Abstract
We developed a protein-proximity assay in yeast based on fusing a histone lysine methyltransferase onto a bait and its substrate onto a prey. Upon binding, the prey is stably methylated and detected by methylation-specific antibodies. We applied this approach to detect varying interaction affinities among proteins in a mitogen-activated protein kinase pathway and to detect short-lived interactions between protein phosphatase 2A and its substrates that have so far escaped direct detection.
Methods for the investigation of protein-protein interactions (PPIs) in living cells1 are suboptimal for the analysis of dynamic and short-lived enzyme-substrate interactions. Therefore, data on kinase-substrate and phosphatase-substrate interactions are mostly absent from current PPI databases2. To overcome some of the limitations we developed M-Track (for 'methyl-tracking'), an assay that uses an enzyme-catalyzed methylation of a specific substrate lysine for the detection of PPIs in yeast (Fig.1a). A biotinylation-based enzyme-substrate approach has been described for mammalian cells3, but it is inappropriate for yeast studies because of their high biotinylation background. In addition, this approach appeared unsuitable for detection of short-lived PPIs (Supplementary Fig. 1 and Supplementary Note 1). In M-Track, the bait protein is expressed as a fusion protein with the H320R mutant of the human histone lysine (K) methyltransferase (HKMT) SUV39H1, which possesses a more than 20-fold higher catalytic activity than the wild-type enzyme4 and, despite lacking the chromodomain for substrate recruitment, is sufficient for histone H3 Lys 9 (K9) trimethylation4. The prey protein is a fusion protein with three or four tandemly arranged copies of amino acids 1–21 of histone H3 (H3 tag), followed by hemagglutinin (HA) epitope tags. As a readout system we used western blot analysis of whole-cell lysates, for which we generated monoclonal antibodies with high specificity for the different H3K9 methylation states (Supplementary Fig. 2).
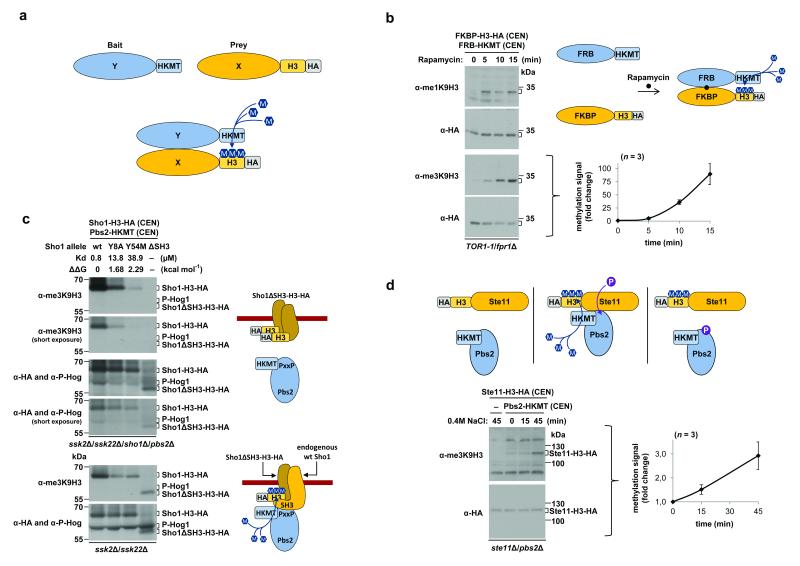
Figure 1. The M-Track assay for detection of stable and transient PPIs in the HOG pathway.
