Abstract
A quantitative bioluminescence assay for rapid and sensitive microRNA (miRNA) expression analysis was developed. The assay utilizes miRNA directly as a primer for binding to a circular single-stranded DNA template, followed by rolling circle amplification. The detection of inorganic pyrophosphate (PPi) molecules released during the DNA polymerization and amplification process is performed by a multi-enzyme system. PPi is converted to ATP by ATP-sulfurylase, which provides energy for luciferase to oxidize luciferin and produce light. Experimental results show that the assay has a dynamic range exceeding three orders of magnitude and the ability to discriminate miRNAs with high-homology sequences. Quantification of nine miRNAs in human heart tissues demonstrated a high cross-platform consistency between this assay and the TaqMan real time PCR assay with R2 = 0.941. The assay requires fewer reagents, can be performed at an isothermal condition without thermal cycling, and is capable of detecting miRNAs in less than an hour. Compared to the real time PCR and microarray-based detection methods, this assay provides a simpler, faster, and less expensive platform for miRNA quantification in life science research, drug discovery, and clinical diagnosis.
Keywords: Bioluminescence assay, microRNA, Quantification, Rolling circle amplification
Introduction
MiRNAs are short, noncoding RNAs of 18–25-nt in length that are processed from longer primary transcripts with hairpin and terminal loop regions through sequential enzymatic cleavage by RNase III proteins Drosha and Dicer [1]. Mature miRNAs form an RNA-induced silencing complex (RISC) with Argonaute proteins and mediate post-transcriptional gene silencing through translational inhibition or mRNA degradation in a sequence-specific manner. Since the discovery of the first miRNA lin-4 in Caenorhabditis elegans [2], miRNAs have been widely identified in plants and animals and play important roles in diverse biological processes including developmental timing, cell proliferation, cell death, apoptosis, angiogenesis, fat metabolism, and neuronal differentiation [3–8]. Changes in miRNA expression levels have been implicated in the etiology of abnormal cellular processes and progression of many human diseases, including cardiovascular and muscle diseases, abnormal inflammatory response, multiple sclerosis, retinal degeneration, Alzheimer’s and Parkinson’s diseases, and various forms of cancer [9–14]. Recent studies have shown that upregulated miRNAs may serve as oncogenes by promoting tumor development, while downregulated miRNAs may serve as tumor suppressor genes [15]. MiRNA profiling may classify poorly differentiated tumors [16]. In addition, miRNAs in body fluids have the potential as novel noninvasive biomarkers for early cancer screening [17].
To date, about 15000 miRNA entries from 142 species including 1212 mature human miRNAs have been collected by the Sanger miRBase database Release 16 (http://www.mirbase.org/). To investigate the biological functions of miRNAs, numerous techniques have been developed. Most initial miRNA expression analyses were carried out by cloning, Northern blots [18–19], or primer extension [20]. Later, to improve throughput and sensitivity of miRNA detection, the oligo-based microarrays and real-time PCR were developed, which are currently recognized as the two dominant miRNA detection techniques. Microarrays provide simultaneous profiling of hundreds of miRNAs in a multiplexed format, however, its sensitivity and specificity are relatively low [21,22]. In contrast, real-time PCR provides highly sensitive and specific miRNA detection, but it is costly and time consuming (> 4 hours for sample preparation and thermal cycling) [23]. Here, we present a novel enzymatic luminescence assay for sensitive and specific miRNA detection. This assay is based on isothermal rolling circle amplification (RCA) using miRNAs as primers. It eliminates a separate step for miRNA reverse transcription and allows for the direct and rapid detection of miRNA in less than an hour, which significantly simplifies the procedures and reduces the assay cost.
Materials and methods
Reagents
The custom-synthesized oligonucleotides and miRNAs were purchased from Integrated DNA Technologies (Coralville, IA). All oligonucleotides were purified by poly-acrylamide gel electrophoresis (PAGE). The synthetic miRNAs were purified by RNase-free high-performance liquid chromatography (HPLC). Natural 2′-deoxynucleoside-5′-triphosphates (dNTPs including dGTP, dCTP, and dTTP) were purchased from Promega (Madison, WI). The 2′-deoxyadenosine-5′-O-(1- thiotriphosphate) Sp-isomer (Sp-dATPαS) was purchased from Axxora, LLC (San Diego, CA). Adenosine 5′-phosphosulfate sodium salt (APS), sodium pyrophosphate decahydrate (PPi), apyrase, hexokinase, and the adenosine 5′-triphosphate (ATP) bioluminescent assay kit were purchased from Sigma (St. Louis, MO). NTPhos™ thermolabile phosphatase and CircLigase™ ssDNA ligase were purchased from Epicentre Biotechnologies (Madison, WI). The QIAquick nucleotide removal kit was purchased from Qiagen (Valencia, CA). ATP sulfurylase and Bst DNA polymerase large fragment were purchased from New England Biolabs (Ipswich, MA). Inorganic pyrophosphatase (PPase) was purchased from Roche Applied Science (Indianapolis, IN). Pre-cast 15% polyacrylamide gels were purchased from Jule, Inc. (Milford, CT). SYBR Green II was purchased from Lonza (Rockland, ME). E. coli total RNA, human heart total RNA, the TaqMan microRNA reverse transcription kit, TaqMan microRNA forward and reverse primers, and TaqMan universal PCR master mix were purchased from Applied Biosystems (Austin, TX). O’GeneRuler™ 1 kb DNA ladder, O’GeneRuler™ ultra low range DNA ladder, and orange loading dye were purchased from Fermentas (Glen Burnie, MD). All solutions were prepared in nuclease-free water (Ambion, Austin, TX).
