2011;39:9061–71. doi: 10.1093/nar/gkr613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–23. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 22.Mansfield RE, Musselman CA, Kwan AH, Oliver SS, Garske AL, Davrazou F, Denu JM, Kutateladze TG, Mackay JP. Plant homeodomain (PHD) fingers of CHD4 are histone H3-binding modules with preference for unmodified H3K4 and methylated H3K9. Journal of Biological Chemistry. 2011;286:11779–91. doi: 10.1074/jbc.M110.208207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chakravarty S, Zeng L, Zhou MM. Structure and site-specific recognition of histone H3 by the PHD finger of human autoimmune regulator. Structure. 2009;17:670–9. doi: 10.1016/j.str.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X, Gozani O, Cheng X, Shi Y. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–22. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chignola F, Gaetani M, Rebane A, Org T, Mollica L, Zucchelli C, Spitaleri A, Mannella V, Peterson P, Musco G. The solution structure of the first PHD finger of autoimmune regulator in complex with non-modified histone H3 tail reveals the antagonistic role of H3R2 methylation. Nucleic Acids Research. 2009;37:2951–61. doi: 10.1093/nar/gkp166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–12. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie T, Graveline R, Kumar GS, Zhang Y, Krishnan A, David G, Radhakrishnan I. Structural Basis for Molecular Interactions Involving MRG Domains: Implications in Chromatin Biology. Structure. 2012;20:151–160. doi: 10.1016/j.str.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Patel DJ. Combinatorial readout of dual histone modifications by paired chromatin-associated modules. Journal of Biological Chemistry. 2011;286:18363–8. doi: 10.1074/jbc.R111.219139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayakawa T, Ohtani Y, Hayakawa N, Shinmyozu K, Saito M, Ishikawa F, Nakayama J. RBP2 is an MRG15 complex component and down-regulates intragenic histone H3 lysine 4 methylation. Genes to Cells. 2007;12:811–26. doi: 10.1111/j.1365-2443.2007.01089.x. [DOI] [PubMed] [Google Scholar]
- 30.van Oevelen C, Wang J, Asp P, Yan Q, Kaelin WG, Jr, Kluger Y, Dynlacht BD. A role for mammalian Sin3 in permanent gene silencing. Molecular Cell. 2008;32:359–70. doi: 10.1016/j.molcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malovannaya A, Lanz RB, Jung SY, Bulynko Y, Le NT, Chan DW, Ding C, Shi Y, Yucer N, Krenciute G, Kim BJ, Li C, Chen R, Li W, Wang Y, O’Malley BW, Qin J. Analysis of the human endogenous coregulator complexome. Cell. 2011;145:787–99. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–84. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaadige MR, Ayer DE. The polybasic region that follows the plant homeodomain zinc finger 1 of Pf1 is necessary and sufficient for specific phosphoinositide binding. Journal of Biological Chemistry. 2006;281:28831–6. doi: 10.1074/jbc.M605624200. [DOI] [PubMed] [Google Scholar]
- 34.Yochum GS, Ayer DE. Pf1, a novel PHD zinc finger protein that links the TLE corepressor to the mSin3A-histone deacetylase complex. Mol Cell Biol. 2001;21:4110–8. doi: 10.1128/MCB.21.13.4110-4118.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yochum GS, Ayer DE. Role for the mortality factors MORF4, MRGX, and MRG15 in transcriptional repression via associations with Pf1, mSin3A, and Transducin-Like Enhancer of Split. Molecular and Cellular Biology. 2002;22:7868–76. doi: 10.1128/MCB.22.22.7868-7876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Org T, Chignola F, Hetenyi C, Gaetani M, Rebane A, Liiv I, Maran U, Mollica L, Bottomley MJ, Musco G, Peterson P. The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Reports. 2008;9:370–6. doi: 10.1038/embor.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Musselman CA, Ramirez J, Sims JK, Mansfield RE, Oliver SS, Denu JM, Mackay JP, Wade PA, Hagman J, Kutateladze TG. Bivalent recognition of nucleosomes by the tandem PHD fingers of the CHD4 ATPase is required for CHD4-mediated repression. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:787–92. doi: 10.1073/pnas.1113655109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Exp Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 39.Kumar GS, Xie T, Zhang Y, Radhakrishnan I. Solution structure of the mSin3A PAH2-Pf1 SID1 complex: a Mad1/Mxd1-like interaction disrupted by MRG15 in the Rpd3S/Sin3S complex. Journal of Molecular Biology. 2011;408:987–1000. doi: 10.1016/j.jmb.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–26. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 41.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 42.Goddard TD, Kneller DG. Sparky. 2004;3 http://www.cgl.ucsf.edu/home/sparky/ [Google Scholar]
- 43.Bax A, Grzesiek S. Methodological advances in protein NMR. Accounts Chem Res. 1993;26:131–138. [Google Scholar]
- 44.Moseley HN, Sahota G, Montelione GT. Assignment validation software suite for the evaluation and presentation of protein resonance assignment data. Journal of Biomolecular NMR. 2004;28:341–55. doi: 10.1023/B:JNMR.0000015420.44364.06. [DOI] [PubMed] [Google Scholar]
- 45.French M, Swanson K, Shih SC, Radhakrishnan I, Hicke L. Identification and characterization of modular domains that bind ubiquitin. Methods Enzymol. 2005;399:135–57. doi: 10.1016/S0076-6879(05)99009-5. [DOI] [PubMed] [Google Scholar]
- 46.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.