Abstract
Caspase inhibition is a promising approach for treating multiple diseases. Using a reconstituted assay and high-throughput screening, we identified a group of non-peptide caspase inhibitors. These inhibitors share common chemical scaffolds, suggesting same mechanism of action. They can inhibit apoptosis in various cell types induced by multiple stimuli; they can also inhibit caspase-1-mediated interleukin generation in macrophages, indicating potential anti-inflammatory application. While these compounds inhibit all the tested caspases, kinetic analysis indicates they do not compete for the catalytic sites of the enzymes. The co-crystal structure of one of these compounds with caspase-7 reveals that it binds to the dimerization interface of the caspase, another common structural element shared by all active caspases. Consistently, biochemical analysis demonstrates that the compound abates caspase-8 dimerization. Based on these kinetic, biochemical, and structural analyses, we suggest that these compounds are allosteric caspase inhibitors that function through binding to the dimerization interface of caspases.
Introduction
Apoptosis is critical for multicellular organisms to maintain tissue homeostasis and to eliminate unwanted or damaged cells. It is involved in development, immune responses and many other physiological processes (Vaux and Korsmeyer, 1999). Molecularly, apoptosis is executed by a subfamily of cysteine proteases known as caspases (Pop and Salvesen, 2009; Riedl and Shi, 2004). In mammals, there are two well-characterized caspase activation pathways: the intrinsic mitochondria-mediated pathway and the extrinsic death receptor-mediated pathway (Jiang and Wang, 2004; Peter and Krammer, 2003).
In the mitochondria-mediated pathway, caspase activation is initiated by cytochrome c release from mitochondria, a process closely regulated by the Bcl-2 family of proteins (Garrido et al., 2006; Green and Reed, 1998; Youle and Strasser, 2008). Released cytochrome c binds to the essential mediator Apaf-1, activates the nucleotide binding/exchanging activity of Apaf-1 (Jiang and Wang, 2000; Kim et al., 2005), and consequently triggers the assembly of a multimeric protein complex, the apoptosome (Srinivasula et al., 1998; Zou et al., 1999). The apoptosome recruits and activates the initiator caspase, caspase-9. Caspase-9 needs to associate with the apoptosome to be active (Jiang and Wang, 2000; Rodriguez and Lazebnik, 1999) and it subsequently activates downstream executioner caspases, caspase-3 and caspase-7, which mediate apoptotic cell death by cleaving a variety of cellular substrates. Caspase activation can be further regulated by inhibitory IAP proteins and the IAP antagonist Smac/Diablo.
Deregulation of the intrinsic apoptotic pathway is involved in various human diseases, such as cancer and autoimmune disorders (when apoptosis is defective), and neurodegenerative diseases and strokes (when apoptosis is improperly activated) (Hotchkiss and Nicholson, 2006; Reed, 2003; Yuan and Yankner, 2000). Conversely, targeting apoptotic components by both enhancing and attenuating apoptosis represents important therapeutic approaches. For example, several promising targeted compounds, including small molecule inhibitors of Bcl-2 (Oltersdorf et al., 2005) and Smac mimetics (Li et al., 2004), are designed to activate or potentiate this pathway. On the other hand, inhibition of this pathway should also be effective in treating symptoms with pathologically enhanced apoptosis. Notably, caspase inhibition can also be used for treating inflammation, which requires caspase-1-mediated interleukin maturation (Martinon and Tschopp, 2007; Talanian et al., 2000). Much effort in developing potential anti-apoptotic agents focused on caspase inhibition. To date, although caspase inhibitors have been developed with some degree of specificity and are used for basic research, most of them are peptide-based molecules possessing poor potency and are rapidly degraded in vivo.
In this study, we reconstituted cytochrome c-mediated caspase activation in vitro and employed a high-throughput screening approach to identify small molecule inhibitors of this pathway. Four structurally similar compounds were identified as reversible caspase inhibitors. These compounds are not peptide-based, and are able to inhibit apoptosis and caspase-1-mediated interleukin generation in cells. Further kinetic and crystallization studies revealed that the compounds most likely inhibit caspases via a common allosteric mechanism, by binding to the caspase dimerization interface and subsequently altering the conformation of the catalytic site of the enzyme.
Results
Identification of Inhibitors of Cytochrome c-Mediated Caspase Activation
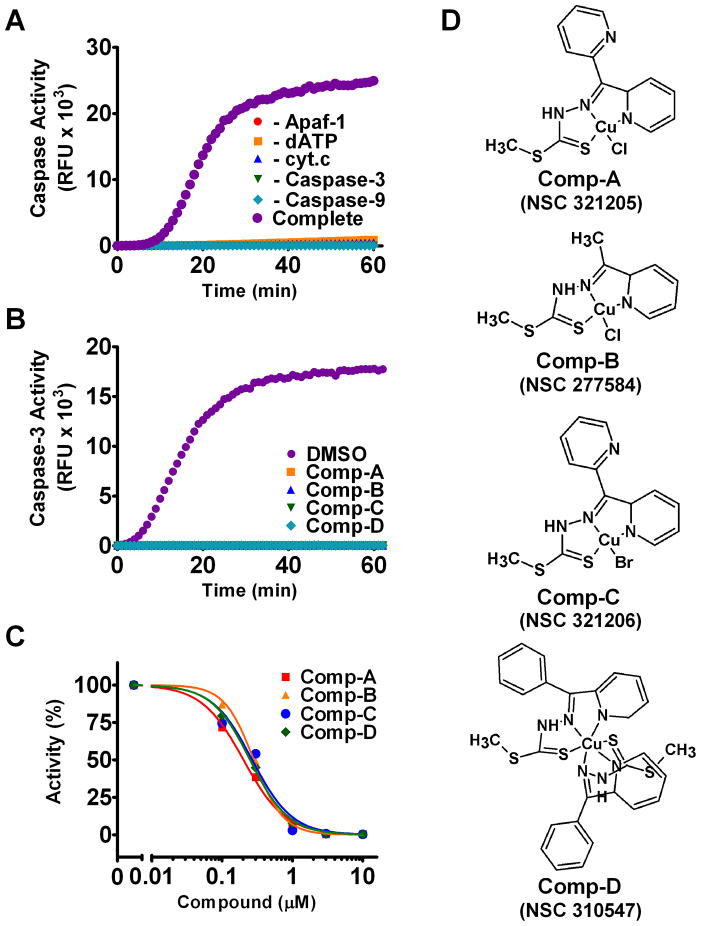
We reconstituted the cytochrome c-mediated caspase activation pathway in vitro using purified recombinant proteins at their near-physiological concentrations (Jiang and Wang, 2000; Kim et al., 2005; Zou et al., 1999). In the presence of Apaf-1, cytochrome c, caspase-9, procaspase-3 and dATP, robust caspase-3 activation was achieved, monitored using a fluorogenic substrate of caspase-3 (Figure 1A). As expected, omission of any component in the reaction completely abated caspase-3 activation (Figure 1A).
Figure 1. Identification of Inhibitors for Cytochrome c-Mediated Caspase Activation.
(A) Time-course of the in vitro reconstituted cytochrome c-mediated caspase activity assay. For the reaction labeled “Complete,” Apaf-1, cytochrome c, dATP, procaspase-3, caspase-9, and a fluorogenic caspase-3 DEVD substrate were incubated. For other reactions, individual components were omitted as indicated.
(B) Inhibition of caspase activation by 10 μM of each compound.
(C) Dose response of compound inhibition of caspase activation. Compounds were added at the concentrations indicated. Activity is shown relative to DMSO control at the 20 minute time-point.
(D) Structure of Compounds A, B, C, and D with NSC numbers.
See also Figure S1.
After adapting this assay to an automated high-throughput screening (HTS) format, we screened a collection of 317,856 chemical compounds at a single compound concentration of 10 μM. The optimized assay exhibited a high signal-to noise ratio of 19:1, as well as good reproducibility with Z′-values ranging from 0.59 to 0.75. We identified 34 initial hits with a threshold of at least 30% inhibition.
