Abstract
Since their discovery over twenty years ago, eukaryotic-like transmembrane receptor Ser/Thr protein kinases (STPKs) have been shown to play critical roles in the virulence, growth, persistence and reactivation of many bacteria. Information regarding the signals transmitted by these proteins, however, remains scarce. To enhance understanding of the basis for STPK receptor signaling, we determined the 1.7-Å-resolution crystal structure of the extracellular sensor domain of the Mycobacterium tuberculosis receptor STPK, PknH (Rv1266c). The PknH sensor domain adopts an unanticipated fold containing two intramolecular disulfide bonds and a large hydrophobic and polar cleft. The residues lining the cleft and those surrounding the disulfide bonds are conserved. These results suggest that PknH binds a small-molecule ligand that signals by changing the location or quaternary structure of the kinase domain.
Keywords: orphan receptor, virulence, Rv1266c
Introduction
Mycobacterium tuberculosis (Mtb) is a persistent human pathogen that currently infects one-third of the world's population and causes over 1.7 million deaths per year.1 The pathogenicity of Mtb stems from its ability to alter its developmental programs and metabolism in different host niches. After phagocytosis by alveolar macrophages, Mtb slows growth and alters the composition of cell wall mycolic and fatty acids to survive the nutrient poor phagocytic environment and resist microbicides such as nitric oxide and reactive oxygen species.2 However, little is known about the developmental programs and molecular signals that trigger these adaptive responses. Candidate sensor molecules for transmitting environmental signals into adaptive responses include the 11 eukaryotic-like Ser/Thr protein kinases (STPKs) encoded in the Mtb genome, nine of which have an intracellular N-terminal kinase domain linked via a single transmembrane helix to an extracellular C-terminal sensor domain.3 Recent sequencing projects indicate eukaryotic-like STPKs exist in many prokaryotes, including a wide range of pathogenic bacteria.4 Since their discovery, STPKs have been shown to regulate diverse cellular functions, such as exit from dormancy,5,6 protein secretion,7 cell division,8 sporulation,9,10 and cell-wall biosynthesis.11
The first bacterial STPK kinase domain (KD) structures, which revealed nucleotide complexes of the Mtb PknB KD, demonstrated that bacterial and eukaryotic STPKs share close structural similarities and common modes of substrate recognition and regulation.12,13 Despite advances in understanding the kinase domains of STPKs, only two of the Mtb STPK sensor domains have been structurally characterized. The PknD sensor domain structure was found to form a rigid, six-bladed beta-propeller with a flexible linker to the transmembrane helix,14 while the PknB sensor domain was found to have four PASTA domains15 that bind peptidoglycan fragments and localize the kinase to sites of peptidoglycan turnover to regulate cell growth and division.5,16 To further understanding of STPK receptor signaling, we determined the X-ray crystal structure of the extracellular sensor domain of the Mtb STPK PknH (Rv1266c).
Protein production and structure determination
To characterize the PknH sensor, we expressed the extracellular domain (ECD; residues 435–626) beginning eight residues after the predicted transmembrane helix. This N-terminally His6-tagged protein was largely insoluble in E. coli. The presence of four conserved Cys residues suggested that the structure may be stabilized by one or two disulfide bonds. To test this idea, we isolated inclusion bodies, denatured the protein in 6 M guanidine-hydrochloride (GuHCl), and re-folded it on a Ni-NTA column by stepwise dilution of the GuHCl in the presence of a 10:1 mixture of reduced-to-oxidized glutathione. This procedure yielded soluble ECD that was sensitized to proteolysis and precipitation when reduced, suggesting the disulfides are essential for the stability of the fold. The oxidized protein crystallized after removal of the purification tag. The crystal structure was determined at 1.7-Å resolution by single-wavelength anomalous diffraction (SAD) analysis of a terbium derivative (Table 1). The entire sequence from residues 435 to 626 was ordered.
Table 1.
X-ray data collection, analysis and refinement statistics for the crystal structure of the PknH ECD.