Purification of dNTPs and buffer reagents
The RCA miRNA assay is based on the bioluminescence detection of PPi released during primer extension. Any endogenous PPi and ATP in the dNTPs and other reagents may contribute to undesirable background luminescence. Indeed, it was reported that dNTPs available from commercial sources often contain PPi and ATP contaminants at relatively high levels, which may limit the sensitivity of the bioluminometric assays [24,25]. To reduce the background luminescence, contaminants were enzymatically removed from dNTPs. In this study, 2 μl of 100 mM dCTP/dGTP/dTTP were mixed with 100 mM Tris-HCl (pH 7.5), 2 mM MgCl2, 10 mM D-glucose, 1 U of PPase, and 0.3 U of hexokinase in a total volume of 200 μl and incubated at room temperature for 20 minutes. Hexokinase selectively degrades contaminating ATP to ADP in the presence of D-glucose [25]. The PPase and hexokinase were subsequently removed by centrifugation on a Microcon YM-10 column (Millipore, Billerica, MA) at 14,000 g for 12 min.
Synthesis of circular oligonucleotide template
A linear oligonucleotide about 70 nt long was circularized by CircLigase™ ssDNA ligase, which catalyzes the formation of a phosphodiester bond between the 5′-phosphate and 3′-hydroxyl ends of the linear single-stranded DNA (ssDNA). In contrast to previously reported T4 DNA ligase and Ampligase DNA ligase that ligate the 5′ and 3′ DNA ends annealed adjacent to each other on a hybridized complementary DNA sequence [26], the CircLigase™ covalently joins the two ends of ssDNA without binding to the complementary DNA bridge. Avoiding the use of a DNA bridge is important here since the bridge may act as an unwanted primer in the downstream RCA reaction. The ligation process was performed following the manufacturer’s protocol. In brief, 20 pmol of 5′-phosphorylated oligonucleotide probe was mixed in 20 μl of reaction solution containing CircLigase™ 1x reaction buffer, 50 μM of ATP, 2.5 mM of MnCl2, and 200 U of CircLigase. The reaction was incubated at 60°C for 1 hour, heated to 80°C for 10 min to inactivate the CircLigase, and chilled on ice. The remaining linear ssDNA template and linear single-stranded adenylated-intermediate were subsequently digested by incubating the reaction solution with 10 U of Exonuclease I and 100 U of Exonuclease III at 37°C for 45 min. The synthesized circular ssDNA probe was resistant to exonuclease activity and therefore kept intact. The circularized oligonucleotide was further purified to remove enzymes, salts, and short linear DNA segments using the QIAquick nucleotide removal kit following the manufacturer’s protocol. Formation of ligation product was confirmed by running a denaturing 15% PAGE-urea gel stained with SYBR Green II.
MiRNA-primed rolling circle amplification
In addition to dNTPs, the oligonucleotide probes and RNA samples were cleaned up from the endogenous ATP and PPi before proceeding with RCA reaction. This was performed by adding 2 U of NTPhos™ phosphatase into an 8 μl reaction solution containing 1.5 μl of circularized DNA template, synthetic miRNA target or total RNA, 10 ng of primer P2, and 1x reaction buffer solution (50 mM NaCl, 10 mM Tris-HCl, 2 mM MgCl2, 1 mM DTT, pH 8) and incubated at 37°C for 20 min. The NTPhos™ enzyme removes 5′-phosphates from modified and unmodified NTPs, dNTPs, and degrades PPi, which significantly reduces the background luminescence. The NTPhos™ enzyme was then irreversibly heat-inactivated by incubating the reaction solution at 65°C for 15 min. The solution was cooled on ice for 10 min for primer annealing, followed by incubating at 50°C for 15 min in a 20 μl reaction volume containing 200 μM dNTP (dGTP, dCTP, dTTP, and dATPαS), 8 U Bst DNA polymerase, 1x reaction buffer solution, and 0.1% Triton X-100.
Enzymatic luminescence detection of pyrophosphate
The PPi generated from miRNA extension on circularized DNA was detected using a multi-enzyme reaction system. The ATP assay mix from the Sigma ATP bioluminescent assay kit was dissolved in 5 ml of sterile water and kept on ice for at least one hour for complete dissolution. A 100 μl volume of ATP detection solution was then mixed with 30 mU of ATP-sulfurylase, 4 mU of PPase, 1.5 mU of apyrase and kept on ice for at least 20 min. The detection solution was activated immediately before PPi detection by adding 0.2 nmol of APS substrate and mixing well by pipetting. For PPi quantification, 1 μl of the DNA amplification solution was mixed with 10 μl of detection reagent. The luminescence signal was measured immediately with a Sirius luminometer (Berthold Detection Systems, Pforzheim, Germany).
Real-time PCR
The TaqMan miRNA assay, a two-step real-time PCR assay from Applied Biosystems, was used in validation studies for miRNA quantitation in total RNA samples. Human heart total RNA (5 ng) was first reversed transcribed to cDNA using reagents and specific reverse transcription primers from the TaqMan microRNA assay. The cDNA was subsequently quantified by real time PCR using specific forward and reverse primers following manufacturer’s protocols. The real time PCR analysis was performed on an ABI Prism® 7000 Sequence Detection System (Applied Biosystems).