(a) Cartoon depiction of the M-Track assay. Bait protein 'Y' is fused to the catalytic domain of a human histone lysine (K) methyltransferase (HKMT), and prey protein 'X' is fused to the N terminus of histone H3 and the hemagglutinin epitope tag (HA). Upon interaction with the bait, the prey is stably marked by methylation (M, methyl group). (b) M-Track analysis of rapamycin-induced dimerization of FKBP12-rapamycin binding (FRB)-HKMT and FK506 binding protein (FKBP)-H3-HA in a TOR1-1 fpr1Δ yeast strain. A schematic of the assay and immunoblots with the indicated antibodies are shown. The plot shows fold changes of the trimethylation signal normalized to HA levels; plotted values are the mean ± s.d. (n = 3). Basal signal at t = 0 min after rapamycin treatment was set to 1. CEN = centromeric, low copy number yeast vector. (c) M-Track analysis of Pbs2-HKMT and Sho1-H3-HA mutants. In vitro dissociation constants of the interactions are indicated. Immunoblots with the indicated antibodies are shown for samples from an ssk2Δ/ssk22Δ (bottom) or ssk2Δ/ssk22Δ/sho1Δ/pbs2Δ (top) strain. The cartoons (right) depict the interaction between Pbs2 and the Sho1ΔSH3 mutant in the absence (top) or presence (bottom) of endogenous Sho1. (d) M-Track analysis of Pbs2-HKMT and Ste11-H3-HA. Schematics depicting the interaction (P, phosphate group) and immunoblots of samples from the ste11Δ/pbs2Δ strain in the absence or presence of NaCl-induced osmotic stress are shown. Fold changes of the trimethylation signal are plotted as in b; n = 3.
To assess the performance of M-Track, we first analyzed the rapamycin-induced dimerization of FK506 binding protein (FKBP) and FKBP12-rapamycin binding (FRB)5. In the absence of rapamycin, we detected hardly any methylation of the prey FKBP-H3-HA (Fig. 1b). Upon rapamycin addition, the methylation levels increased, with monomethylation peaking after 5 min and trimethylation increasing steadily between minutes 5 and 15. Next, we determined the influence of stress conditions on our assay system (Supplementary Fig. 3). The methylation rates rose substantially with increasing temperatures, but they were not affected by osmotic or oxidative stress.
Because we were interested in the detection of stress-induced PPIs, we studied the high osmolarity glycerol response (HOG) pathway in which Sho1, a transmembrane protein, interacts stably via an SH3 domain with the MAPKK Pbs2 that gets activated by the upstream MAPKKK Ste116. After determining that the tagged proteins are functional in vivo (Supplementary Fig. 4), we used M-Track to monitor binding to Pbs2 of Sho1-SH3 mutants known to have increasing dissociation constants for this interaction7. We expressed H3-tagged Sho1 and HKMT-tagged Pbs2 in strains either lacking or expressing the endogenous proteins, which additionally lack signaling through the Sln1 branch (ssk2Δ/ssk22Δ) (Fig. 1c). In the sho1Δ/pbs2Δ strain, the methylation signal decreased with increasing dissociation rates of the mutants and was undetectable for the ΔSH3 mutant allele (Fig. 1c). In comparison with wild-type Sho1, we observed a 70% and 92% reduction in methylation signal for the Y8A and Y54M mutants of Sho1, respectively. Furthermore, we observed less Hog1 phosphorylation with the SH3 mutants, indicating that the results of the M-Track methylation assay correlated with biological outcome.
When endogenous untagged Sho1 and Pbs2 were present (Fig. 1c), however, we could no longer observe the Kd-dependent differences in methylation of Sho1 mutants, although wild-type Sho1 was still more strongly methylated than the mutants. Even the ΔSH3 mutant was methylated by HKMT-Pbs2 when endogenous Sho1 was present, an observation that can be explained by the assumed homo-oligomerization of Sho1 at the plasma membrane (Fig. 1c)8. We conclude that methylation levels in M-Track can reveal binding affinities but also that close proximity of proteins in a membrane or in a complex could generate a methylation signal. We could also use M-Track to detect the short-lived Ste11-Pbs2 interaction and to track the changes of this MAPKKK-MAPKK interaction in response to osmotic stress in vivo, as indicated by a threefold increase in the methylation signal (Fig. 1d).