| Data Collection | PknH ECD Tb3+ Complex |
|---|---|
| Wavelength (Å) | 1.12 |
| Temperature (K) | 100 |
| Space group | P21 |
| Unit cell parameters | |
| a, b, c (Å) | 47.46, 35.92, 49.31 |
| β(°) | 98.36 |
| Resolution (Å) a | 50.0-1.70 (1.76–1.70) |
| Number of unique reflections | 34200 (3328) |
| Rsym (%) | 6.1 (25.3) |
| I/σI | 17.4 (4.9) |
| Completeness (%) | 98.8 (98.1) |
| Redundancy | 3.8 (3.8) |
| SAD Solution | |
| Proteins per a.u. | 1 |
| Terbium Sites per a.u. | 2 |
| Mean figure of merit | 0.424 |
| Refinement | |
| Resolution (Å) | 48.78-1.70 |
| Number of reflections | 34190 |
| Rwork/Rfree (%) | 16.30 / 19.74 |
| Number of atoms | |
| Protein | 1467 |
| Solvent | 221 |
| Average B factors | |
| Protein (Å2) | 17 |
| Solvent (Å2) | 25 |
| Rmsd | |
| Bond lengths (Å) | 0.013 |
| Bond angles (°) | 0.96 |
| Ramachandran plot | |
| Favored (%) | 96 |
| Allowed (%) | 4 |
| PDB ID | 4ESQ |
Notes on Table 1: Data were collected at 100 K at Beamline 8.3.1 at the Lawrence Berkeley National Laboratory Advanced Light Source.39 Data were reduced and scaled with HKL2000.40 The structure was determined using PHENIX41 and the model was adjusted manually using Coot42. Phenix.autosol found two terbium sites per asymmetric unit, and phenix.autobuild produced a model with 191 residues and an Rfree of 24% after seven cycles of automatic building and refinement. Building and refinement were completed with phenix.refine and Coot and included addition of a single ordered molecule of BIS-TRIS buffer that coordinated one of the two terbium ions. The final model was validated using MolProbity.43
PknH sensor domain structure
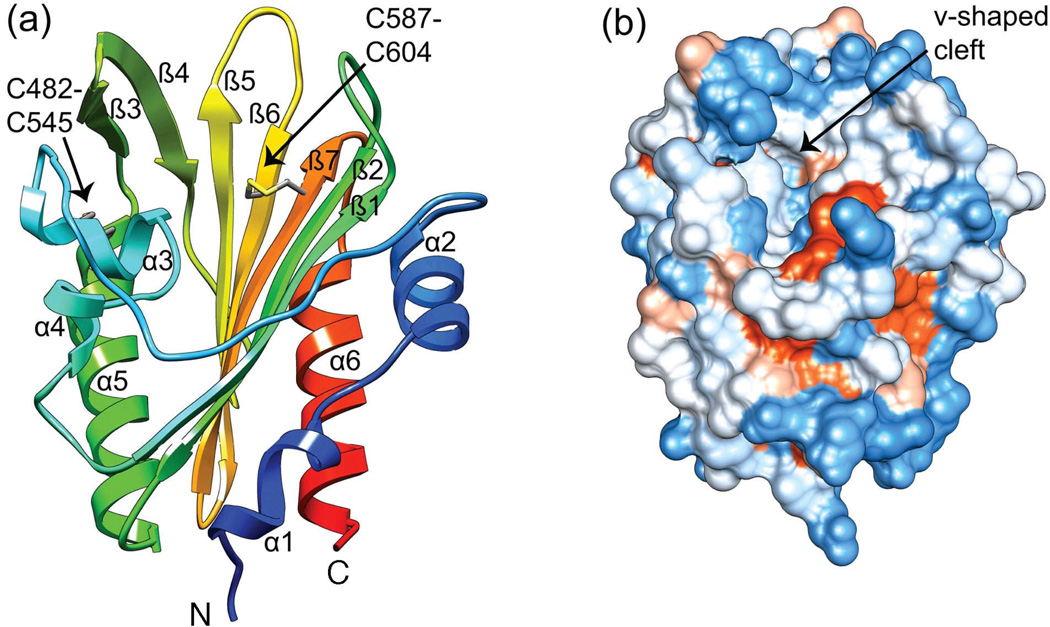
The PknH sensor domain contains six alpha helices and seven anti-parallel beta strands with α1-α2-α3-α4-β1-β2-α5-β3-β4-β5-β6-β7-α6 topology (Fig. 1a). Two intramolecular disulfide bonds link α3 to α5 (C482–C545) and β6 to β7 (C587–C604). A 22-residue irregular loop connects α2 and α3. The most prominent feature is a large v-shaped central cleft (Fig. 1b). Five of the seven anti-parallel beta-strands (β1/β2 and β5–β7) make up one side of this cleft, while alpha helices α3 to α5 and beta strands β3 and β4 comprise the other side. The α2-α3 loop forms the rim of the cleft, and residues 486–490 in the α3-α4 loop line the cleft inner wall (Fig. 1). The cleft has a calculated17 surface area of 1134 Å2 and volume of 2,768 Å3.
Fig. 1. Crystal structure of the PknH extracellular sensor domain.