We focus on 4 of the 34 compounds after further analysis. These compounds, named Comp-A, B, C, D thereafter, are also included in the chemical repository of the Developmental Therapeutics Program of the National Cancer Institute, with the codes NSC321205, NSC277584, NSC321206, and NSC310547, respectively. At 10 μ, these compounds completely inhibited cytochrome c-mediated caspase activation (Figure 1B). Further titration shows sub-micromolar IC50 values (Figure 1D). The decrease in fluorescence intensity by these compounds was not due to direct quenching of the fluorescent substrate (Figure S1A). These compounds share a similar molecular scaffold, suggesting a common structure-function relationship. They are pyridinyl, copper-containing molecules with a multi-ring structure (Figure 1C). Although Comp-D is more complex structurally, it appears to be a covalent dimer of the other complexes. Mass spectrometry yielded a molecular mass of each compound consistent with their molecular formulas (Figure S1B).
The Identified Inhibitors Target Caspases
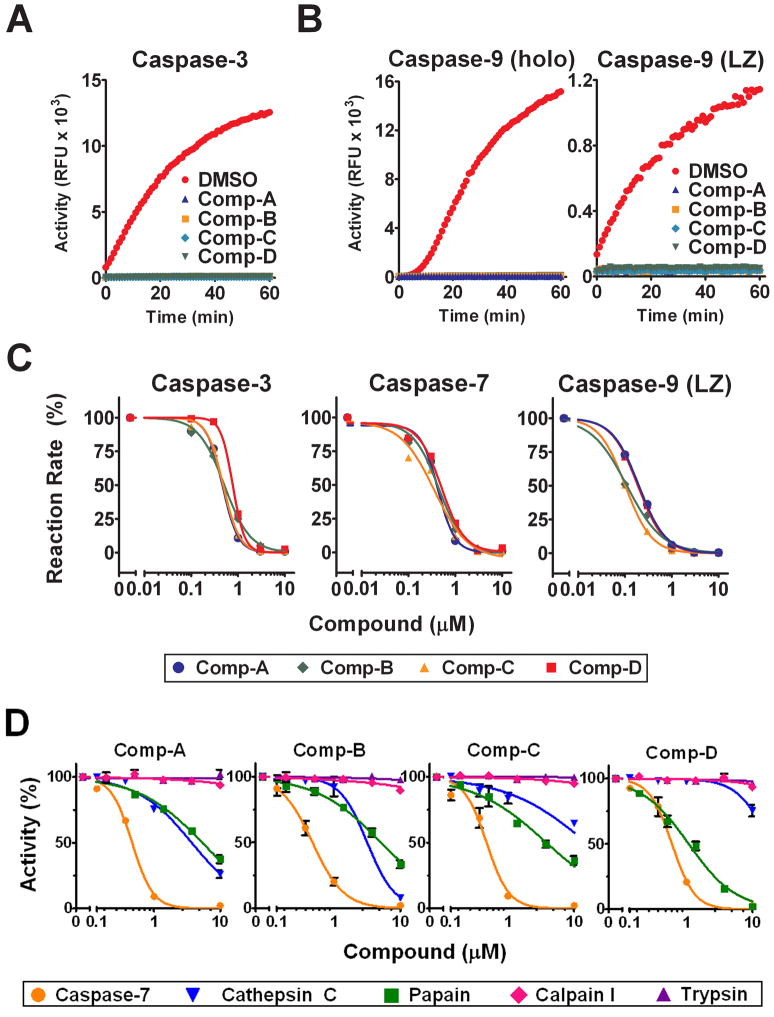
We subsequently performed deconvolution analysis to identify the direct target of the inhibitors and found they can directly inhibit caspase-9 and caspase-3. As shown in Figure 2, the activity of recombinant caspase-3 (fully activated when expressed in bacteria) was completely inhibited by 10 μM of each compound (Figure 2A), and these compounds inhibit caspase-3 activity with sub-micromolar IC50 values (Figure 2C). The effect of these compounds on caspase-9 activity was measured using an engineered caspase-9 dimer that contains a leucine-zipper dimerization domain (Caspase-9-LZ) (Yin et al., 2006). Unlike native caspase-9 whose activity requires an active apoptosome complex, caspase-9-LZ is constitutively active. As with caspase-3, caspase-9-LZ was completely inhibited by 10 μM of each compound (Figure 2B) with sub-micromolar IC50 values (Figure 2C). Similarly, apoptosome-activated caspase-9 was also inhibited by these compounds (Figure 2B).
Figure 2. The Identified Compounds Are Pan-caspase Inhibitors.
(A) Inhibition of caspase-3. 2 nM of active recombinant caspase-3 were incubated with 10 μM compounds or DMSO.
(B) Inhibition of caspase-9. Left panel: 20 nM of recombinant caspase-9 were incubated with Apaf-1, cytochrome c, dATP, and either 10 μM compounds or DMSO. Right panel: 200 nM of caspase-9 Leucine-Zipper (LZ) recombinant protein were incubated with 10 μM compounds or DMSO.
(C) Normalized compound dose-response curves for caspases-3, -7, and -9-LZ. Recombinant caspase proteins (15 nM caspase-3, 20 nM caspase-7, 100 nM caspase-9-LZ) were incubated with compounds at the indicated concentrations in the presence of their corresponding fluorogenic substrates. Reaction rates are expressed relative to the DMSO control.
(D) Protease specificity of compound inhibition. Recombinant caspase-7 (20 nM), cathepsin C (20 nM), papain (20 nM), calpain I (100 nM), and trypsin (20 nM) were incubated with compounds at the indicated concentrations in the presence of their corresponding fluorogenic substrates. Reaction rates are expressed relative to the DMSO control.
See also Figure S2.
We tested other members of the caspase family as well. Caspase-7, an effector caspase that is highly similar to caspase-3, was inhibited by the compounds with sub-micromolar IC50 values (Figure 2C). Caspase-8 and caspase-2, both initiator caspases that share only limited similarity with caspase-9 or caspase-3, and caspase-1, which is involved in the inflammatory response, were all inhibited by the compounds (Figure S2). Therefore, these compounds are pan-caspase inhibitors.
We also tested the inhibitory activity of these compounds on a panel of other proteases that are not caspases. Cathepsin C and papain are cysteine proteases. Although they were inhibited by the compounds in a dose-dependent manner, the IC50 values were mostly 5 μM or greater, which is over 10-fold higher than for caspases (Figure 2D). For calpain I (a cysteine protease) or trypsin (a serine protease), these compounds had marginal, if any, inhibitory effect even at 10-μM (Figure 2D). These results indicate that the compounds preferentially inhibit caspases. One exception is Comp-D, which inhibits papain and caspase-7 with similar potency. This might be due to the more bulky and complex structure of Comp-D compared to the other three compounds.
Inhibition of Cellular Caspase Activation by the Compounds
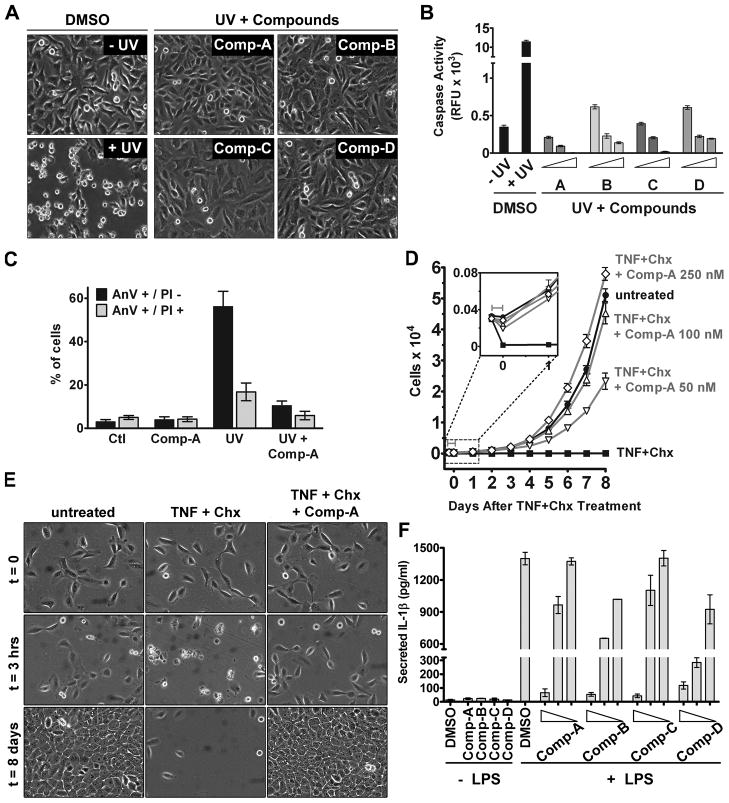
We determined whether these compounds were able to block cellular apoptosis in various mammalian cell lines. The intrinsic apoptotic pathway was triggered by UV radiation in HeLa cells. Treatment with the compounds greatly diminished apoptotic morphology of the cells (Figure 3A). Remarkably, even at 100 nM, the compounds still possess apparent activity in preventing UV-induced apoptosis (Figure 3A). Because in the in vitro assays 100 nM of compounds only yielded mild inhibition of caspases, we suggest cells might actively take in the compounds from culture medium, resulting in a higher cellular concentration of the compounds. Measurement of caspase-3 activity in cell extracts confirmed that caspase activity was inhibited in a compound dose-dependent manner (Figure 3B). Annexin V and propidium iodide (PI) staining also confirmed the inhibition of UV-induced apoptosis in HeLa cells by the compounds (Figure 3C). Similarly, UV-induced caspase activation in mouse embryonic fibroblast (MEF) cells was inhibited by these compounds (Figure S3D). We also tested the effect of the compounds on the extrinsic pathway in U937 cells, a human leukemic monocyte lymphoma cell line. The extrinsic pathway was induced by tumor necrosis factor-α (TNF-α) plus cellular protein synthesis inhibitor cycloheximide. The compounds were able to inhibit such extrinsic apoptosis in a dose-dependent manner as well (Figures S3A–C).