Other short-lived interactions that are notoriously difficult to detect are the ones between protein serine/threonine phosphatases, such as PP2A, and their substrates. In yeast two regulatory B-subunits of PP2A, Cdc55 and Rts1, exert non-redundant functions, presumably by targeting distinct substrates9,10, although the identities of these substrates have remained elusive. A probable PP2A–Cdc55 specific substrate is the nucleolar protein Net1 11, but there is no evidence for direct interaction of these proteins12. We used M-Track to investigate this question (Fig. 2a). After testing that the fusion proteins were functional (Fig. 2b and Supplementary Fig. 5a,b), we probed for an interaction between Myc-HKMT-Cdc55 and H3-HA-Net11-600 (Net1 amino acids 1-600) (Fig. 2c). Despite their overexpression these proteins did not co-immunoprecipitate at substantial levels (Supplementary Fig. 6). Upon galactose-induced bait expression, Myc-HKMT-Cdc55 reached a steady-state level after 2 h, whereas the prey level was constant over the entire time course. Concomitant with increasing Myc-HKMT-Cdc55 levels, we observed a time-shifted curve progression of the mono-, di- and trimethylated prey species. This indicated that the reaction mechanism is probably non-processive and suggested that Myc-HKMT-Cdc55 targeted H3-HA-Net11-600 several times, possibly at several sites.
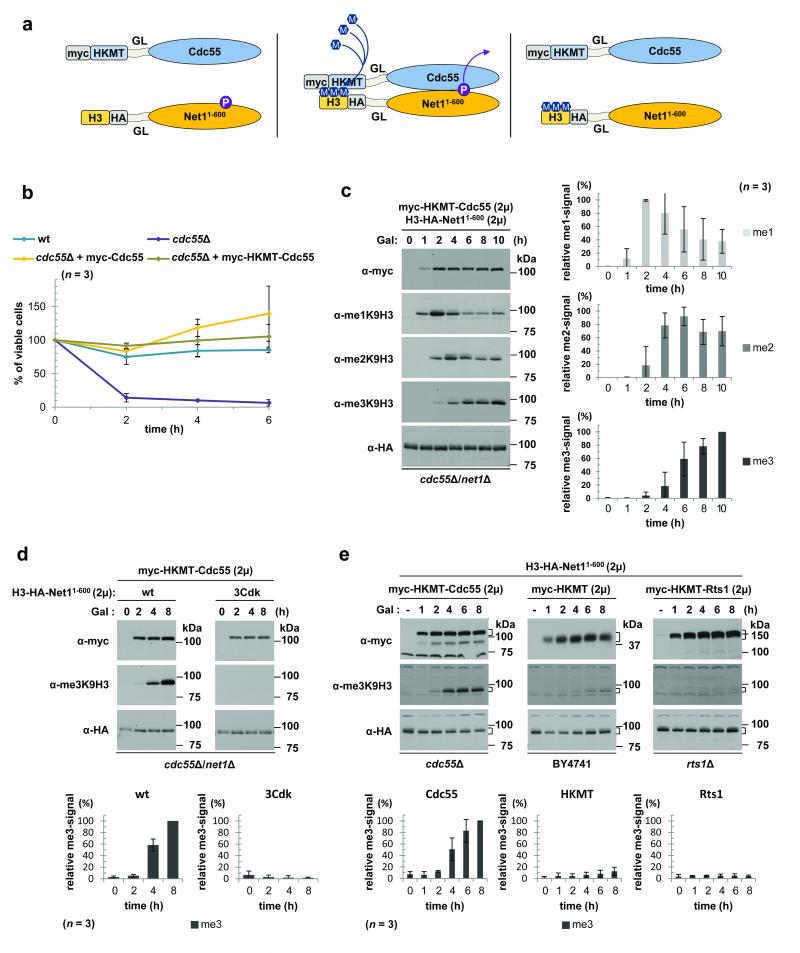
Figure 2. M-Track detection of the short-lived PPI between PP2A-Cdc55 and its substrate Net1.