(a) Ribbon diagram color-coded from the N-terminus (blue) to the C-terminus (red). The v-shaped cleft is surrounded by the β-sheet, α3-α5, and the long α2-α3 loop. PknH residues 435–626 were PCR-amplified from Mycobacterium tuberculosis H37Rv genomic DNA and cloned into the pET28- based destination vector pHGWA using Gateway enzymes (Invitrogen). The pHGWA vector has an N-terminal His6 tag followed by a TEV protease cleavage site 39 that leaves Gly-His at the N-terminus of the protein after TEV cleavage. The protein was expressed in BL21 (DE3) CodonPlus cells (Stratagene) using auto-induction.40 Cells were grown at 37 °C for six hours and then at 30 °C for 18 hours. Cell paste was resuspended in Buffer A (200 mM NaCl, 5% v/v glycerol and 20 mM Bis-Tris propane at pH 8.5) plus protease inhibitors E-64 and leupeptin, and lysed by sonication on ice. The lysate was centrifuged for 40 minutes at 16000 rpm and the resulting supernatant was discarded. The pellet was resuspended in Buffer A plus 0.1% Triton X-100 and this mixture was sonicated and centrifuged at 9000 rpm for 20 minutes. The supernatant was discarded and the detergent extraction step was repeated twice. The pellet was resuspended at 4 °C overnight in Buffer A plus 6 M guanidine hydrochloride (GuHCl), 1 mM reduced glutathione, and 0.1 mM oxidized glutathione. The GuHCl solution was sonicated and centrifuged at 17000 rpm for 30 minutes. The supernatant was incubated with Ni-NTA agarose beads (Qiagen) for 4 hours, and step-wise washes that reduced the GuHCl concentration in 1 M increments were used to refold the protein in the presence of the 10:1 ratio of reduced to oxidized glutathione. The protein was eluted with Buffer A plus 300 mM imidazole. After dialysis into Buffer A to remove the imidazole, the protein was incubated with TEV protease overnight at 4 °C. Cleaved PknH protein was purified by a second IMAC step and further purified by size exclusion chromatography using a HiLoad 26/60 Superdex 75 column in Buffer B (100 mM NaCl, 5% v/v glycerol and 20 mM PIPES pH 6.5). A single peak corresponding to the PknH sensor domain monomer was concentrated to 5 mg/mL and was crystallized at 4 °C by vapor diffusion in a 1:1.5 ratio of protein to initial crystallization buffer (0.1 M Bis-Tris (pH 5.5), 0.2 M ammonium acetate, 25% PEG 3350). Additive screening of the initial crystal hit led to improved crystals with Tb(NO3)3, which were used for SAD phasing. Optimal crystals were grown overnight at 18 °C using 250 nL of the initial crystallization buffer, 500 nL of 5 mg/mL PknH in Buffer B, and 100 nL of 100 mM Tb(NO3)3. Crystals were cryoprotected with mother liquor containing 15% v/v glycerol. Images of the structure were created using Chimera v1.5.3.41 (b). PknH sensor domain surface colored according to the Kyte and Doolittle hydrophobicity scale.42 The most hydrophobic areas are in orange and the most polar areas are in blue. The cleft (arrow) has a surface area of 1134 Å2 and a volume of 2,768 Å3 calculated using CASTp.17
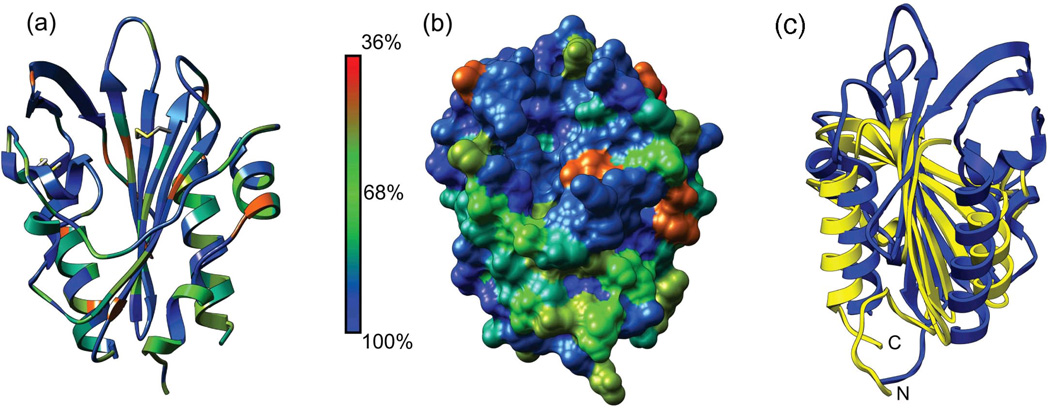
A BLAST search reveals that PknH orthologs occur only in pathogenic mycobacteria. Homologous sensor domains in STPKs generally show >50 % sequence identity. In addition, the PknH sensor domain shows 27 – 40 % sequence identity to mycobacterial LppH proteins, which contain an N-terminal lipid attachment sequence, but no kinase domain. More remote sequence homologs in Mtb include LppR, Rv3705c, LpqA, LpqQ, LprH and the PknJ sensor domain. Plotting the sequence conservation in PknH orthologs on the sensor-domain structure reveals that residues surrounding the disulfide bonds and lining the bottom of the cleft have a high degree of conservation (Fig. 2). In contrast, the residues forming the edges and surface-exposed sides of the cleft are less conserved. These results point to the recent emergence of this fold in mycobacteria and suggest that the sensor-domain homologs are adapted to bind different related ligands in different species.