Figure 3. The Identified Compounds Inhibit Cellular Caspase Activation.
(A) Compounds diminish UV-induced apoptotic morphology in HeLa cells. Cells were irradiated with UV and either DMSO or 100 nM of each compound was added to the culture medium. Cells were imaged 6 hours following irradiation.
(B) Inhibition of caspase activation in HeLa cells. Cells were treated as in (A) and collected after 6 hours. Caspase activity in cell extracts was measured. Compound concentrations were 0.5, 1, and 3 μM, as indicated from left to right. Error bars represent the SEM from triplicate experiments.
(C) Annexin V/PI double-staining of UV-treated HeLa cells. Cells were irradiated with UV in the presence or absence of Comp-A (100 nM). Cells were collected 6 hours following irradiation, stained with Annexin V-FITC and PI, and subjected to FACS analysis. Error bars represent the SEM of five experiments.
(D) MCF10A cell growth following treatment with TNF-α and cycloheximide (Chx). Cells were treated with TNF-α and Chx in the presence of either DMSO or Comp-A at the indicated concentrations. After 3 hours (denoted by gray bar), TNF-α, cycloheximide, DMSO and Comp-A were removed and growth medium was replaced, and the number of viable cells was determined at the indicated time points using Resazurin dye. Error bars represent the SEM from triplicate experiments.
(E) Long-term survival of MCF10A cells following TNF-α + cycloheximide induction. Cells were treated with TNF-α and Chx in the presence of either DMSO or 100 nM Comp-A as in (D). Cells were imaged before induction, 3 hours later, and 8 days later.
(F) Inhibition of IL-1β secretion from J774 cells following LPS stimulation. J774 cells were stimulated with 1 μg/ml LPS in the presence of DMSO or compounds. After 24 hours, IL-1β in medium was measured by ELISA. Compound concentrations were 2, 1, and 0.5 μM, as indicated from left to right. Error bars represent the SEM from triplicate experiments.
See also Figure S3.
We next tested the ability of these compounds to enhance long-term survival of cells upon transient apoptotic activation. Although inhibition of caspase activation often fails to prevent eventual cell death due to loss of mitochondrial integrity (for intrinsic apoptosis) or switching to programmed necrosis (for extrinsic apoptosis), under certain conditions, caspase inhibition can restore cell viability. Indeed, it has been established that intrinsic apoptosis-associated mitochondrial outer membrane permeability can be incomplete, and under such conditions, caspase inhibition can restore cell viability (Tait et al., 2010). Further, in cells with specific genetic backgrounds, such as lack of RIP3 expression, TNF-α-induced programmed necrosis can be defective (He et al., 2009; Oberst et al., 2011; Vandenabeele et al., 2010). We found that MCF10A cells, a human mammary epithelial cell line, are defective in TNF-α-induced necrosis because caspase inhibition completely restored long-term viability. As shown in Figures 3D and 3E, when apoptosis was transiently induced in MCF10A cells by TNF-α and cycloheximide, TNF-α-treated cells in the absence of Comp-A all died, while Comp-A recovered cell viability. Therefore, under certain contexts, inhibition of apoptosis by these compounds can significantly restore long-term cell viability.
To investigate whether the compounds would be effective in inhibiting cellular caspase activation beyond the context of apoptosis, we treated murine macrophage J774 cells with lipopolysaccharide (LPS). LPS can activate inflammatory caspase-1, which results in the cleavage and subsequent secretion of the proinflammatory cytokine Interleukin (IL)-1β (Martinon et al., 2002). When added to the culture medium during LPS stimulation, the compounds were able to decrease the secretion of IL-1β in a dose-dependent manner (Figure 3F).
It should be noted that when higher concentrations of these compounds were used, noticeable cell toxicity was observed. In culture dishes, concentrations up to 5 μM of these compounds caused attached cells (such as HeLa and MEF cells) to detach in a short period of time. If the compounds were removed rapidly, cells were still able to re-attach and grow. However, longer treatment caused the cells to lose viability. We suggest that such toxicity is due to their chemical nature as transition metal complexes. Alternatively, the compounds might exert the toxicity by targeting other biological molecules. For example, it has been reported that Comp-C can target Cdc25B (Vogt et al., 2003) and X-IAP (Glover et al., 2003), although both with a lower potency (IC50 over 10 μM). It should also be noted that at the concentrations sufficient for apoptosis inhibition, the compounds do not significantly affect cell viability (Figure S3E).
Non-competitive Inhibition of Caspases by the Compounds
In order to understand the mechanism of these compounds in inhibiting caspase activity, we performed kinetic analysis. We first focused on Comp-A, and examined its inhibition of caspase-7 (we chose caspase-7 because its crystal structure can be solved both unliganded and in complex with Comp-A, as described later). We later expanded the kinetic analysis to all 4 compounds and multiple caspases (Figures 4 and S4; Tables S1 and S2).
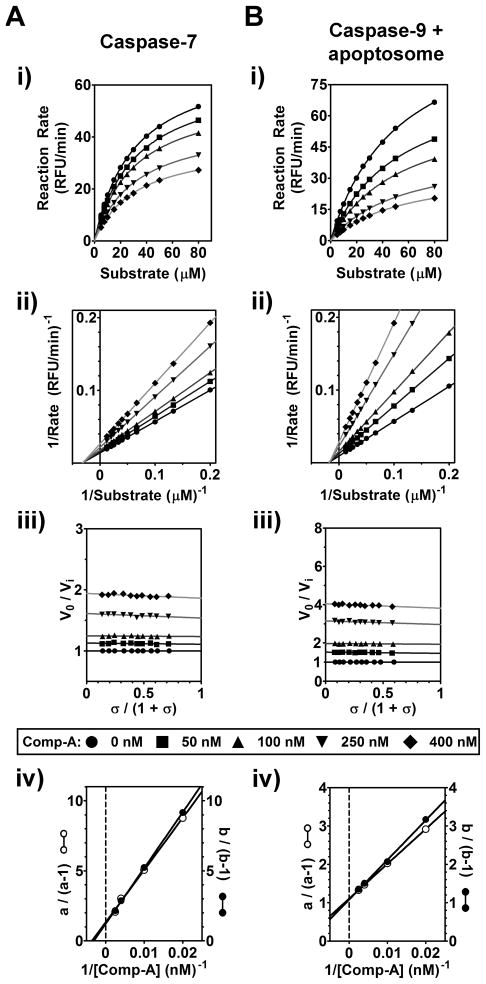
Figure 4. Kinetic Analysis of Inhibition of Caspase-7 and Caspase-9 by Comp-A.
Activity of caspase-7 (20 nM) (A) and caspase-9 (20 nM, with Apaf-1, cytochrome c, and dATP) (B) was determined in the presence of Comp-A at the indicated concentrations.
(i) Substrate concentration curves of caspase activity in the presence of the indicated amount of Comp-A. The curves and numerical values of Vmax and Km represent nonlinear fitting of the data to the Michaelis-Menten equation using Prism software.