(a) Cartoon depiction of the assay (GL, glycine-linker; P, phosphate group). (b) Graph showing the percentage of surviving cells ± s.d. of the indicated strains after nocodazole treatment relative to untreated cells (t = 0 h; n = 3). (c) M-Track analysis of Myc-HKMT-Cdc55 and H3-HA-Net11-600 in a cdc55Δ/net1Δ strain. The immunoblots show Myc, methylation and HA signals over time after galactose (Gal) induction of HKMT-Cdc55 expression. The plots show methylation signals normalized to HA levels and plotted as a percentage of the highest normalized methylation signal (100%) ± s.d. (n = 3). 2μ = high copy number yeast vector with 2μ origin of replication (d) Immunoblots showing M-Track analysis of Myc-HKMT-Cdc55 and either H3-HA-Net11-600 (left) or H3-HA-Net11-600(3Cdk) (right) in a cdc55Δ/net1Δ strain. The plots (below) were generated as in c (n = 3). (e) Immunoblots showing M-Track analysis of H3-HA-Net11-600 and either Myc-HKMT-Cdc55 in a cdc55Δ strain (left), a Myc-HKMT in a BY4741 strain (center) or a Myc-HKMT-Rts1 in an rts1Δ strain (right). The plots were generated as in c; n = 3.
Consistent with this finding, Net1 is phosphorylated by cyclin-dependent kinase (Cdk) at several sites; these represent potential PP2A-Cdc55 targets13. We conducted an M-Track assay with a Net1 mutant, 3Cdk, that lacks three of the mapped Cdk phosphorylation sites13. Myc-HKMT-Cdc55 was unable to trimethylate the 3Cdk Net1 mutant (Fig. 2d), indicating that efficient methylation depended on the presence of phosphorylatable Cdk sites. These results strongly suggest that Net1 is an in vivo substrate of PP2A-Cdc55 holoenzymes. The HKMT domain alone or an HKMT fusion protein with the second regulatory subunit of PP2A, Rts1, showed very little trimethylation of H3-HA-Net11-600 (Fig. 2e), in contrast to the results with Myc-HKMT-Cdc55. The inability of Rts1 to target Net1 was not due to a general impairment of its function by the HKMT fusion (Supplementary Fig. 5). Moreover, we could detect an interaction between Myc-HKMT-Rts1 and its putative substrate Kin414 (Supplementary Fig. 7). These findings indicated that M-Track detection of the hybrid protein interaction between PP2A and Net1 depended on the substrate specificity provided by the specific B-subunit rather than the HKMT.
For M-Track to efficiently detect fast catalytic PPIs, the methyltransferase reaction requires an HKMT mutant with high catalytic activity that also lacks the substrate recruitment domain, which reduces the HKMT's affinity for its endogenous substrate and minimizes false-positive results. Notably, the methylation level reflects the integrated sum of multiple transient or stable interaction events minus the protein turnover rate as demethylation is absent in yeast, which limits the use of M-Track the analysis of dissociation kinetics. Furthermore, the methylation rate will not only depend on the binding affinities and duration of protein interactions but also on steric parameters. The positions of the HKMT and H3 tags as well as the design and flexibility of the linkers may influence the readout of the system and must be empirically evaluated.
High-throughput discovery and phosphoproteome screens have identified large numbers of putative enzyme-substrate interactions that require validation by a method such as M-Track. The ability to discriminate between a direct and an indirect enzyme target will help determine the hierarchical structure of signaling cascades within existing data sets and will lead to a better understanding of signaling networks. In addition, the methylation signature left on proteins that have interacted can be used to define differences in biochemical profiles (for example, presence of post-translational modifications) between newly synthesized proteins or participants in different protein complexes. Moreover, the three-step enzymatic reaction catalyzed by the HKMT could be used as an indicator for PPI duration. The system may serve as a structural ruler within stable complexes by using differently sized spacers between the protein and the enzymatic domain. Finally, we envision several future developments of the system such as substrate identification screens using mass spectrometry or the quantitative analysis of dynamic PPI changes by microwestern arrays15.
METHODS
Yeast strains
S. cerevisiae strains used in this study are summarized in Supplementary Table 1. Mutant strains were generated by PCR-based gene targeting.
Plasmids
Plasmids used in this study are summarized in Supplementary Table 2. DNA constructs were generated using conventional PCR, restriction and ligation methods. Detailed cloning strategies and information on the individual constructs can be obtained upon request.
Antibodies.
Antibodies used in this study are summarized in Supplementary Table 3.
Sequences.
Sequences of the HKMT SET domain mutant H320R (amino acids 82–412) and the H3 tag are shown in Supplementary Note 2.