Fig. 2. Sequence conservation among PknH sensor domain orthologs identifies the disulfide bonds and the cleft as critical functional elements while structural alignment suggests similarities to the Mog1p fold.
(a) Ribbon representation of the PknH sensor domain with residues colored according to conservation among 11 unique orthologous sequences from different mycobacterial species. The orthologs are: M. tuberculosis H37Rv (GI: 15608406), M. marinum DL240490 (GI: 169245959), M. tuberculosis C (GI: 254231523), M. bovis BCG Tokyo 172 (GI: 224989669), M. bovis AF2122/97 (GI: 31792458), M. tuberculosis K85 (GI: 289573926), M. marinum M (GI: 183984142), M. parascrofulaceum ATCC BAA-614 (GI: 296170175), M. avium 104 (GI: 118465657), M. colombiense CECT 3035 (GI: 342862145), and M. intracellulare ATCC 13950 (GI: 254820140). The alignment for the first nine sequences was downloaded from PhyloFacts43 family bpg0185066 (redundant sequences removed), and the two additional sequences were identified using BLAST and added to the alignment using Chimera Multalign Viewer. (b) Surface representation of residue conservation. Conservation was mapped onto the PknH sensor domain and imaged using version 1.5.3 of Chimera.42 (c) Ribbon reperesentation of the PknH ECD (blue) and DIP2269 (yellow), a Mog1p-fold protein from Corynebacterium diphtheriae (PDB 3V7B). The PknH ECD shares a central anti-parallel beta-sheet and two alpha helices with DIP2269, but PknH has an extra β3-β4 hairpin and a significantly different N-terminus.
Structural comparisons
No structures of proteins with similar sequences have been reported, making the structure of PknH ECD the first for its Pfam18 family (PF 14032). A search for similar structures using the DALI server19 found DIP2269, a hypothetical protein from Corynebacterium diphtheriae (PDB ID: 3V7B, Z = 9.6, rmsd = 3.6 Å over 118 aligned residues), TM1622, a GTP-binding regulator from Thermotoga maritima (PDB ID: 1VR8, Z = 8.8, rmsd = 3.5 Å over 120 aligned residues) and BT1490, an uncharacterized protein from Bacteroides thetaiotaomicron (PDB ID: 3HLZ, Z = 8.8, rmsd = 3.2 Å over 119 aligned residues). Structural alignment of PknH ECD with these three proteins using POSA20 revealed a shared core of two alpha helices and six antiparallel beta-strands (β1-β2-α5-β4 extension-β5-β6-β7-α6 in PknH, Fig. 2c) that comprises 60% of the structure. The longer, unique N-terminal segment, and an extra beta-hairpin (β3-β4) between the core elements α5 and β5 in PknH provide evidence for the new fold. Nonetheless, presence of the common core also is consistent with the idea that the PknH ECD may be a distant variant of the SCOP Mog1p fold.21 In any case, the modest Z-scores, partial alignments, distinct secondary structures, high Cα rmsds and uncharacterized activities led us to conclude that direct structural comparisons between the PknH sensor domain and the DALI search results do not provide insights into the possible functions of this STPK.
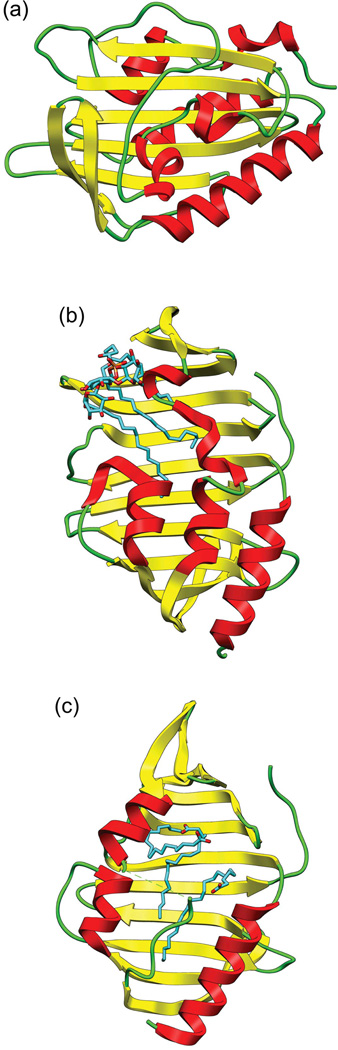
Despite the lack of close structural homologs, visual comparison between the PknH sensor domain and the glycolipid-binding Mtb lipoproteins LprG (Rv1411c22,23 and LppX (Rv2945c24 shows that all three have a large central cleft bordered on one side by a beta sheet and on the other by two to three alpha helices (Fig. 3). The cleft of PknH has a surface area of 1134 Å2 and volume of 2,768 Å3 compared to the clefts for LprG (1549 Å2 and 2679 Å3) and LppX (1375 Å2 and 2835 Å3).17
Fig. 3. Structures qualitatively resembling the PknH sensor domain.