Results
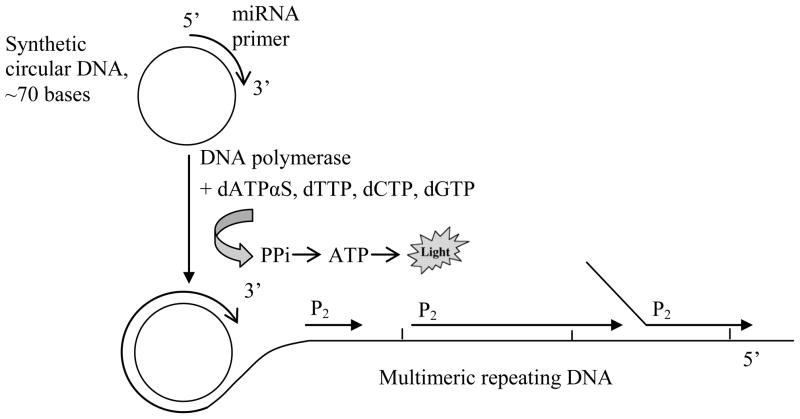
A diagram illustrating the RCA luminescence assay for miRNA detection is shown in Figure 1. A short, single-stranded DNA circular template is designed as a probe to recognize and hybridize with high specificity to a target miRNA molecule. After hybridization, the miRNA serves as a primer for strand extension by Bst DNA polymerase, which facilitates the rolling circle amplification by extending the miRNA primer around the circular DNA template, displacing the downstream non-template strand, and generating a long multimeric DNA product that consists of consecutive repeats of the circular template sequence. Another primer P2 (reverse primer) bearing a sequence identical to a second binding site on the circular probe subsequently initiates a second round of extension, which uses the multimeric DNA product as a template. Once an upstream primer encounters a bound downstream primer, the polymerase displaces the downstream bound primer along with its extended sequence. With the multiple primer extensions taking place simultaneously, the ramification amplification (RAM) generates a large ramified complex, during which the PPi molecules are released as the result of nucleotide incorporation by the polymerase. The released PPi are converted into ATP by ATP-sulfurylase, and subsequently, the ATP provides energy for luciferase to oxidize luciferin and generating light:
Figure 1.
Schematic representation of the enzymatic luminescence miRNA assay. The quanitification of miRNAs includes two steps: rolling circle amplification and detection of released PPi molecules. The miRNA molecule specifically binds to its complementary circular DNA probe and serves as a primer for rolling circle amplification by Bst DNA polymerase. Thousands of PPi molecules are released during the isothermal amplification, which are subsequently quantified by the bioluminescence reaction with ATP-sulfurylase/luciferase enzymes.
Per each dNTP incorporated by the DNA polymerase into the elongated strand, one PPi molecule is released. In the RCA miRNA assay, the synthesis of the resulting ramified products requires the incorporation of thousands of deoxynucleotides, which results in the production of thousands of PPi molecules and the emission of large numbers of photons that can be detected by a luminometor. Therefore, the RCA miRNA assay implements both the target amplification, i.e., production of a large number of copies of target miRNA sequence, and signal amplification, i.e., generation of a large number of reporter PPi molecules per each copy of miRNA sequence produced. Accordingly, the assay could be extremely sensitive. What is reducing assay sensitivity is the background luminescence. One source of background luminescence is from endogenous contaminants that could be addressed as was discussed in the previous section. Another source is dATP, which is required for the RCA reaction. It was reported that dATP can serve as a luciferase substrate with light emission efficiency about 1.7% relative to ATP [25]. Fortunately, dATP can be substituted by a nucleotide analogue dATPαS in PCR and RCA reactions. As has been demonstrated in our previous work, dATPαS was efficiently incorporated into the extended DNA strand by Bst DNA polymerase, while it had orders of magnitudes less efficiency to be used as a substrate for the luciferase [27].
Sensitivity of bioluminescence detection of PPi molecules
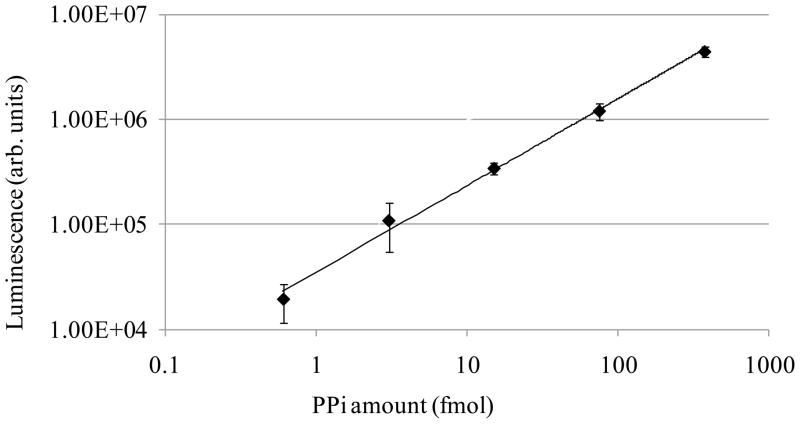
The bioluminescence detection of PPi at different concentrations is illustrated in Figure 2. The PPi stock solution of 3.76 mM was prepared by dissolving 0.05 g of sodium pyrophosphate decahydrate (Na4P2O7·10H2O) in 30 ml of 10 mM Tris-HCl (pH 8) buffer. The stock solution was then serially diluted 1:5 in the buffer to concentrations of 0.6 nM, 3 nM, 15 nM, 75 nM, and 376 nM followed by the luminescence detection. The PPi calibration curve in Fig. 2 shows an excellent linearity, demonstrating a dynamic range exceeding three orders of magnitude with a detection sensitivity as low as 0.6 fmol of PPi.
Figure 2.
PPi calibration curve. One μl of serially diluted PPi solution with concentrations ranging from 0.6 nM to 376 nM was mixed with 10 μl of PPi detection solution. The light generated from the enzymatic reaction was immediately quantified using a Sirius luminometer.