M-Track time courses and yeast cell extracts
For FKBP-FRB and HOG pathway studies, overnight cultures in selective medium were diluted and grown at 30 °C up to exponential phase. For the rapamycin experiments, a zero-time-point (t = 0) sample was taken, and the rest of the culture was treated with 100 μg l1 of rapamycin (LC-Laboratories). For all experiments, cells equivalent to OD600 × ml = 10 were pelleted, and protein extracts were obtained by a post-alkaline extraction method16.
For PP2A time course studies, overnight cultures in selective medium with raffinose were diluted and grown at 30 °C up to early exponential phase. Then either the t= 0 sample was taken and the remaining culture was incubated with 2% galactose (Figure 2c,d), or the culture was split and incubated with 2% glucose for 2 h (Figure 2e and Supplementary Figure 7) or 2% galactose for the indicated periods of time. Cells were pelleted, resuspended in 50 μl/OD600 cold lysis buffer (1.95 M NaOH, 7.5% β-mercaptoethanol in H2O) and incubated for 10 min on ice; 50 μl/OD600 cold 50% trichloroacetic acid was then added and the lysates incubated for 10 min on ice. Samples were centrifuged and pellets were resuspended in Laemmli protein sample buffer and analyzed by SDS-PAGE.
Immunoblotting
Protein samples were separated by SDS-PAGE, transferred to nitrocellulose membranes (Amersham Pharmacia for FKBP-FRB and Hog1 pathway experiments or Whatman® Protran® for PP2A experiments) and incubated with the antibodies summarized in Supplementary Table 3.
For the detection of rapamycin-induced dimerization of FKBP and FRB in Figure 1b, samples were analyzed on different blots for mono- (anti-me1K9H3, clone 7E7-H12) or trimethylation (anti-me3K9H3, clone 6F12-H4). For the detection of protein interactions in the HOG pathway in Figure 1b–d, membranes were blocked with 1% nonfat dry milk (NFDM) in phosphate buffered saline (pH 7.4) with 0.1% Tween 20 (PBS-T 0.1%) for 1 h at 25°C and incubated for 1.5 h at 4 °C with the primary antibody (anti-me1K9H3 or anti-me3K9H3) and a 1:10,000 dilution of the secondary antibody (anti-mouse–HRP, Jackson ImmunoResearch, catalog number: 115-035-008) in a yeast extract solution (1 g of yeast mechanically disrupted using a Freezer/Mill, diluted in 10 ml PBS-T 0.1% supplemented with one tablet of Complete mini EDTA-free protease inhibitor (Roche)). Blots were then washed three times for 20 min (3 × 20 min) in PBS-T 0.1% before being developed with ECL (West Pico Chemiluminescent kit, Thermo Scientific). After being probed with the methylation antibodies, blots were stripped for 10 min at 25°C in stripping buffer (2 M MgCl2, 0.1 M acetic acid), and reprobed with HA antibody. For this purpose blots were blocked with 5% NFDM in PBS-T 0.1% for 1 h at 25°C and afterwards incubated for 1 h at 25°C with a 1:10,000 dilution of both primary (anti-HA clone 12CA5 or 16B12, Covance, catalog number MMS-101R) and secondary (anti-mouse–HRP, Jackson ImmunoResearch, catalog number: 115-035-008) antibodies in 0.5% NFDM PBS-T 0.1%. After the membrane was washed for 3 × 10 min in PBS-T 0.1%, the protein bands were detected by ECL.
For the detection of the varying interaction affinities between Pbs2 and Sho1 mutants in Figure 1c, blots were stripped after methylation detection and blocked with Tris-buffered saline–0.1% Tween (TBS-T 0.1%) containing 4% BSA. For detection of phospho-Hog1, blots were incubated at 4°C overnight, with 1:4,000 anti-P-p38 MAPK T180/Y182 (Cell Signaling 9211S) and 1:6,000 anti-rabbit-HRP (GE Healthcare, catalog number NA934V) in TBS-T 0.1% containing 2% BSA. Blots were then washed briefly in PBS-T 0.1% and reprobed for HA (anti-HA clone 12CA5 or 16B12) as described above and both protein bands (phospho-Hog1 and Sho1-H3-HA) were detected by ECL.