Ribbon representation of (a) the PknH sensor domain, (b) Mtb lipoprotein LprG bound to the glycolipid cell wall precursor molecule AC1PIM2 (PDB 3MHA), and (c) Mtb lipoprotein LppX bound to two molecules of cis-vaccenic acid (C18:1) and one docosanoic acid (C22:0) molecule (PDB 2BYO).
LprG is a widely distributed and conserved Mtb lipoprotein with TLR2 agonist activity that has been shown to bind the cell wall precursor molecule Ac1PIM2.22 Phosphatidylmyoinositol mannosides (PIMs) are glycolipids found in the inner and outer membrane of the cell envelopes of all mycobacteria. They consist of a phosphatidylmyoinositol lipid anchor that carries one to six mannose residues with up to four acyl chains.25 In addition to being critical structural components of the cell envelope, PIMs such as Ac1PIM2 are precursors for lipomannan and lipoarabinomannan, which modulate host-pathogen interactions over the course of a tuberculosis infection.26 LppX is another conserved Mtb lipoprotein that shares 31% sequence identity with LprG and is predicted to bind to phthiocerol dimycocerosates and transport them to the outer layer of the mycobacterial cell envelope.24
PknH function
The PknH STPK sensor domain adopts a distinctive fold with two intramolecular disulfide bonds and a large v-shaped cleft. Possible functions for the PknH sensor domain can be inferred from its genomic location and reported substrates. The pknH gene (Rv1266c) is adjacent to the gene for the EmbR transcriptional regulator (Rv1267) on the Mtb chromosome. In vitro phosphorylation by PknH enhances EmbR binding to the promoter of the embCAB arabinosyltransferase genes and leads to increased transcription of these enzymes.27 EmbA and EmbB are glycosyltransferases that create the terminal hexaarabinoside motif in Mtb cell wall arabinogalactan,28 while EmbC synthesizes the arabinan portion of lipoarabinomannan.29 Deletion of PknH in Mtb results in a hypervirulent phenotype in mice and decreased transcription of embB and embC in cultures treated with sublethal concentrations of ethambutol.30 By phosphorylating EmbR, PknH may control the ratio of lipoarabinomannan to lipomannan, a critical determinant of Mtb virulence.
In addition, PknH and several other Mtb STPKs have been shown to phosphorylate KasA, KasB, and MtFabH. KasA and KasB are β-ketoacyl-ACP synthases that elongate mycolic acid precursors.31 Mycolic acids are long-chain (C60–C90) α-alkyl-β-hydroxy fatty acids that promote bacterial resistance to antibiotics and environmental stress32 and thus increase Mtb virulence33 and persistence32. Phosphorylation-induced inhibition of KasA activity presumably leads to immature mycolic acids while phosphorylation-induced stimulation of KasB activity is thought to ensure production of the full-length mycolates required for bacterial survival and virulence.34,35 MtFabH is the β-ketoacyl-ACP synthase III enzyme that catalyzes the condensation of FAS-I derived acyl-CoAs with malonyl-AcpM, thus linking the FAS-I and FAS-II systems in Mtb.36 MtFabH is phosphorylated in vitro by PknH, PknA, and PknF.37 If PknH functions as a feedback regulator, the sensor domain may be responsive to signals generated in the complex Mtb cell wall. Alternatively, PknH may regulate cell-wall production in response to environmental cues, including compounds that are unrelated to the mycobacterial cell wall.
The novel structure of the sensor domain affords few clues about the signaling ligand(s). The putative recognition cleft is narrow, deep, and conserved, as expected for small molecule binding sites in proteins.38 Visual comparison between the cleft in PknH and the glycolipid-binding clefts of the Mtb lipoproteins LprG and LppX indicates that the PknH binding site is less hydrophobic, making it unlikely that cell-wall glycolipids are the signals. The mixed hydrophobic and polar character of the PknH cleft is consistent with a more polar signal. The stabilizing disulfides impart rigidity to the fold, and the N-terminus, which connects the domain to the TM helix, extends away from the structure. These general characteristics of structural stiffness and loose tethering to the TM helix are shared by the PknD and PknB sensor domains, implying a common signaling mechanism in which ligands may modulate the localization or oligomerization of the kinase.
Coordinates
The coordinates and structure factors were deposited in the PDB under the accession number 4ESQ.
Highlights.
Structures of two bacterial Ser/Thr kinase sensor domains have been reported.
We determined the crystal structure of the sensor domain of M. tuberculosis PknH.
The structure reveals a conserved ligand-binding cleft.
PknH likely binds a molecule that regulates kinase localization or oligomerization.