Evaluation of the bioluminescence-based miRNA assay
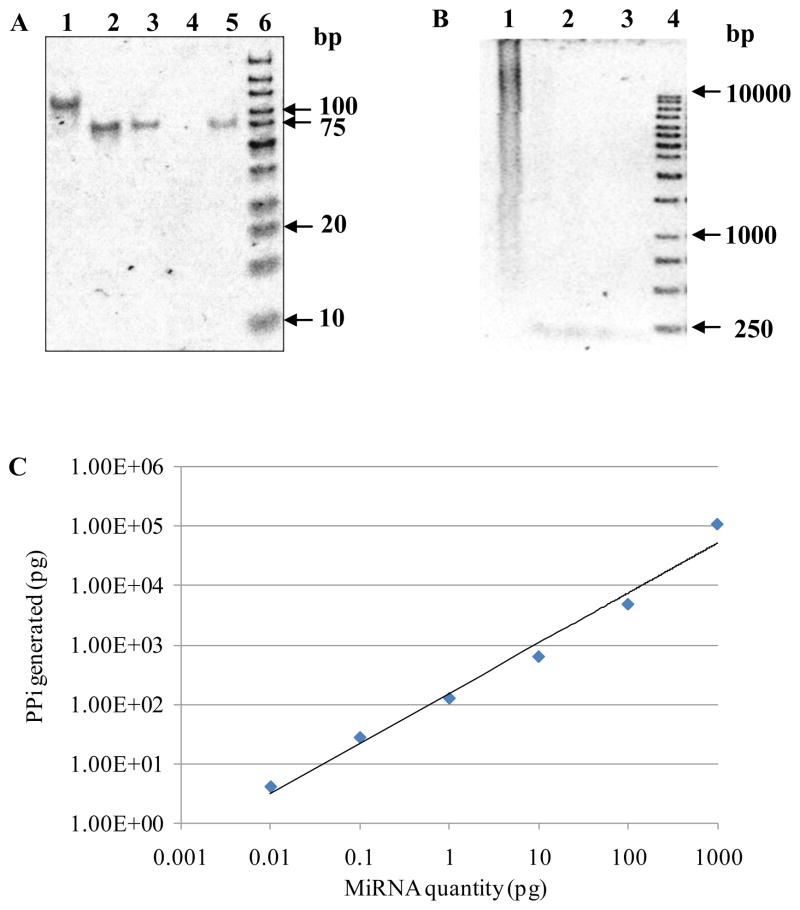
The dynamic range and sensitivity of the miRNA assay were evaluated using a synthetic 71-base-long oligonucleotide probe and a synthetic 23-base-long RNA target. The sequence of the 5′-phosphorylated linear probe is: 5′-CAGAACAGCACAAGACAGGACAAGACACACGCCGA AGAAACCCAGCAGACAACGCAGCCCCAGACAGACGA-3′. The probe is homologous to a synthetic miRNA target with the sequence of GGCUGCGUUGUCUGCUGGGUUUC. The linear probe was circularized and purified following the protocol as described above. Formation of the circular probe was verified by analyzing purified ligation product on a 15% denaturing PAGE gel as shown in Figure 3A. From the gel image, the circularized DNA migrated slower than the linear ssDNAs (Figure 3A Lane 1 vs. Lanes 2, 3, and 5). This is consistent with the fact that supercoiled DNA migrates the most, linear in the middle, and open circular migrates the least on an electrophoresis gel. According to the migration time for the products in Fig. 3A, line 1 indicates the formation of an open circular DNA product.
Figure 3.
Sensitivity and dynamic range of the enzymatic luminescence assay. (A) 15% acrylamide/7M urea denaturing gel electrophoresis of closed circular ssDNA converted from a linear ssDNA by CircLigase. Lane 1: closed circular ssDNA reaction product. Lane 2: linear oligonucleotide probe hsa-miR-26a, 50 ng. Lane 3: linear oligonucleotide probe hsa-miR-26a, 10 ng. Lane 4: blank. Lane 5: 71-base-long linear oligonucleotide probe, 8 ng. Lane 6: O’GeneRuler™ ultra low range DNA ladder, 1 μl. (B) 1% agarose gel electrophoresis of RCA reaction. Lane 1: RCA reaction products from synthetic circular ssDNA and 1 ng complementary RNA target. Lane 2: circular ssDNA. Lane 3: duplex of circular ssDNA and its complementary RNA target. Lane 4: O’GeneRuler™ 1 kb DNA ladder. (C) Sensitivity and dynamic range of the enzymatic luminescence assay for miRNA detection.
Assay sensitivity and dynamic range have been evaluated in complex RNA samples. Six RNA samples were prepared by spiking known amounts of synthetic miRNA target (0.01pg–1000 pg) to 1 ng of total E. coli RNA. The amplification products from the sample containing 1 ng of synthetic miRNA target were analyzed on 1% agarose gel and stained by SYBR Green II (Figure 3B Lane 1). As shown in Figure 1, the rolling circle amplification is a process bearing miRNA-primed DNA polymerization, binding and extension of multiple reverse primers from the initial RCA ssDNA product, and the strand displacement of downstream primers. The reaction end-products were multimeric dsDNAs of various lengths, including smaller units such as monomers, dimers, trimers, etc., which yielded a broad smear of DNA bands with continuous distribution as indicated by lane 1 in Figure 3B. The PPi molecules produced from the RCA reactions can be accurately quantified by the luminescence PPi assay as illustrated in Figure 3C. The log plot of the luminescence intensity, Ilum, vs. the quantity of miRNA target, [miRNA], is approximated by a straight line, which corresponds to power dependence Ilum [miRNA]0.8 and a dynamic range over 5 orders of magnitude. The assay sensitivity estimated from Fig. 3C is 0.01 pg of miRNA target.
Recognition of closely related miRNAs
In our previous study, the specificity of microRNA-primed DNA polymerization reaction by Bst DNA polymerase was investigated using linear oligonucleotide templates [27]. It was found out that thermophilic Bst DNA polymerase large fragment was capable of highly specific miRNA target recognition, which can discriminate two miRNAs with single-base mismatch located at the 6th base from the 3′ end. It was reported that Bst and Taq DNA polymerases have a more than five-fold lower efficiency in extending a mismatched primer than a matched primer [28]. The extension efficiency of single-mismatch primers was strongly influenced by the mismatch location [29]. It was observed that single-base mismatch present at the last five positions from the 3′ end of the primer almost totally block the primer extension (extension efficiencies is zero or <3.9%). When the mismatch position is located in the internal regions, i.e., positions 7–13 from the 3′ end of the primer, the extension efficiency is in the range from 1.8% to 28.5% affected mainly by the primer-target binding stability. When the single mismatch occurred at the positions of 14 to 19 from the 3′ end, the extension efficiencies were distributed between 11.9% and 69.3%.