(ii) Lineweaver-Burk double reciprocal transformation of the concentration-response curves in (i). Lines represent a linear least-squares fitting of the data.
(iii) Specific velocity plot for the inhibition of caspases by Comp-A. The ratio of caspase activation rate in the absence of compound (V0) to the caspase activation rate in the presence of varying concentrations of Comp-A (Vi) was plotted as a function of the specific velocity σ/(1+σ), where σ= [S]/K m.
(iv) Replot of the specific velocity plot. Two sets of intercepts on the ordinate axes of (iii) at abscissa value = 0 (defined as a) and 1 (defined as b) results in the resolution of kinetic parameters α, β, and Ki. Lines represent a linear least-squares fitting of the data.
See also Figure S4 and Tables S1 and S2.
Because these inhibitors are pan-caspase inhibitors, we predicted that they must act upon a common functional moiety of all caspases. The catalytic center of caspases shares a similar conformation and is a common site of inhibition for most known pharmacological caspase inhibitors (Ivachtchenko et al., 2009). The catalytic site contains a substrate binding groove shaped by four peptide loops (L1, L2, L3, L4) that harbor the catalytic cysteine and determine substrate specificity (Shi, 2002). Although individual caspases show different protein substrate preferences, aspartate at the P1 position is universally required for all caspases (Pop and Salvesen, 2009). If Comp-A binds to the catalytic site of caspase-7, it should then inhibit the enzyme by competitively inhibiting substrate binding to the enzyme, thus Michaelis-Menten analysis would yield a constant Vmax but increased Km values. Surprisingly, kinetic analysis showed that Comp-A caused a drastic decrease of Vmax values in a Comp-A concentration- dependent manner, whereas no significant change in Km values was observed (Figure 4A panel i and Table S1). Such pattern of kinetic parameters was also confirmed by the double reciprocal analysis (Figure 4A, panel ii). This result suggests that Comp-A is not a competitive inhibitor of caspase-7, but is rather likely to be a non-competitive inhibitor.
The mechanism of inhibition was investigated in greater detail using the specific velocity plot (Baici, 1981). This method offers many advantages for the analysis of non-tight-binding, reversible inhibitors, and has been previously used to elucidate the mechanism of a caspase-2 inhibitor (Schweizer et al., 2007). The specific velocity was plotted versus the ratio V0/Vi at different concentrations of Comp-A, yielding a series of linear curves parallel to the abscissa (Figure 4A, panel iii), indicating no effect of the compound on substrate binding. Using the replots of the specific velocity plots (Figure 4A, panel iv), the values of α and β and an approximate value of Ki were calculated graphically (Table S2). For caspase-7 and Comp-A, α = 1.01 and β = 0.06, and Ki = 0.34 μM. These values suggest pure non-competitive inhibition (for which α = 1 and β = 0, theoretically). In agreement with this, the compound appears to bind to caspase-7 with similar affinity regardless of the presence of substrate (Figure S4C).
We measured kinetic parameters for other caspases with the four compounds (Figure S4, Tables S1 and S2). Consistently, caspase-3, caspase-9-LZ, caspase-1, caspase-2, and caspase-8 also showed a decrease in Vmax and a relatively constant Km upon the addition of increasing concentrations of each compound. Specific velocity replots yielded α values close to 1 and β values between 0 and 1. The deviation of β from 0 (indicative of pure non-competitive inhibition) to a value between 0 and 1 suggests partial non-competitive inhibition: a hyperbolic system in which the inhibitor will convert the enzyme into an enzyme-substrate-inhibitor complex with a decreased rate of product formation (Baici, 1981). This partial non-competitive inhibition is especially evident in the β values for caspase-1 and caspase-8.
We also measured kinetic parameters for caspase-9 that is activated by the apoptosome or engineered dimerization (Caspase-9-LZ). It is under debate whether the apoptosome activates caspase-9 by triggering its dimerization (Jiang and Wang, 2004; Renatus et al., 2001). The kinetic profiles of Comp-A inhibition of caspase-9-LZ and apoptosome-activated caspase-9 are almost identical (Figure 4B and Figure S4B, Tables S1 and S2). This lends kinetic support that caspase-9 dimerization is required for the activity of the apoptosome-caspase-9 holoenzyme, in agreement with a model proposed previously (Renatus et al., 2001).
Crystal Structure of Caspase-7 in Complex with Comp-A
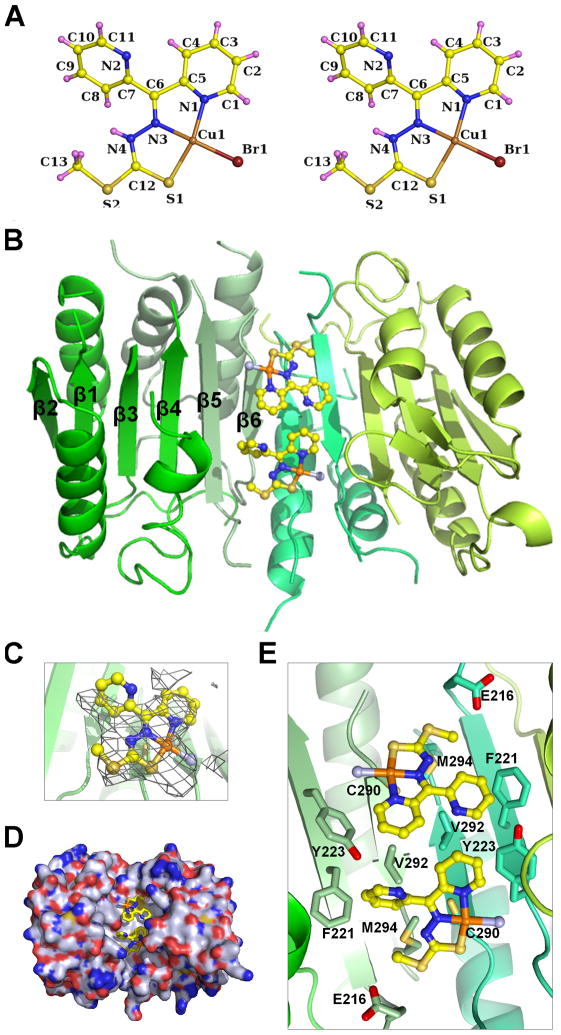
To identify the exact nature of the caspase-compound interaction, we determined crystal structures of Comp-C at 0.81 Å resolution (Table S3 and Figure 5A), unliganded caspase-7 at 2.8 Å resolution, and caspase-7 in complex with Comp-A at 3.8 Å resolution (Table 1 and Figures 5B). Because Comp-A is identical to Comp-C except the replacement of Cl for Br, we substituted the Br atom with a Cl atom in the crystal structure of Comp-C to generate the structure of Comp-A. The complex crystals contain a caspase-7 dimer per crystallographic asymmetric unit. We used Cu anomalous difference Fourier to locate Comp-A molecules in the soaked caspase-7 crystals. This yielded two high peaks of 11.5 σ and 7.4 σ, respectively, symmetrical with respect to the two chains of the caspase-7 dimer. These difference Fourier peaks were used as Cu positions to model the Comp-A structure rigidly into the Fo-Fc difference electron density map followed by refinement (Figure 5C).
Figure 5. Crystal Structure of Caspase-7 in Complex with Comp-A.
(A) Ball and stick model of the crystal structure of Comp-C. Carbon: yellow; nitrogen: blue; sulfur: gold; copper: orange; bromine: dark red; hydrogen: pink. Atom names are labeled.
(B) Ribbon representation of the structure of caspase-7 in complex with Comp-A. The two p20 and p10 subunits are shown in different shades of green. Comp-A is shown in stick models with chloride atoms in light blue.
(C) Comp-A superimposed with the Fo-Fc difference Fourier density contoured at 3.0 σ.
(D) Surface diagram of caspase-7 (shown with carbon atoms in gray, oxygen atoms in red, nitrogen atoms in blue and sulfur atoms in gold) in complex with stick models of Comp-A bound at the dimerization interface.
(E) Detailed interaction between caspase-7 and Comp-A. The different caspase-7 subunits are shown in shades of green. Carbon atoms are shown in the same shades of green as the subunits.
See also Tables S3.
Table 1.