Acknowledgments
We thank James Holton, George Meigs, and Jane Tanamachi at Beamline 8.3.1 at Lawrence Berkeley National Laboratory for crystallographic assistance. This work was supported by NIH grants R01 GM70962 and P01 AI095208 to T.A.
Abbreviations
- STPK
Ser/Thr protein kinase
- Mtb
Mycobacterium tuberculosis
- KD
kinase domain
- ECD
extracellular domain
- SAD
single-wavelength anomalous diffraction
- GuHCl
guanidine hydrochloride
- TEV
tobacco etch virus
- a.u.
asymmetric unit
- rmsd
root mean square deviation
- PDB
Protein Data Bank
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.World Health Organization. Global tuberculosis control report 2011. (WHO/HTM/TB/2011.16) [Accessed 2012 April 4];2011 http://www.who.int/tb/publications/global_report/2011/gtbr11_full.pdf.
- 2.Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, Schoolnik GK. Transcriptional Adaptation of Mycobacterium tuberculosis within Macrophages: Insights into the Phagosomal Environment. J. Exp. Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Av-Gay Y, Everett M. The eukaryotic-like Ser/Thr protein kinases of Mycobacterium tuberculosis. Trends Microbiol. 2000;8:238–244. doi: 10.1016/s0966-842x(00)01734-0. [DOI] [PubMed] [Google Scholar]
- 4.Galperin MY, Higdon R, Kolker E. Interplay of heritage and habitat in the distribution of bacterial signal transduction systems. Mol. Biosyst. 2010;6:721–728. doi: 10.1039/b908047c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shah IM, Laaberki MH, Popham DL, Dworkin J. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135:486–496. doi: 10.1016/j.cell.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shah IM, Dworkin J. Induction and regulation of a secreted peptidoglycan hydrolase by a membrane Ser/Thr kinase that detects muropeptides. Mol. Microbiol. 2010;75:1232–1243. doi: 10.1111/j.1365-2958.2010.07046.x. [DOI] [PubMed] [Google Scholar]
- 7.Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat. Cell Biol. 2007;9:797–803. doi: 10.1038/ncb1605. [DOI] [PubMed] [Google Scholar]
- 8.Fiuza M, Canova MJ, Zanella-Cleon I, Becchi M, Cozzone AJ, Mateos LM, Kremer L, Gil JA, Molle V. From the characterization of the four serine/threonine protein kinases (PknA/B/G/L) of Corynebacterium glutamicum toward the role of PknA and PknB in cell division. J. Biol. Chem. 2008;283:18099–18112. doi: 10.1074/jbc.M802615200. [DOI] [PubMed] [Google Scholar]
- 9.Madec E, Laszkiewicz A, Iwanicki A, Obuchowski M, Seror S. Characterization of a membrane-linked Ser/Thr protein kinase in Bacillus subtilis, implicated in developmental processes. Mol. Microbiol. 2002;46:571–586. doi: 10.1046/j.1365-2958.2002.03178.x. [DOI] [PubMed] [Google Scholar]
- 10.Madec E, Stensballe A, Kjellstrom S, Cladiere L, Obuchowski M, Jensen ON, Seror SJ. Mass spectrometry and site-directed mutagenesis identify several autophosphorylated residues required for the activity of PrkC, a Ser/Thr kinase from Bacillus subtilis. J. Mol. Biol. 2003;330:459–472. doi: 10.1016/s0022-2836(03)00579-5. [DOI] [PubMed] [Google Scholar]
- 11.Fiuza M, Canova MJ, Patin D, Letek M, Zanella-Cleon I, Becchi M, Mateos LM, Mengin-Lecreulx D, Molle V, Gil JA. The MurC ligase essential for peptidoglycan biosynthesis is regulated by the serine/threonine protein kinase PknA in Corynebacterium glutamicum. J. Biol. Chem. 2008;283:36553–36563. doi: 10.1074/jbc.M807175200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Young TA, Delagoutte B, Endrizzi JA, Falick AM, Alber T. Structure of Mycobacterium tuberculosis PknB supports a universal activation mechanism for Ser/Thr protein kinases. Nat. Struct. Biol. 2003;10:168–174. doi: 10.1038/nsb897. [DOI] [PubMed] [Google Scholar]
- 13.Ortiz-Lombardia M, Pompeo F, Boitel B, Alzari PM. Crystal structure of the catalytic domain of the PknB serine/threonine kinase from Mycobacterium tuberculosis. J. Biol. Chem. 2003;278:13094–13100. doi: 10.1074/jbc.M300660200. [DOI] [PubMed] [Google Scholar]
- 14.Good MC, Greenstein AE, Young TA, Ng HL, Alber T. Sensor domain of the Mycobacterium tuberculosis receptor Ser/Thr protein kinase, PknD, forms a highly symmetric beta propeller. J. Mol. Biol. 2004;339:459–469. doi: 10.1016/j.jmb.2004.03.063. [DOI] [PubMed] [Google Scholar]