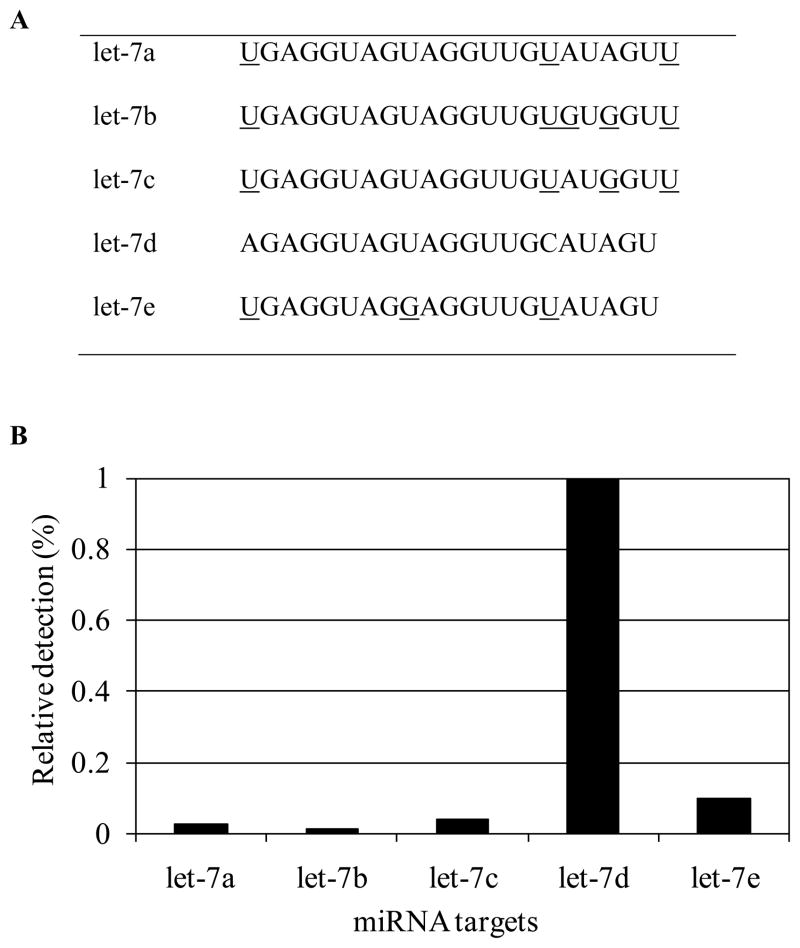
In this study, the ability of the enzymatic luminescence assay to discriminate miRNAs with a high degree of homology was evaluated using five synthetic miRNAs let-7a, let-7b, let-7c, let-7d, and let-7e (sequences shown in Figure 4A). The similarities between two miRNA sequences in the let-7 family were calculated using Hamming distances (Table 2). The Hamming distance is the number of differing letters between two equal-length RNA sequences aligned end-to-end [30]. The similarity is defined as the ratio of the difference of the miRNA length and the Hamming distance to the miRNA length. As listed in Table 2, the similarities between two miRNAs in the let-7 family vary from 77% to 95%. The let-7a/c and let7b/c pairs have the highest similarities of 95% with their single-base mismatches located at the 4th and 6th positions from the 3′ end, respectively. As demonstrated in our previous study, Bst DNA polymerase effectively discriminated the single-base mismatches located as far as the 6th positions from the 3′ end [27,29]. To illustrate the specificity of the luminescence assay, we performed a series of rolling circle amplification experiments by hybridizing an oligonucleotide (0.71 pmol) with perfect-match sequence to let-7d as the template with each of the five miRNAs (0.016 pmol) as a target individually. The similarities of let-7d to other miRNAs are in the range of 77% to 91%. The single-base mismatch for let-7d and let-7e occurs at the 6th positions from the 3′ end. In addition, two base mismatches exist between let-7a and let-7d in the 3′ end from positions 1–7. By analyzing the sequence differences of the five highly homologous miRNAs, we identified that the cross-hybridization experiments described above could effectively demonstrate the capability of the assay to distinguish miRNAs with differences as little as one single nucleotide.
Figure 4.
Discrimination of the high-homology miRNA let-7 family. (A) Sequences of miRNA let-7 family. Bases that are different from let-7d are underlined. (B) Relative detection efficiency calculated by the ratio of PPi amount generated between the perfectly matched let-7d target and other mismatched miRNA targets, assuming 100% efficiency for the perfectly matched let-7d target.
Table 2.
Similarities between two miRNA sequences in the let-7 family
| let-7a | let-7b | let-7c | let-7d | let-7e | |
|---|---|---|---|---|---|
| let-7a | 100 | 91 | 95 | 91 | 91 |
| let-7b | 91 | 100 | 95 | 77 | 82 |
| let-7c | 95 | 95 | 100 | 82 | 82 |
| let-7d | 91 | 77 | 82 | 100 | 86 |
| let-7e | 91 | 82 | 82 | 86 | 100 |
For rolling circle amplification, the probe was circularized and reaction was performed as described in previous section for each of the five synthetic miRNAs individually. The relative detection efficiency was calculated by the ratio of PPi amount generated between perfectly matched and mismatched miRNA targets, assuming 100% efficiency for the perfectly matched let-7d target (Figure 4B). The non-specific signals for miRNAs with 2–4 base differences from let-7d were as low as 1%–3.8%. The mismatched signal reached 9.8% for let-7e, which differs from perfectly matched let-7d by only a single nucleotide at the 3′ end.