For the detection of the interaction between Cdc55 and Net11-600 in Figure 2c,d, samples were analyzed by SDS-PAGE on separate gels, immunoblotted and incubated individually with the different antibodies indicated in the figures. For the experiment in Figure 2e an initial 10% SDS-PAGE western blot analysis was performed for each biological replicate to assess the levels of prey expression in the three strains (TKY103, YBB19, YBB26), and HA levels were quantified using ImageQuant. Subsequently, lysate amounts normalized to the HA levels were separated on 10% SDS-PAGE gels and blotted on two membranes. One was first incubated with antibody against trimethylation (anti-me3K9H3) and then re-probed with Myc tag–specific antibody (clone 4A6, Millipore), and the other blot was incubated with HA tag–specific antibody (clone 16B12). For the experiment in the figure, twice the amount of whole-cell protein was analyzed for the rts1Δ strain as for the other two strains.
For the two experiments in Figure 2d,e, samples from M-Track assays with different interaction partners were separated on the same gel and blotted on the same membrane. Thus, in these figures, panels originate from the same exposure of the same membrane.
In all PP2A experiments, membranes were blocked with 3% NFDM in PBS-T 0.05% and sodium azide (0.002%) (Blocking Solution 1) for 1 h at 25°C and incubated with primary antibody in 0.5% NFDM/PBS-T 0.05% with Thiomersal (0.001%) (Blocking Solution 2) overnight at 4 °C. Membranes were then washed for 3 × 5 min with PBS-T 0.05% and incubated with HRP-conjugated anti-mouse secondary antibody (AffiniPure Goat Anti Mouse IgG [Fcγ fragment] specific antibody, Jackson ImmunoResearch, catalog number: 115-035-008) 1:10,000 in Blocking Solution 2 for 1 h at 25°C followed by 3 × 10 min washing with PBS-T 0.05%. Protein bands were detected by Western Lightning ECL solution (PerkinElmer).
In cases where blots were reincubated with a second primary antibody (Figure 2e and Supplementary Figures 5a and 6), they were washed in PBS-T 0.05% and treated as described before, except that sodium azide (0.005%) was added to Blocking Solution 1 and the primary antibody mix.
Immunoprecipitations and protein phosphatase assay.
For immunoprecipitation assays, yeast whole-cell extracts were prepared as described previously17 except that lysis was performed using Fastprep (MP Biomedicals, 1 × 40 s, 6 m s-1), and for the experiment in Supplementary Figure 6, an additional washing step (with 0.5 M LiCl, 50 mM Tris, pH 7.5) was performed before the TBS washing steps. HA- or Myc-tagged proteins were immunoprecipitated with anti-HA (clone 12CA5) or anti-Myc (clone 4A6) antibodies, cross-linked to BSA-coated protein A–Sepharose beads (GE Healthcare). Immunoprecipitates were analyzed by 7.5% or 10% SDS-PAGE, immunoblotted and incubated with specific antibodies against Myc tag (clone 4A6), HA tag (clone 16B12), me3K9H3 (clone 6F12-H4), Pph21 (rabbit pAb), Cdc55 (clone 9D3-H6), Rts1 (rabbit pAb) or Tpd3 (clone 5G2).
Phosphatase activity of PP2A immunoprecipitates was assayed toward 32P-labeled phosphorylase a as described previously17. Immunoprecipitates of Myc-Cdc55/Rts1 containing PP2A complexes were twofold serially diluted and their phosphatase activity was plotted against the respective amount of co-immunoprecipitated PP2A catalytic subunit Pph21 (activity-plot) as determined by immunoblot analysis and quantification by ImageQuant. This standard curve was used to calculate the specific activity of Pph21 co-immunoprecipitated with Myc-Cdc55/Rts1. The specific activities of Pph21 co-immunoprecipitated by Myc-HKMT-Cdc55/Rts1 are presented as percent activity with respect to the specific activity of Myc-Cdc55/Rts1–associated Pph21, which was set to 100%.