Crystallographic Statistics of Unliganded Caspase-7 and the Complex of Caspase-7 with Comp-A
| Caspase7 | Caspase7 with Comp-A | |
|---|---|---|
| Data Collection | ||
| Space group | P3221 | P3221 |
| Cell dimensions (Å) | 88.4, 88.4, 185.5 | 88.7, 88.7, 185.7 |
| Wavelength (Å) | 1.000 | 1.378 |
| Resolution (Å) | 37.0 – 2.8 | 45.0 – 3.8 |
| Rsym (%) | 8.3 (46.2) | 10.8 (70.0) |
| I /σI | 30.5 (2.0) | 11.9 (2.8) |
| Completeness (%) | 88.7 (46.0) | 94.1 (96.0) |
| Redundancy | 3.6 (2.5) | 5.1 (5.1) |
|
| ||
| Refinement | ||
| Resolution (Å) | 20.0 – 2.8 | 20 – 3.8 |
| No. reflections | 19,773 | 8,256 |
| Rwork / Rfree (%) | 19.4 / 23.9 | 23.7 / 28.6 |
| No. atoms | ||
| Protein | 3,759 | 2,753 |
| Cu | 0 | 2 |
| R.m.s. Deviations | ||
| Bond lengths (Å) | 0.009 | 0.01 |
| Bond angles (0 ) | 1.20 | 1.35 |
| Ramachandran plot | ||
| Most favored (%) | 96.1 | 89.2 |
| Allowed (%) | 3.9 | 10.8 |
Values in parenthesis are for highest-resolution shell. One crystal was used for each data set.
Comp-A is bound to the solvent-exposed dimer interface in an edge-to-edge fashion (Figures 5B, 5D, 5E), and is away from the catalytic active site Cys186. It lays essentially flat on β5 and β6 of the central β-sheet at the dimerization interface (Figures 5B, 5E). Comp-A contacts only the p10 subunits, including residues Tyr223 and Cys290 from one subunit and Glu216, Phe221, Tyr223, Val292 and Met294 from the neighboring subunits (Figure 5E). Interestingly, previously reported allosteric caspase-7 inhibitors DICA and FICA interact with the caspase dimerization interface and form covalent bonds with the thiol of Cys290 (Hardy et al., 2004). In our structure, the thiols of Cys290 in both monomers of the caspase-7 dimer are too far away from any atoms in Comp-A to allow covalent bonding (Figure 5E).
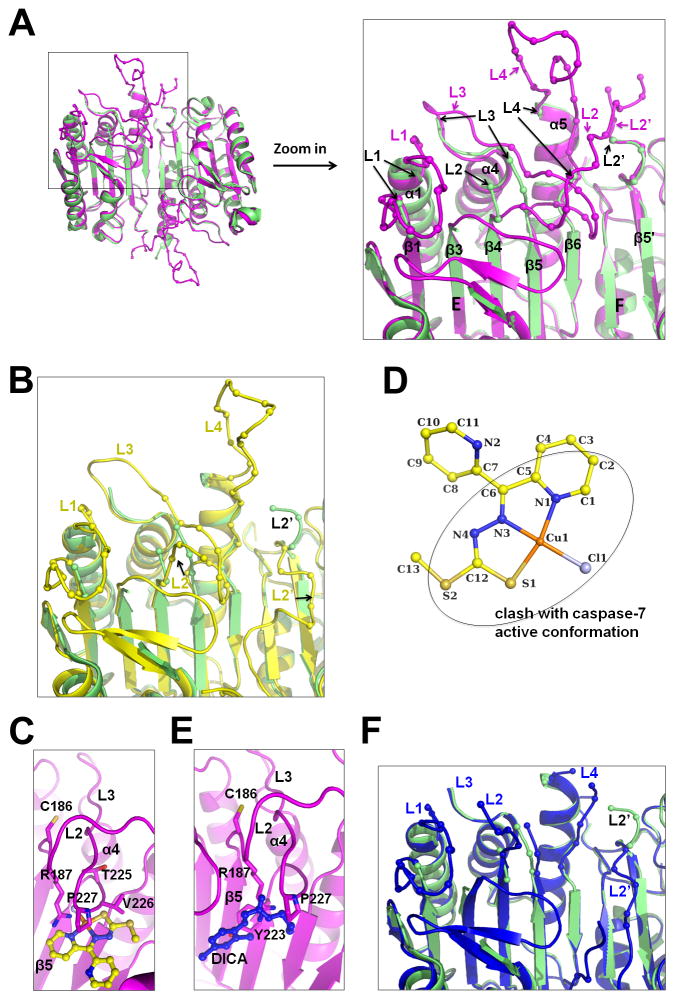
Comp-A Binding Induces Conformational Changes and Disordering of Active Site Loops
The substrate binding groove of caspase-7 is composed of flexible surface loops that include L1, L2, L3, and L4 from one chain (A), and L2′ from the other chain (B), among which L2 harbors the catalytic Cys186 (Chai et al., 2001; Riedl et al., 2001). While L3 and L4 form the base and one side of the catalytic groove, respectively, the interaction between L4, L2 and L2′ is important for maintaining a stable active conformation for substrate binding and catalysis. Two unliganded caspase-7 structures were reported (accession codes 3IBF and 1K86) (Agniswamy et al., 2009; Chai et al., 2001). Our unliganded structure is highly similar to the 3IBF structure: both are essentially in a catalytically productive conformation resembling to the DEVD-CHO–bound conformation (accession code 1F1J) (Figure S5), but is different from the 1K86 structure, which has a reversed conformation for the L2′ loop and an unproductive conformation for the L2 loop.
Extensive conformational changes and disorder of caspase-7 were induced in all loops upon Comp-A binding (Figure 6A). L2 has only two residues with the catalytic Cys186 as the last ordered residue; L3 has a few residues at the beginning and the disorder continues to residue 236; L4 has a few residues each at the beginning and the end of the loop; and L2′ is two residues shorter. In one of the monomers of the caspase-7 dimer, even L1, which is usually conserved in conformation in different forms of caspase-7 structures, has only two ordered residues at the beginning of the loop, and a neighboring region from residues 144–155 is also invisible. In the other monomer of the caspase-7 dimer, L1 and its neighboring region are involved in crystal packing and are ordered in the crystal data set we used. The tendency of the region to become disordered upon Comp-A soaking explains deterioration of diffraction quality and the need to compromise soaking conditions to preserve some diffraction and to allow reasonable occupancy of the compound.
Figure 6. Conformational Changes and Disordering in the Caspase-7 Structure in Complex with Comp-A.
(A) Superposition of the structure in complex with Comp-A (green) with that in complex with DEVD-CHO in the active conformation (magenta, accession code 1F1J). The L1, L3 and L4 loops and the L2 and L2′ regions are labeled in magenta for 1F1J. For caspase-7 in complex with Comp-A, end residues in these loops are labeled in black and with arrows to indicate the breaking points or last residues in them. Relevant secondary structures are also labeled.
(B) Superposition of the structure in complex with Comp-A (green) with a procaspase-7 structure (yellow, accession code 1K88).
(C) Comp-A would have been in clash with active conformation of caspase-7. Active site Cys186 and caspase-7 residues in direct clash with Comp-A are shown and labeled.
(D) Atoms in Comp-A that would have been in clash with caspase-7 are shown within the oval.
(E) Previously reported inhibitor DICA (blue, accession code 1SHJ) would have caused similar clash with the active conformation of caspase-7.
(F) Superposition of the structure in complex with Comp-A (green) with DICA-bound caspase-7 (blue, accession code 1SHJ).
See also Figure S5.
Structural comparison showed that the ordered parts of L2, L3 and L4 of the Comp-A bound caspase-7 are more similar to procaspase-7 structures (1K88 and 1GQF) (Chai et al., 2001; Riedl et al., 2001) (Figure 6B), with the exception that L2 of Comp-A bound caspase-7 is in an open conformation instead of an inverted conformation as in procaspase-7 structures. This change of conformation upon Comp-A binding is induced by steric clash. In particular, Comp-A is in direct clash with Arg187 of L2 and Thr225, Val226 and Pro227 of L3 in the active conformation (Figure 6C), pushing these residues away to assume a conformation more similar to procaspase-7. L2 and the end of L3 coming from the β5 strand are both almost 90° away from the conformation in active caspases (Figure 6A). Although the key inducing event in procaspase-7, which is the linkage between L2 and L2′, is different from Comp-A binding, the consequence is similar as they both disrupt the highly interdependent conformations of the active site loops. Strikingly, Comp-A atoms that are in clash with active caspase-7 conformation are all from the common core of the compound scaffold (Figure 6D), which supports that all four compounds would interact with caspase-7 in a similar manner.