The factors that contribute to the high specificity of the enzymatic luminescence assay are: (1) MiRNA molecules have a high 3′-end heterogeneity variation as demonstrated in Northern blotting and cloning studies [31]. Bst DNA polymerase has a high capability to discriminate 3′ mismatched. (2) The thermophilic characteristics of Bst DNA polymerase allows the reaction to be carried out at the optimum temperature of 65°C instead of 37°C for mesophilic DNA polymerases, which significantly reduces nonspecific priming and DNA strand extension. (3) The utilization of a second primer (reverse primer) complementary to a second binding site in the circular ssDNA template greatly improves the sequence-specific amplification to distinguish closely related mRNAs.
MiRNA quantification in human total RNA samples
Nine miRNAs with high (hsa-miR-1, hsa-miR-26a, hsa-miR-133b), medium (hsa-miR-195, hsa-miR-21, hsa-miR-221), and low (hsa-miR-191, hsa-miR-186, hsa-miR-103) abundance were selected as representative targets for expression analysis based on a previous study on miRNA profiling in normal human tissue samples [32]. MiRNA cel-miR-2 (a miRNA from Caenorhabditis elegans) was used as a negative control. The sequences of the designed linear oligonucleotide probes, negative control, and reverse primer P2 are listed in Table 1. The miRNA-primed RCA reaction was performed using 500 ng of human heart total RNA with three replicates for each miRNA target. The luminescence signals were calibrated by subtracting the signals from the negative control sample.
Table 1.
Sequences of linear oligonucleotide probes
| No. | Symbol | Linear oligonucleotide probe sequences |
|---|---|---|
| 1 | hsa-miR-1 | /5Phos/CAG AAC AGC ACA AGA CAG GAC AAG ACA CAC GCC GAA ATA CAT ACT TCT TTA CAT TCC ACC AGA CAG ACG A |
| 2 | hsa-miR-26a | /5Phos/CAG AAC AGC ACA AGC AAG AGC AGA CAA CAC GCC GAA AGC CTA TCC TGG ATT ACT TGA ACC AGA CAG ACG A |
| 3 | hsa-miR-133b | /5Phos/CAG AAC AGC ACA AGA CAG GAC AAG ACA CAC GCC GAA TAG CTG GTT GAA GGG GAC CAA ACC AGA CAG ACG A |
| 4 | hsa-miR-195 | /5Phos/CAG AAC AGA GAG CAC CAG GAC AAG ACA CAC GCA GAC GCC AAT ATT TCT GTG CTG CTA CAG ACA GAC GA |
| 5 | hsa-miR-21 | /5Phos/CAG AAC AGC ACA AGA CAG GAC AAG ACA CAC GCC GAA TCA ACA TCA GTC TGA TAA GCT ACC AGA CAG ACG A |
| 6 | hsa-miR-221 | /5Phos/CAG AAC AGC ACA AGA CAG GAC AAG ACA CAC GCC GAA GAA ACC CAG CAG ACA ATG TAG CTC CAG ACA GAC GA |
| 7 | hsa-miR-191 | /5Phos/CAG AAC AGC ACA GGA CAA GAC ACA CGC CGA ACA GCT GCT TTT GGG ATT CCG TTG CAG ACA GAC GA |
| 8 | hsa-miR-186 | /5Phos/CAG AAC AGC ACA AGA CAG GAC AAG ACA CAC GCC GAA AGC CCA AAA GGA GAA TTC TTT GCC AGA CAG ACG A |
| 9 | hsa-miR-103 | /5Phos/CAG AAC AGC AAG ACA ACA AGA CAC ACG CCG AAT CAT AGC CCT GTA CAA TGC TGC TCA GAC AGA CGA |
| 10 | cel-miR-2 | /5Phos/CAG AAC AGC ACA AGA CAG GAC AAG ACA AGG ACA GGA GCA CAT CAA AGC TGG CTG TGA TAC CAG ACA GAC GA |
| 11 | P2 | GACAGACGACAGAACAG |
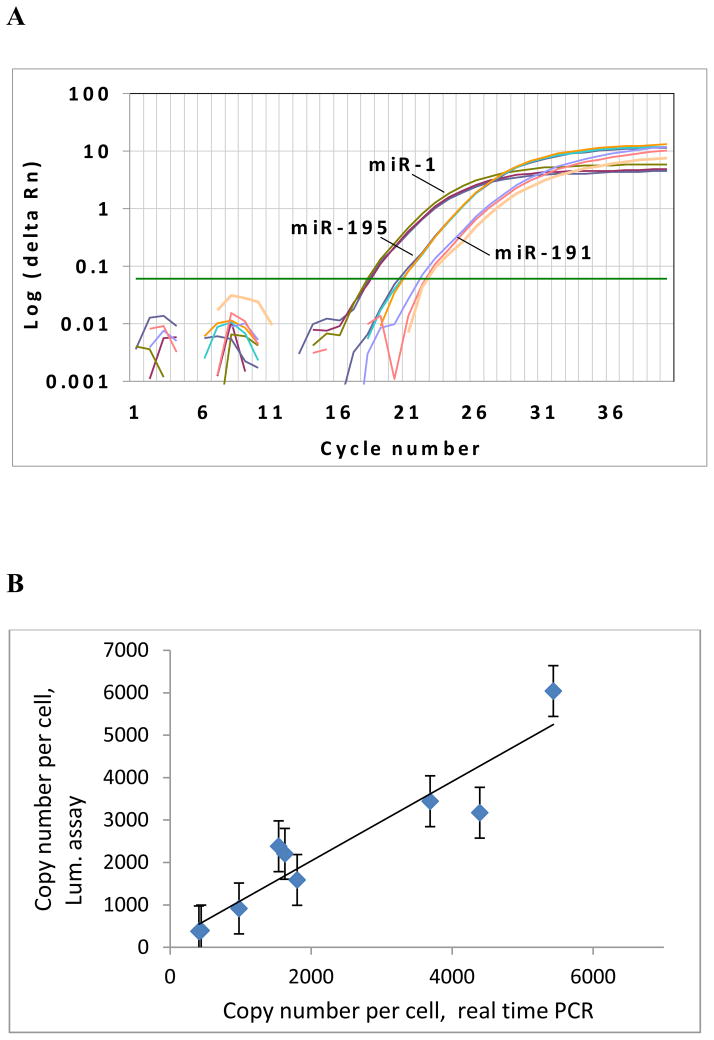
The experimental data above were compared with the TaqMan real-time PCR assay developed by Applied Biosystems. The amplification plot of the real-time PCR assay for miRNA targets of miR-1, miR-195, and miR-191 were illustrated in Figure 5A. The consistency of miRNA quantification by real-time PCR and the bioluminescence assay is illustrated in Figure 5B. The copy number per cell was calculated based on the standard curve of lin-4 synthetic miRNA (assuming 15 pg of total RNA per cell). The miRNA quantification data of the PPi-based luminescence assay is consistent with that of the real-time PCR assay with R2 = 0.941. The experimental data successfully classified the nine miRNAs into three groups of high, medium, and low expression levels with three miRNAs in each group, which are consistent with previous studies using real-time PCR for miRNA quantification [32].