Image gathering
For FKBP-FRB and HOG pathway studies, developed Fuji medical X-ray films were scanned on a Canon CanoScan N640P scanner using Scancraft (Canocraft) CS-P (3.8.2) with a resolution of 600 d.p.i. in grayscale mode. For PP2A and Biotin-ligase studies, developed Fuji medical X-ray films were scanned on a Canon CanoScan 4200F scanner using Adobe Photoshop CS3 with a resolution of 600 d.p.i. in color mode. After cropping, the images were changed to grayscale, saved in TIFF format (16-bit) and were imported to Microsoft Office PowerPoint 2007 where panels from different scans were combined and labels were added. All figures were then exported as an Adobe Acrobat PDF and again imported into Adobe Photoshop CS3. Subfigures were combined into final figures and exported in TIFF format (600 d.p.i., 8bit, CMYK, LZW-compression).
Quantification of western blots.
Signal intensities of protein bands detected by the specific antibodies (Lys9 methylation–specific antibodies (clone 7E7-H12, cone 5E5-G5 and clone 6F12-H4), anti-HA– (clone 16B12) and anti-Myc–epitope tag (clone 4A6) antibodies) from three biological replicates were quantified using ImageQuant. Methylation signals were normalized to the prey expression levels (anti-HA signal intensities). The normalized values are graphically presented in Figure 1b,d as fold change with respect to the methylation signal set to one at t = 0. In Figure 2c–e, values are presented as percent relative methylation signal with respect to the highest methylation signal, which was normalized to 100%. Results are shown as mean ± s.d.
Nocodazole assay.
The assay is based on the rapid death assay for nocodazole sensitivity. S. cerevisiae strains were inoculated in Glu-CM or Glu-CM L− medium and grown overnight at 30 °C. We inoculated 1 × 108 cells into 50 ml of fresh medium containing 1% DMSO and grown for 3 h at 30 °C up to exponential phase. We plated 300 cells on YPD plates (t = 0) and then added nocodazole (Acros Organics) to the remaining cultures for a final concentration of 15 μg/ml. After 2, 4 and 6 h, we again plated 300 cells on YPD. After incubating the plates at 30 °C for 2 d, we determined, the number of colony-forming units and calculated the fraction of viable colonies based on the number of viable colonies at t = 0.
Benomyl assay.
The assay is based on the plate assay for benomyl sensitivity. S. cerevisiae strains were inoculated in Glu-CM or Glu-CM L− medium and grown overnight at 30 °C. The yeast cultures were then diluted to an OD600 of 0.5 and allowed to grow at 30 °C for 3 h. The logarithmically growing cultures were diluted to an OD600 of 0.8, and a tenfold serial dilution series was spotted on YPD plates containing 0, 20 and 25 μg/ml benomyl (Sigma). Plates were incubated at 30 °C for 2–3 d and scored for yeast cell growth to measure their sensitivity to benomyl.
Peptide dot blot.
For the dot plot procedure, 50 pmol and 10 pmol of peptides corresponding to amino acid 1–20 of histone H3 and bearing either unmodified Lys9 or mono-, di- or trimethylated Lys9 (piCHEM and Thomas Jenuwein lab) were spotted onto a PVDF membrane (Amersham), and equal loading was confirmed by Ponceau S staining. Membranes were then treated according to the immunoblotting protocol for PP2A experiments (see "Immunoblotting").
Generation of Lys9 methylation–specific antibodies.
Female Balb/c mice were immunized with keyhole limpet hemocyanin (KLH)-coupled branched peptides corresponding to amino acids 5–15 of histone H3 ([QTARKSTGGKA]2KC)18, which contained either mono-, di- or trimethylated Lys9. Three days after the final boost, the mice were sacrificed, the splenocytes were fused to X63-Ag8.653 mouse myeloma cells by polyethylene glycol cell fusion, and hybridoma cells were grown under hypoxanthin-aminopterin-thymidine (HAT) selection19. One week after fusion, hybridoma supernatants were screened by ELISA against linear peptides corresponding to amino acids 1–20 of histone H3 and bearing either unmodified or mono-, di- or trimethylated Lys9 for the presence of the desired antibodies. Positive clones were subcloned and weaned from HAT selection to obtain stable antibody-secreting single clones, namely anti-me1K9H3 clone 7E7-H12, anti-me2K9H3 clone 5E5-G5 and anti-me3K9H3 clone 6F12-H4. For the detection of methylated prey proteins, crude cell culture supernatants were used.