- 15.Barthe P, Mukamolova GV, Roumestand C, Cohen-Gonsaud M. The structure of PknB extracellular PASTA domain from mycobacterium tuberculosis suggests a ligand-dependent kinase activation. Structure. 2010;18:606–615. doi: 10.1016/j.str.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 16.Mir M, Asong J, Li X, Cardot J, Boons GJ, Husson RN. The extracytoplasmic domain of the Mycobacterium tuberculosis Ser/Thr kinase PknB binds specific muropeptides and is required for PknB localization. PLoS Pathog. 2011;7:e1002182. doi: 10.1371/journal.ppat.1002182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye Y, Godzik A. Multiple flexible structure alignment using partial order graphs. Bioinformatics. 2005;21:2362–2369. doi: 10.1093/bioinformatics/bti353. [DOI] [PubMed] [Google Scholar]
- 21.Xu Q, Krishna SS, McMullan D, Schwarzenbacher R, Miller MD, Abdubek P, Agarwalla S, Ambing E, Astakhova T, Axelrod HL, Canaves JM, Carlton D, Chiu HJ, Clayton T, DiDonato M, Duan L, Elsliger MA, Feuerhelm J, Grzechnik SK, Hale J, Hampton E, Han GW, Haugen J, Jaroszewski L, Jin KK, Klock HE, Knuth MW, Koesema E, Kreusch A, Kuhn P, Morse AT, Nigoghossian E, Okach L, Oommachen S, Paulsen J, Quijano K, Reyes R, Rife CL, Spraggon G, Stevens RC, van den Bedem H, White A, Wolf G, Hodgson KO, Wooley J, Deacon AM, Godzik A, Lesley SA, Wilson IA. Crystal structure of an ORFan protein (TM1622) from Thermotoga maritima at 1.75 A resolution reveals a fold similar to the Ran-binding protein Mog1p. Proteins. 2006;65:777–782. doi: 10.1002/prot.21015. [DOI] [PubMed] [Google Scholar]
- 22.Drage MG, Tsai HC, Pecora ND, Cheng TY, Arida AR, Shukla S, Rojas RE, Seshadri C, Moody DB, Boom WH, Sacchettini JC, Harding CV. Mycobacterium tuberculosis lipoprotein LprG (Rv1411c) binds triacylated glycolipid agonists of Toll-like receptor 2. Nat. Struct. Mol. Biol. 2010;17:1088–1095. doi: 10.1038/nsmb.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sieling PA, Hill PJ, Dobos KM, Brookman K, Kuhlman AM, Fabri M, Krutzik SR, Rea TH, Heaslip DG, Belisle JT, Modlin RL. Conserved mycobacterial lipoglycoproteins activate TLR2 but also require glycosylation for MHC class II-restricted T cell activation. J. Immunol. 2008;180:5833–5842. doi: 10.4049/jimmunol.180.9.5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sulzenbacher G, Canaan S, Bordat Y, Neyrolles O, Stadthagen G, Roig-Zamboni V, Rauzier J, Maurin D, Laval F, Daffe M, Cambillau C, Gicquel B, Bourne Y, Jackson M. LppX is a lipoprotein required for the translocation of phthiocerol dimycocerosates to the surface of Mycobacterium tuberculosis. EMBO J. 2006;25:1436–1444. doi: 10.1038/sj.emboj.7601048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guerin ME, Kordulakova J, Alzari PM, Brennan PJ, Jackson M. Molecular basis of phosphatidyl-myo-inositol mannoside biosynthesis and regulation in mycobacteria. J. Biol. Chem. 2010;285:33577–33583. doi: 10.1074/jbc.R110.168328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mishra AK, Driessen NN, Appelmelk BJ, Besra GS. Lipoarabinomannan and related glycoconjugates: structure, biogenesis and role in Mycobacterium tuberculosis physiology and host-pathogen interaction. FEMS Microbiol. Rev. 2011;35:1126–1157. doi: 10.1111/j.1574-6976.2011.00276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma K, Gupta M, Pathak M, Gupta N, Koul A, Sarangi S, Baweja R, Singh Y. Transcriptional control of the mycobacterial embCAB operon by PknH through a regulatory protein, EmbR, in vivo. J. Bacteriol. 2006;188:2936–2944. doi: 10.1128/JB.188.8.2936-2944.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Escuyer VE, Lety MA, Torrelles JB, Khoo KH, Tang JB, Rithner CD, Frehel C, McNeil MR, Brennan PJ, Chatterjee D. The role of the embA and embB gene products in the biosynthesis of the terminal hexaarabinofuranosyl motif of Mycobacterium smegmatis arabinogalactan. J. Biol. Chem. 2001;276:48854–48862. doi: 10.1074/jbc.M102272200. [DOI] [PubMed] [Google Scholar]
- 29.Zhang N, Torrelles JB, McNeil MR, Escuyer VE, Khoo KH, Brennan PJ, Chatterjee D. The Emb proteins of mycobacteria direct arabinosylation of lipoarabinomannan and arabinogalactan via an N-terminal recognition region and a C-terminal synthetic region. Mol. Microbiol. 2003;50:69–76. doi: 10.1046/j.1365-2958.2003.03681.x. [DOI] [PubMed] [Google Scholar]