Figure 5.
Comparison of the enzymatic luminescence assay and TaqMan miRNA assay. (A) Detection of miR-1, miR-195, and miR-191 by the TaqMan real-time PCR. Three replicates were performed for each miRNA. (B) Copy number per cell for each miRNA using the enzymatic luminescence assay and TaqMan real-time PCR assay.
Discussion
Since the discovery of the first miRNA in 1993, a surge of interest in miRNA characterization has facilitated the development of highly sensitive and specific miRNA detection techniques to identify disease-related miRNAs and their regulation mechanism. Current methods for miRNA quantification are largely based on real-time PCR and microarrays, which are time-consuming and expensive. The enzymatic luminescence assay presented in this paper provides a fast, simple, and cost effective miRNA detection technique. The assay utilizes miRNA molecules directly as primers for sequence recognition, which eliminates the reverse transcription procedure required by real-time PCR and the cy-dye labeling required for microarray-based assay. The RCA reaction is performed at isothermal conditions without the need for a thermocycler. The isothermal nature makes it readily adaptable to routine clinical use with reduced instrument setup cost and fewer issues concerning quality control of the instrument. The presented assay takes less than one hour, which is much faster than real-time PCR (~ 4 hr) and microarrays (overnight). In addition, the luminescence signals are collected using a luminometer, which costs much less than a real time RT-PCR system. The assay can be performed in a multiplex format using a microplate with 96, 384 or even 1536 sample wells. Considering that there are only a few hundred mature miRNAs per organism reported in the most recently updated miRBase release 18, the microplate-based bioluminescence assay fits very well for miRNA expression profiling, which in most typical applications requires a throughput of a few hundred miRNA targets per assay.
The combination of rolling circle amplification and sulfurylase/luciferase-based PPi detection provides high sensitivity for miRNA quantification. In this type of amplification, two primers are used. One is the miRNA target (forward primer) and another is a 15–20 base long oligonucleotide (reverse primer) with an identical sequence to a second binding site in the circular ssDNA template. Compared to the one-primer linear amplification, the two-primer ramification amplification is an exponential amplification process, which can be expressed as 2U (U is the number of repeats generated from the initial circular DNA template) [33]. The utilization of two primers in the amplification process not only provides an equivalent power of amplification as PCR, but also highly improves the sequence-specific recognition of the assay to distinguish closely related mRNAs. Another factor that contributes to the highly sensitive miRNA detection is the release of one PPi molecule per each dNTP incorporated into the DNA strand. Therefore, with the multiple primer extensions taking place simultaneously in the ramification amplification, thousands of PPi molecules are generated in a short period of time. In addition, the PPi molecules are detected by a luminometer, which operates in a photon-counting mode. Due to its “digital nature”, the photon-counting detection technique has very high sensitivity and dynamic range [34]. Our experimental data indicate that the Sirius single-tube luminometer from Berthold is able to detect at least 0.6 fmol PPi molecules with a dynamic range of 3 logs. To detect reaction solutions with PPi concentration of more than 600 fmol/μl, dilution of the solution may be required to meet the detection limit of the luminometer. Low abundance miRNAs in 500 ng total RNA were quantified in a 15-min amplification reaction. The miRNA quantification data were consistent with the data from real-time PCR assay with R2 = 0.941. Higher sensitivity may be achieved by extending the DNA amplification time, which will generate more PPi molecules during the DNA polymerization process.
A number of DNA polymerases are known to have strand-displacement activity and are able to perform RCA reactions, including ø29 DNA polymerase, 3′ 5′ exo− Klenow fragment, Sequenase, and Bst DNA polymerase [35]. Among these enzymes, Bst DNA polymerase was selected due to its capability of extending RNA primers and discriminating homologous miRNAs for highly specific sequence recognition [27]. In addition, Bst DNA polymerase possesses stronger RNA strand displacement activity and thermal stability that allows the incubation at 65°C for at least 30 min [36]. The novel miRNA detection system presented in this paper can detect as low as 0.01 pg of miRNA in a 15-min reaction with a dynamic range of at least 5 orders of magnitude. The cross-reaction signal between homologous sequences that differ by only a single nucleotide at the 3′ end was 9.8% for let-7d and let-7e, which is much lower than 39% reported by the microarray-based assay [22]. It indicates that the miRNA-primed assay has a high specificity to discriminate high-homology miRNA targets. It was reported that miRNAs are frequently altered by editing, i.e., by a site-specific modification of a miRNA sequence, which yields a product differing from that encoded by the DNA template. Most RNA editing in human cells is adenosine to inosine (A-to-I) RNA editing (identified as A-to-G changes) and 3′ terminal A and U addition [37]. The 3′-end miRNA editing, including A and U addition, can be detected by the bioluminescence assay using Bst DNA polymerase. The assay has high capability to discriminate the 3′-end modifications, which has been illustrated by the detection of let-7a and let-7d in Figure 4. Detection of miRNA editing at the 5′ end could be more challenging and may require the design of probes with carefully placed mismatches at the 5′ end. Mismatches in the miRNA sequence are able to control the stringency of the probe-target binding and increase the probe specificity for the recognition of 5′-end edited miRNAs.