Interestingly, analysis of the previously reported caspase-7 structures inhibited by DICA and FICA (1SHJ and 1SHL) (Hardy et al., 2004) revealed a similar pattern of clashes. DICA clashes with Arg187 at the L2 loop, and Tyr223 and Pro227 of the L3 loop (Figure 6E). FICA clashes with exact the same residues, except that one FICA molecule clashes with Pro227 of the monomer it is bound to, and Arg187 and Tyr223 of the neighboring monomer. Similar to Comp-A-inhibited structure, distortion of the L2 and L3 loops by inhibitor binding leads to massive conformational changes and disordering of the active site loops (Figure 6F). In DICA and FICA-inhibited structures, L2′ is inverted towards the dimerization interface and interacts with both DICA and FICA directly.
Mutagenesis Analysis of the Reversible Inhibition of Caspase-7 by Comp-A
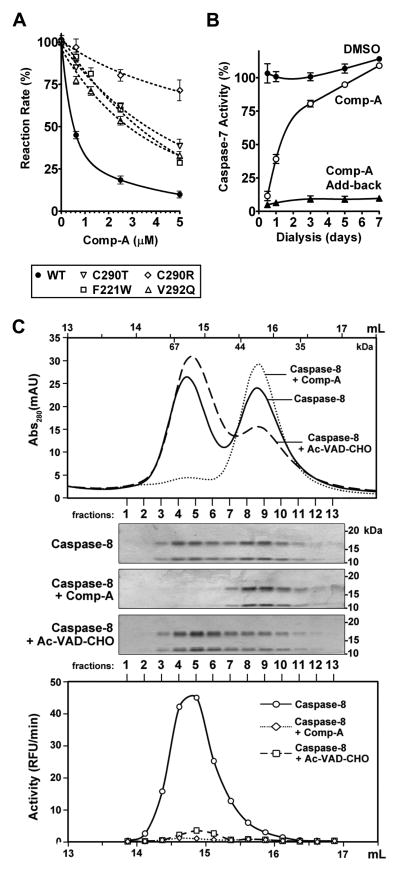
Although alignment of the amino acid sequences that construct the dimerization interface of individual caspases does not yield obvious conserved residues across the board, this region is highly conserved for vertebrate caspase-3 and caspase-7 (Figure S6). Mutagenesis of caspase-7 residues proximal to Comp-A (Figure 5E and Table S4) indicates that some of them are involved in inhibition by Comp-A. Notably, some of the mutations yield inactive caspase, confirming that proper dimerization is essential for caspase activity (Table S4). Of the mutants that yielded functional caspase-7, F221W, C290T, C290R, and V292Q showed a significantly lower sensitivity to Comp-A when compared to wild-type caspase-7 (Figure 7A), indicating that interactions of these residues with Comp-A contribute to the inhibitory function of Comp-A.
Figure 7. Comp-A Inhibits Effective Dimerization of Caspases in a Noncovalent, Reversible Manner.
(A) Mutation of C290, F221, and V292 affect caspase-7 inhibition by Comp-A. Initial reaction rates of wild-type and mutant caspase-7 were measured in the presence of Comp-A at the indicated concentrations. Reaction rates are expressed relative to the DMSO control. Error bars represent the SEM from triplicate experiments.
(B) Inhibition of caspase-7 by Comp-A is reversible. 200 nM of caspase-7 were incubated with either DMSO (labeled “DMSO”) or 10 μM Comp-A (labeled “Comp-A”). The samples were dialyzed against buffer ASC. Aliquots were removed at the indicated time-points and caspase-7 activity was measured with or without the addition of 10 μM of exogenous Comp-A (labeled “Comp-A Add-back”). Activity is expressed relative to un-dialyzed Caspase-7. Error bars represent the SEM from triplicate experiments.
(C) Comp-A disrupts dimerization of caspase-8. 100 nM of recombinant caspase-8 were incubated with DMSO, 10 μM Comp-A, or 10 μM Ac-VAD-CHO for 15 minutes at 30°C and then subjected to Superdex 200 gel filtration. Collected fractions were subjected to SDS-PAGE stained with Coomassie Blue (middle). Activity in each fraction was measured (bottom plot). The volume (mL) labels on the top and bottom plots indicate the elution volume of the chromatographic run (starting from sample injection). The elution positions of protein mass standards are labeled inside of the top Abs280 plot.
The Cys290 residue of caspase-7 is of particular interest. It is conserved in vertebrates (Figure S6) and is in contact with Comp-A in the complex structure (Figure 5E). Caspase-1 also contains a cysteine at the dimer interface, Cys331, structurally positioned on a different β-strand but located in a position close to Cys290 of caspase-7 (Scheer et al., 2006). Likewise, Cys390 of caspase-2 coordinates the interaction between the two dimers (Schweizer et al., 2003). Although caspase-8 and caspase-9 do not have a cysteine at the dimer interface, dimerization is essential for their activation (Acehan et al., 2002; Boatright et al., 2003). Comp-A contacts Cys290 in a noncovalent manner, since the physical distance revealed by the crystal structure is too big for a covalent bond, and mutating the Cys290 residue did not fully abate the ability of Comp-A to inhibit caspase-7 (Figure 7A). On the other hand, both the C290T and C290R mutants show significantly decreased inhibition by Comp-A compared to WT caspase-7, indicating that Cys290, though not covalently interacting with Comp-A, is involved in the inhibitor binding. Additionally, inhibition by Comp-A can be reversed upon dialysis (Figure 7B), further demonstrating a noncovalent, reversible inhibitory mechanism.
Inhibition of Caspase-8 Dimerization by Comp-A
Because the dimerization interfaces of caspases are not generally similar to each other at the sequence level, is the interference of proper caspase dimerization a common mechanism for these compounds to inhibit other caspases? To test this, we subjected recombinant caspase-8 to gel filtration analysis. Active caspase-8 is a dimer, which is in equilibrium with inactive monomer. The caspase-8 dimer and monomer can be resolved as two distinct peaks by gel filtration, with enzymatic activity present only in the dimeric peak (Figure 7C). Using this assay, we found that Comp-A caused diminishment of dimerized caspase-8, yielding only the monomeric species with enzymatic activity completely abrogated. In contrast, incubation of caspase-8 with the competitive caspase inhibitor Ac-VAD-CHO resulted in a shift to the dimeric species, as the enzyme is “locked” in the active conformation upon inhibitor binding (Figure 7C). Interestingly, while Comp-A binding completely disrupted the physical dimerization of caspase-8, Comp-A-associated caspase-7 can still maintain dimerized (albeit a distorted, non-productive conformation). This difference might reflect the dissimilarity of the dimerization interfaces of the two caspases at the level of primary sequence.
Discussion
We identified four pan-caspase inhibitors that share a similar chemical scaffold and are distinct from the commonly used peptide inhibitors. Our kinetic, biochemical, and structural analyses strongly support a common allosteric mechanism underpinning the inhibition of caspases by these compounds: by binding to the dimerization interface, the compounds alter the productive conformation of caspases, thus ablating their catalytic activity. It should be noted that without direct evidence to validate the interaction of each compound with each caspase (ideally co-crystal structures), it remains a hypothesis to suggest that all these inhibitions follow a common mechanism. It should also be noted that the dimerization interfaces of individual caspases share limited sequence homology. The sequence diversity of the caspase dimerization interfaces might present a unique opportunity: specific inhibitors might be developed to target this moiety of each individual caspase. If successful, this will be a significant advancement, because non-competitive inhibitors specific to individual caspases will be important research and therapeutic tools.
Mechanistically, binding of Comp-A to the dimerization interface of caspase-7 is reminiscent of the interaction of DICA and FICA with caspase-7 (Hardy et al., 2004; Hardy and Wells, 2009). However, unlike Comp-A which is a reversible inhibitor, DICA and FICA are irreversible inhibitors that covalently modify a cysteine residue located within the cavity of caspase-7 and caspase-3 (Hardy et al., 2004; Scheer et al., 2006). Because this cysteine residue is not conserved, DICA and FICA can only inhibit caspase-3 and caspase-7, while the four compounds identified in this study are pan-caspase inhibitors. Further, the irreversible nature of DICA and FICA and lack of evidence that they can function in cells limited their potential as pharmacological tools or therapeutic leads. In contrast, the inhibitors identified in this study are reversible inhibitors that inhibit cellular apoptosis at sub-micromolar levels. They are also able to inhibit the cellular activation of caspase-1, a critical player in the immune response and various inflammatory diseases.