Ethics statement
The mice maintenance and all experimental procedures involved in the generation of the various anti–methyl-K9H3 and PP2A antibodies were conducted according to the Austrian Animal Experiments Act and approved by the Austrian Federal Ministry of Science and Research (GZ-66.009/0101-BrGT/2005 and BMWF-66.009/0091-II/3b/2011) and the animal experiments ethics committee of the Medical University of Vienna.
Supplementary Material
ACKNOWLEDGMENTS
We thank K. Nasmyth for scientific input; T. Jenuwein (Max Planck Institute of Immunobiology and Epigenetics, Freiburg) for materials crucial in setting up the assay; H. Charbonneau (Purdue University, West Lafayette), F. Uhlmann (London Research Institute, London), R. Deshaies (California Institute of Technology, Pasadena) and D. Brautigan (University of Virginia School of Medicine, Charlottesville) for providing plasmids and materials; and H. Hombauer, S. Kuderer and M. Roblek for technical assistance. This work was supported by grants from the Austrian Science Foundation (FWF P21712 and doctoral program APW01220FW) to E.O. G.A. was supported by FP6 (QUASI) and FP7 (UNICELLSYS) European Commission Program grants.
Footnotes
COMPETING FINANCIAL INTERESTS The authors declare competing financial interests: EO serves as a consultant to Merck/Millipore Corporation. The EO lab receives royalties from several biotech companies for monoclonal antibodies αme3K9H3 clone 6F12-H4 and α-myc clone 4A6. None of the other co-authors have a competing interest.
REFERENCES
- 1.Piehler J. Curr. Opin. Struct. Biol. 2005;15:4–14. doi: 10.1016/j.sbi.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 2.Bodenmiller B, et al. Sci. Signal. 2010;3:rs4. doi: 10.1126/scisignal.2001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandez-Suarez M, Chen TS, Ting AY. J. Am. Chem. Soc. 2008;130:9251–9253. doi: 10.1021/ja801445p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rea S, et al. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 5.Choi J, Chen J, Schreiber SL, Clardy J. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- 6.Hohmann S. FEBS Lett. 2009;583:4025–4029. doi: 10.1016/j.febslet.2009.10.069. [DOI] [PubMed] [Google Scholar]
- 7.Marles JA, Dahesh S, Haynes J, Andrews BJ, Davidson AR. Mol. Cell. 2004;14:813–823. doi: 10.1016/j.molcel.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 8.Hao N, et al. Curr. Biol. 2007;17:659–667. doi: 10.1016/j.cub.2007.02.044. [DOI] [PubMed] [Google Scholar]
- 9.Shu Y, Yang H, Hallberg E, Hallberg R. Mol. Cell. Biol. 1997;17:3242–3253. doi: 10.1128/mcb.17.6.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Y, Boguslawski G, Zitomer RS, DePaoli Roach AA. J. Biol. Chem. 1997;272:8256–8262. doi: 10.1074/jbc.272.13.8256. [DOI] [PubMed] [Google Scholar]
- 11.Queralt E, Lehane C, Novak B, Uhlmann F. Cell. 2006;125:719–732. doi: 10.1016/j.cell.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 12.Stark C, et al. Nucleic. Acids. Res. 2011;39:D698–704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azzam R, et al. Science. 2004;305:516–519. doi: 10.1126/science.1099402. [DOI] [PubMed] [Google Scholar]
- 14.Chan LY, Amon A. Genes Dev. 2009;23:1639–1649. doi: 10.1101/gad.1804609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciaccio MF, Wagner JP, Chuu CP, Lauffenburger DA, Jones RB. Nat. Methods. 2010;7:148–155. doi: 10.1038/nmeth.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kushnirov VV. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 17.Fellner T, et al. Genes Dev. 2003;17:2138–2150. doi: 10.1101/gad.259903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez-Burgos L, et al. Methods Enzymol. 2004;376:234–254. doi: 10.1016/S0076-6879(03)76016-9. [DOI] [PubMed] [Google Scholar]
- 19.Köhler G, Milstein C. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.