- 30.Papavinasasundaram KG, Chan B, Chung JH, Colston MJ, Davis EO, Av-Gay Y. Deletion of the Mycobacterium tuberculosis pknH gene confers a higher bacillary load during the chronic phase of infection in BALB/c mice. J. Bacteriol. 2005;187:5751–5760. doi: 10.1128/JB.187.16.5751-5760.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molle V, Brown AK, Besra GS, Cozzone AJ, Kremer L. The condensing activities of the Mycobacterium tuberculosis type II fatty acid synthase are differentially regulated by phosphorylation. J. Biol. Chem. 2006;281:30094–30103. doi: 10.1074/jbc.M601691200. [DOI] [PubMed] [Google Scholar]
- 32.Daffe M, Draper P. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 1998;39:131–203. doi: 10.1016/s0065-2911(08)60016-8. [DOI] [PubMed] [Google Scholar]
- 33.Dubnau E, Chan J, Raynaud C, Mohan VP, Laneelle MA, Yu K, Quemard A, Smith I, Daffe M. Oxygenated mycolic acids are necessary for virulence of Mycobacterium tuberculosis in mice. Mol. Microbiol. 2000;36:630–637. doi: 10.1046/j.1365-2958.2000.01882.x. [DOI] [PubMed] [Google Scholar]
- 34.Bhatt A, Fujiwara N, Bhatt K, Gurcha SS, Kremer L, Chen B, Chan J, Porcelli SA, Kobayashi K, Besra GS, Jacobs WR., Jr Deletion of kasB in Mycobacterium tuberculosis causes loss of acid-fastness and subclinical latent tuberculosis in immunocompetent mice. Proc. Natl. Acad. Sci. USA. 2007;104:5157–5162. doi: 10.1073/pnas.0608654104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhatt A, Molle V, Besra GS, Jacobs WR, Jr., Kremer L. The Mycobacterium tuberculosis FAS-II condensing enzymes: their role in mycolic acid biosynthesis, acid-fastness, pathogenesis and in future drug development. Mol. Microbiol. 2007;64:1442–1454. doi: 10.1111/j.1365-2958.2007.05761.x. [DOI] [PubMed] [Google Scholar]
- 36.Brown AK, Sridharan S, Kremer L, Lindenberg S, Dover LG, Sacchettini JC, Besra GS. Probing the mechanism of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III mtFabH: factors influencing catalysis and substrate specificity. J. Biol. Chem. 2005;280:32539–32547. doi: 10.1074/jbc.M413216200. [DOI] [PubMed] [Google Scholar]
- 37.Veyron-Churlet R, Molle V, Taylor RC, Brown AK, Besra GS, Zanella-Cleon I, Futterer K, Kremer L. The Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III activity is inhibited by phosphorylation on a single threonine residue. J. Biol. Chem. 2009;284:6414–6424. doi: 10.1074/jbc.M806537200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang J, Edelsbrunner H, Woodward C. Anatomy of protein pockets and cavities: measurement of binding site geometry and implications for ligand design. Protein Sci. 1998;7:1884–1897. doi: 10.1002/pro.5560070905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacDowell AA, Celestre RS, Howells M, McKinney W, Krupnick J, Cambie D, Domning EE, Duarte RM, Kelez N, Plate DW, Cork CW, Earnest TN, Dickert J, Meigs G, Ralston C, Holton JM, Alber T, Berger JM, Agard DA, Padmore HA. Suite of three protein crystallography beamlines with single superconducting bend magnet as the source. J. Synchrotron Radiat. 2004;11:447–455. doi: 10.1107/S0909049504024835. [DOI] [PubMed] [Google Scholar]
- 40.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Busso D, Delagoutte-Busso B, Moras D. Construction of a set Gateway-based destination vectors for high-throughput cloning and expression screening in Escherichia coli. Anal. Biochem. 2005;343:313–321. doi: 10.1016/j.ab.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 42.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 43.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 44.Busso D, Delagoutte-Busso B, Moras D. Construction of a set Gateway-based destination vectors for high-throughput cloning and expression screening in Escherichia coli. Anal. Biochem. 2005;343:313–321. doi: 10.1016/j.ab.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 45.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 46.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 47.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 48.Datta RS, Meacham C, Samad B, Neyer C, Sjolander K. Berkeley PHOG: PhyloFacts orthology group prediction web server. Nucleic Acids. Res. 2009;37:W84–W89. doi: 10.1093/nar/gkp373. [DOI] [PMC free article] [PubMed] [Google Scholar]