Strikingly, all four compounds identified here contain a transition metal atom, copper. Although excess copper is known to be toxic in mammalian tissues, the general consensus is that free copper ions, rather than complexed copper, is the major contributor to this cytotoxicity (Nor, 1987). In fact, complexed copper is present in a variety of native biological enzymes, including cytochrome c oxidase and copper nitrite reductase (MacPherson and Murphy, 2007). Therefore, it is likely that contaminating free copper ion in these complexes is what causes the observed cell toxicity. Although the presence of a transition metal is usually a concern for potential therapeutic applications, there are precedents of transition metal compounds being used therapeutically. For example, cisplatin, a widely-used anticancer agent, contains at its core a transition platinum atom. Other metal complexes, including ruthenium, titanium and gallium, have also been explored for their therapeutic properties, and are currently under clinical trials (Hannon, 2007).
In conclusion, small molecule inhibitors of caspase activation, such as the ones presented here, might be developed into therapeutic agents for treating relevant human diseases, as well as tool compounds to study the mechanisms and functions of caspase activation. Further, given the recent intriguing findings that mitochondrial outer membrane permeabilization is not always the point of no-return for mitochondria-mediated cell death (Tait et al., 2010), mitochondrial caspase activation possesses cell death-independent biological functions (Li et al., 2010), and caspase activity can regulate the necrotic cell death (Oberst et al., 2011), it becomes crucial to develop in vivo and therapeutically amenable caspase inhibitors in order to understand apoptotic and non-apoptotic functions of caspase activation and to fight the relevant diseases.
Experimental Procedures
For more detailed experimental procedures, see the Supplemental Information.
Cytochrome c-Mediated Caspase Activation Assay
A fluorogenic assay was used to measure cytochrome c-mediated caspase activation in vitro. Recombinant procaspase-3 (50 nM) and caspase-9 (20 nM) were mixed with Apaf-1 (5 nM), cytochrome c (0.5 μM) and the nucleotide dATP (10 μM) in buffer ASC (see supplement for detail). The conversion of a fluorogenic caspase-3 Rhodamine-DEVD substrate (15 μM) was measured at 30°C.
High Throughput Screening
Compounds from the SKI corporate compound library collection were pre-plated in 1536-well microtiter plates using the automated pipetting system TPS-384 (Apricot Designs) and were tested at a final concentration of 10 μM in 1% DMSO (v/v). Reaction mix was added to each well using the Flexdrop precision reagent dispensers (Perkin Elmer). The fluorescence signal of the converted Rhodamine-DEVD substrate was recorded using the CCD-based LEADseeker Multimodality Imaging System (GE Healthcare). Screening data files were processed using the ORIS HTS Core Screening Data Management System (ChemAxon). The statistical performance was assessed by calculating the Z′ factor as previously described (Antczak et al., 2007; Seideman et al., 2010; Shelton et al., 2009; Shum et al., 2008). Further detail is described in supplement.
Crystallization and Structure Determination of Caspase-7 in Complex with Comp-A
Crystals of active, unliganded caspase-7 were grown at room temperature using the hanging drop vapor diffusion method from 6–9 mg/mL of caspase-7 with a well solution of 0.1 M sodium citrate buffer at pH 5.0 to 5.7 and 1.9 M sodium formate. Comp-A was soaked into unliganded caspase-7 crystals. All crystals were quickly cryo-protected in 25% glycerol with the mother liquor and flash frozen in liquid nitrogen. Data for the unliganded caspase-7 and the caspase-7 in complex with Comp-A were collected at the Cu edge wavelength at the X4A beam line of BNL and GMCAT beam line of APS and processed and scaled using XDS (Kabsch, 1993) and HKL2000 (Otwinowski and Minor, 1997), respectively. The structures were solved using the program PHASER (McCoy et al., 2007) using a native unliganded caspase-7 structure (3IBF) as the search model without the residues 186–196, 212–217 to minimize any bias.
Supplementary Material
Highlights.
High-throughput screening identified a group of non-peptide caspase inhibitors
These compounds can inhibit apoptotic and inflammatory pathways in cells
They are allosteric inhibitors that bind to the dimer interface of caspases
Acknowledgments
we thank J. Wang and C. Radu for their excellent technical support. This work is partially supported by NIH R01CA136513 (to XJ and HD), U54CA137788/U54CA132378 (to XJ), and R01CA113890 (to XJ). The HTS Facility is partially supported by Goodwin and the Commonwealth Foundation for Cancer Research, the Experimental Therapeutics Center (ETC) of the Sloan-Kettering Cancer Center, the William Randolph Hearst Fund of ETC, and the Lillian S. Wells Foundation.
Footnotes
Accession Numbers
Crystal structure of caspase-7: PDB ID code 4FDL
Crystal structure of caspase-7 in complex with allosteric inhibitor (Comp-A): PDB ID code 4FEA.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acehan D, Jiang X, Morgan D, Heuzer J, Wang X, Akey C. Three-Dimensional Structure of the Apoptosome Implications for Assembly, Procaspase-9 Binding, and Activation. Molecular Cell. 2002;9:423–432. doi: 10.1016/s1097-2765(02)00442-2. [DOI] [PubMed] [Google Scholar]
- Agniswamy J, Fang B, Weber IT. Conformational similarity in the activation of caspase-3 and -7 revealed by the unliganded and inhibited structures of caspase-7. Apoptosis. 2009;14:1135–1144. doi: 10.1007/s10495-009-0388-9. [DOI] [PubMed] [Google Scholar]
- Antczak C, Shum D, Escobar S, Bassit B, Kim E, Seshan VE, Wu N, Yang G, Ouerfelli O, Li YM, et al. High-throughput identification of inhibitors of human mitochondrial peptide deformylase. J Biomol Screen. 2007;12:521–535. doi: 10.1177/1087057107300463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baici A. The specific velocity plot. A graphical method for determining inhibition parameters for both linear and hyperbolic enzyme inhibitors. Eur J Biochem. 1981;119:9–14. doi: 10.1111/j.1432-1033.1981.tb05570.x. [DOI] [PubMed] [Google Scholar]
- Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, Ricci JE, Edris WA, Sutherlin DP, Green DR, et al. A unified model for apical caspase activation. Mol Cell. 2003;11:529–541. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- Chai J, Wu Q, Shiozaki E, Srinivasula SM, Alnemri ES, Shi Y. Crystal structure of a procaspase-7 zymogen: mechanisms of activation and substrate binding. Cell. 2001;107:399–407. doi: 10.1016/s0092-8674(01)00544-x. [DOI] [PubMed] [Google Scholar]
- Garrido C, Galluzzi L, Brunet M, Puig PE, Didelot C, Kroemer G. Mechanisms of cytochrome c release from mitochondria. Cell death and differentiation. 2006;13:1423–1433. doi: 10.1038/sj.cdd.4401950. [DOI] [PubMed] [Google Scholar]
- Glover C, Hite K, DeLosh R, Scudiero D, Fivash M, Smith L, Fisher R, Wu J, Shi Y, Kipp R, et al. A high-throughput screen for identification of molecular mimics of Smac/DIABLO utilizing a fluorescence polarization assay. Anal Biochem. 2003;320:157–169. doi: 10.1016/s0003-2697(03)00389-0. [DOI] [PubMed] [Google Scholar]
- Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- Hannon MJ. Metal-based anticancer drugs: From a past anchored in platinum chemistry to a post-genomic future of diverse chemistry and biology. Pure and Applied Chemistry. 2007;79:2243–2261. [Google Scholar]