Following the wave of the initial discoveries in miRNA field, there is an increasing need for simple, fast, and inexpensive miRNA profiling technology with high sensitivity and specificity. Our bioluminescence assay based on miRNA-primed isothermal amplification addresses some drawbacks of the current existing techniques and provides a new expression profiling tool that can be used in the study of disease-specific miRNA biomarkers, as well as their function in a wide range of biological processes.
Acknowledgments
The authors express gratitude to James Patton at Department of Biological Sciences in Vanderbilt University for his comments and suggestions. This work is supported by the NIH/NIGMS SBIR grant R43CA126647.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- 4.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 5.Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Lee CG. MicroRNA and cancer-focus on apoptosis. J Cell Mol Med. 2009;13:12–23. doi: 10.1111/j.1582-4934.2008.00510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hua Z, Lv Q, Ye W, Wong CK, Cai G, Gu D, Ji Y, Zhao C, Wang J, Yang BB, Zhang Y. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 10.Williams AH, Liu N, van Rooij E, Olson EN. MicroRNA control of muscle development and disease. Curr Opin Cell Biol. 2009;21:461–469. doi: 10.1016/j.ceb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keller A, Leidinger P, Lange J, Borries A, Schroers H, Scheffler M, Lenhof HP, Ruprecht K, Meese E. Multiple sclerosis: microRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS One. 2009;4:e7440. doi: 10.1371/journal.pone.0007440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loscher CJ, Hokamp K, Kenna PF, Ivens AC, Humphries P, Palfi A, Farrar GJ. Altered retinal microRNA expression profile in a mouse model of retinitis pigmentosa. Genome Biol. 2007;8:R248. doi: 10.1186/gb-2007-8-11-r248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson PT, Wang WX, Rajeev BW. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008;18:130–138. doi: 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Kerin MJ. MicroRNAs as novel biomarkers for breast cancer. J Oncol. 2009:950201. doi: 10.1155/2010/950201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 17.Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010;251:499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- 18.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 19.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 20.Seitz H, Royo H, Bortolin ML, Lin SP, Ferguson-Smith AC, Cavaillé J. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 2004;14:1741–1748. doi: 10.1101/gr.2743304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 22.Wang H, Ach RA, Curry B. Direct and sensitive miRNA profiling from low-input total RNA. RNA. 2007;13:151–159. doi: 10.1261/rna.234507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou G, Kamahori M, Okano K, Chuan G, Harada K, Kambara H. Quantitative detection of single nucleotide polymorphisms for a pooled sample by a bioluminometric assay coupled with modified primer extension reactions (BAMPER) Nucleic Acids Res. 2001;29:e93. doi: 10.1093/nar/29.19.e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moyer JD, Henderson JF. Nucleoside triphosphate specificity of firefly luciferase. Anal Biochem. 1983;131:187–189. doi: 10.1016/0003-2697(83)90152-5. [DOI] [PubMed] [Google Scholar]
- 26.Nilsson M, Malmgren H, Samiotaki M, Kwiatkowski M, Chowdhary BP, Landegren U. Padlock probes: circularizing oligonucleotides for localized DNA detection. Science. 1994;265:2085–2088. doi: 10.1126/science.7522346. [DOI] [PubMed] [Google Scholar]
- 27.Sun Y, Gregory KJ, Golovlev V. Efficiency and specificity of microRNA-primed nucleotide analog incorporation by various DNA polymerases. Anal Biochem. 2009;391:85–90. doi: 10.1016/j.ab.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aliotta JM, Pelletier JJ, Ware JL, Moran LS, Benner JS, Kong H. Thermostable Bst DNA polymerase I lacks a 3′-->5′ proofreading exonuclease activity. Genet Anal. 1996;12:185–195. [PubMed] [Google Scholar]
- 29.Wu JH, Hong PY, Liu WT. Quantitative effects of position and type of single mismatch on single base primer extension. J Microbiol Methods. 2009;77:267–275. doi: 10.1016/j.mimet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 30.Hamming RW. Numerical methods for scientists and engineers. 2. Dover; New York: 1987. [Google Scholar]
- 31.Aristarkhov A, Barken KB. Exiqon technical note. http://www.exiqon.com/ls/Documents/Scientific/3%E2%80%99-heterogeneity%20and%20microRNA%20expression%20analysis.pdf.
- 32.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang D, Wu J, Ye F, Feng T, Lee I, Yin B. Amplification of circularizable probes for the detection of target nucleic acids and proteins. Clin Chim Acta. 2006;363:61–70. doi: 10.1016/j.cccn.2005.05.039. [DOI] [PubMed] [Google Scholar]
- 34.Berthold F. In: Luminescence immunoassay and molecular applications. Dyke KV, Dyke RV, editors. CRC Press; Boca Raton, FL: 2000. pp. 11–26. [Google Scholar]
- 35.Demidov VV. Rolling-circle amplification in DNA diagnostics: the power of simplicity. Expert Rev Mol Diagn. 2002;2:542–548. doi: 10.1586/14737159.2.6.542. [DOI] [PubMed] [Google Scholar]
- 36.New England Biolabs 2007–2008 Catalog & technical reference, pp. 89.
- 37.Landgraf P, Rusu M, Sheridan R, Sewer A, Lovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression Atlas based on small RNA library sequencing. cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]