- Hardy J, Lam J, Nguyen J, O’Brien T, Wells J. Discovery of an allosteric site in the caspases. Proc Natl Acad Sci U S A. 2004;101:12461–12466. doi: 10.1073/pnas.0404781101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy JA, Wells JA. Dissecting an allosteric switch in caspase-7 using chemical and mutational probes. J Biol Chem. 2009;284:26063–26069. doi: 10.1074/jbc.M109.001826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Wang L, Miao L, Wang T, Du F, Zhao L, Wang X. Receptor Interacting Protein Kinase-3 Determines Cellular Necrotic Response to TNF-α. Cell. 2009;137:1100–1111. doi: 10.1016/j.cell.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Hotchkiss R, Nicholson D. Apoptosis and caspases regulate death and inflammation in sepsis. Nat Rev Immunol. 2006;6:813–822. doi: 10.1038/nri1943. [DOI] [PubMed] [Google Scholar]
- Ivachtchenko A, Okun I, Tkachenko S, Kiselyov A, Ivanenkov Y, Balakin K. In: Design of Caspase Inhibitors as Potential Clinical Agents. O’Brien T, Linton S, editors. CRC Press; 2009. [Google Scholar]
- Jiang X, Wang X. Cytochrome c Promotes Caspase-9 Activation by Inducing Nucleotide Binding to Apaf-1. J Biol Chem. 2000;275:31199–31203. doi: 10.1074/jbc.C000405200. [DOI] [PubMed] [Google Scholar]
- Jiang XJ, Wang XD. Cytochrome C-mediated apoptosis. Annual Review of Biochemistry. 2004;73:87–106. doi: 10.1146/annurev.biochem.73.011303.073706. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst. 1993;26:795–800. [Google Scholar]
- Kim H, Du F, Fang M, Wang X. Formation of apoptosome is initiated by cytochrome c-induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc Natl Acad Sci U S A. 2005;102:17545–17550. doi: 10.1073/pnas.0507900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Thomas RM, Suzuki H, De Brabander JK, Wang XD, Harran PG. A small molecule Smac mimic potentiates TRAIL- and TNF alpha-mediated cell death. Science. 2004;305:1471–1474. doi: 10.1126/science.1098231. [DOI] [PubMed] [Google Scholar]
- Li Z, Jo J, Jia JM, Lo SC, Whitcomb DJ, Jiao S, Cho K, Sheng M. Caspase-3 activation via mitochondria is required for long-term depression and AMPA receptor internalization. Cell. 2010;141:859–871. doi: 10.1016/j.cell.2010.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson IS, Murphy ME. Type-2 copper-containing enzymes. Cell Mol Life Sci. 2007;64:2887–2899. doi: 10.1007/s00018-007-7310-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F, Burns K, Tschopp J. The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- Martinon F, Tschopp J. Inflammatory caspases and inflammasomes: master switches of inflammation. Cell death and differentiation. 2007;14:10–22. doi: 10.1038/sj.cdd.4402038. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nor Y. Ecotoxicity of copper to aquatic biota: a review. Environ Res. 1987;43:274–282. doi: 10.1016/s0013-9351(87)80078-6. [DOI] [PubMed] [Google Scholar]
- Oberst A, Dillon C, Weinlich R, McCormick L, Fitzgerald P, Pop C, Hakem R, Salvesen G, Green D. Catalytic activity of the caspase-8–FLIPL complex inhibits RIPK3-dependent necrosis. Nature. 2011;471:363–367. doi: 10.1038/nature09852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Peter ME, Krammer PH. The CD95(APO-1/Fas) DISC and beyond. Cell death and differentiation. 2003;10:26–35. doi: 10.1038/sj.cdd.4401186. [DOI] [PubMed] [Google Scholar]
- Pop C, Salvesen G. Human caspases: activation, specificity, and regulation. J Biol Chem. 2009;284:21777–21781. doi: 10.1074/jbc.R800084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JC. Apoptosis-targeted therapies for cancer. Cancer Cell. 2003;3:17–22. doi: 10.1016/s1535-6108(02)00241-6. [DOI] [PubMed] [Google Scholar]
- Renatus M, Stennicke H, Scott F, Liddington R, Salvesen G. Dimer formation drives the activation of the cell death protease caspase 9. Proc Natl Acad Sci U S A. 2001;98:14250–14255. doi: 10.1073/pnas.231465798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl SJ, Fuentes-Prior P, Renatus M, Kairies N, Krapp S, Huber R, Salvesen GS, Bode W. Structural basis for the activation of human procaspase-7. Proc Natl Acad Sci U S A. 2001;98:14790–14795. doi: 10.1073/pnas.221580098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl SJ, Shi YG. Molecular mechanisms of caspase regulation during apoptosis. Nature Reviews Molecular Cell Biology. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- Rodriguez J, Lazebnik Y. Caspase-9 and APAF-1 form an active holoenzyme. Genes Dev. 1999;13:3179–3184. doi: 10.1101/gad.13.24.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer J, Romanowski M, Wells J. A common allosteric site and mechanism in caspases. Proc Natl Acad Sci U S A. 2006;103:7595–7600. doi: 10.1073/pnas.0602571103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer A, Briand C, Grutter MG. Crystal structure of caspase-2, apical initiator of the intrinsic apoptotic pathway. J Biol Chem. 2003;278:42441–42447. doi: 10.1074/jbc.M304895200. [DOI] [PubMed] [Google Scholar]
- Schweizer A, Roschitzki-Voser H, Amstutz P, Briand C, Gulotti-Georgieva M, Prenosil E, Binz H, Capitani G, Baici A, Plückthun A, et al. Inhibition of Caspase-2 by a Designed Ankyrin Repeat Protein: Specificity, Structure, and Inhibition Mechanism. Structure. 2007;15:625–636. doi: 10.1016/j.str.2007.03.014. [DOI] [PubMed] [Google Scholar]
- Seideman JH, Shum D, Djaballah H, Scheinberg DA. A high-throughput screen for alpha particle radiation protectants. Assay Drug Dev Technol. 2010;8:602–614. doi: 10.1089/adt.2010.0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton CC, Tian Y, Shum D, Radu C, Djaballah H, Li YM. A miniaturized 1536-well format gamma-secretase assay. Assay Drug Dev Technol. 2009;7:461–470. doi: 10.1089/adt.2009.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YG. Mechanisms of caspase activation and inhibition during apoptosis. Molecular Cell. 2002;9:459–470. doi: 10.1016/s1097-2765(02)00482-3. [DOI] [PubMed] [Google Scholar]
- Shum D, Radu C, Kim E, Cajuste M, Shao Y, Seshan VE, Djaballah H. A high density assay format for the detection of novel cytotoxic agents in large chemical libraries. J Enzyme Inhib Med Chem. 2008;23:931–945. doi: 10.1080/14756360701810082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasula S, Ahmad M, Fernandes-Alnemri T, Alnemri E. Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Molecular Cell. 1998;1:949–957. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- Tait S, Parsons M, Llambi F, Bouchier-Hayes L, Connell S, Munoz-Pinedo C, Green D. Resistance to caspase-independent cell death requires persistence of intact mitochondria. Developmental cell. 2010;18:802–813. doi: 10.1016/j.devcel.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talanian R, Brady K, Cryns V. Caspases as Targets for Anti-Inflammatory and Anti-Apoptotic Drug Discovery. J Med Chem. 2000;43:3351–3371. doi: 10.1021/jm000060f. [DOI] [PubMed] [Google Scholar]
- Vandenabeele P, Declercq W, Van Herreweghe F, TVB The role of the kinases RIP1 and RIP3 in TNF-induced necrosis. Sci Signal. 2010;3:re4. doi: 10.1126/scisignal.3115re4. [DOI] [PubMed] [Google Scholar]
- Vaux D, Korsmeyer S. Cell death in development. Cell. 1999;96:245–254. doi: 10.1016/s0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- Vogt A, Cooley K, Brisson M, Tarpley M, Wipf P, Lazo J. Cell-active dual specificity phosphatase inhibitors identified by high-content screening. Chem Biol. 2003;10:733–742. doi: 10.1016/s1074-5521(03)00170-4. [DOI] [PubMed] [Google Scholar]
- Yin Q, Park H, Chung J, Lin S, Lo Y, da Graca L, Jiang X, Wu H. Caspase-9 holoenzyme is a specific and optimal procaspase-3 processing machine. Mol Cell. 2006;22:259–268. doi: 10.1016/j.molcel.2006.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nature reviews. Molecular cell biology. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- Yuan JY, Yankner BA. Apoptosis in the nervous system. Nature. 2000;407:802–809. doi: 10.1038/35037739. [DOI] [PubMed] [Google Scholar]
- Zou H, Li Y, Liu X, Wang X. An APAF-1.cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J Biol Chem. 1999;274:11546–11556. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.