Summary
Histone chaperones physically interact with histones to direct proper assembly and disassembly of nucleosomes regulating diverse nuclear processes such as DNA replication, promoter remodelling, transcription elongation, DNA damage, and histone variant exchange. Currently, the best characterised chaperone-histone interaction is that between the ubiquitous chaperone Asf1 and a dimer of H3 and H4. Nucleosome Assembly Proteins (Nap proteins) represent a distinct class of histone chaperone. Using pulsed electron double resonance (PELDOR) measurements and protein cross-linking we show that two members of this class, Nap1 and Vps75, bind histones in the tetrameric conformation also observed when they are sequestered within the nucleosome. Furthermore, H3 and H4 trapped in their tetrameric state can be used as substrates in nucleosome assembly and chaperone mediated lysine acetylation. This alternate mode of histone interaction also provides a potential means of maintaining the integrity of the histone tetramer during cycles of nucleosome reassembly.
Keywords: Nucleosome assembly, Histone Chaperone, Nap1, Vps75, Chromatin
Introduction
Access to the genetic information of eukaryotic organisms is regulated in part by the assembly and disassembly of chromatin. This involves interplay between the action of histone chaperones, ATP-dependent remodelling enzymes and histone modifying enzymes.
There are at least 7 major structural families of histone chaperones related to; nucleoplasmin, chromatin assembly factor 1 (CAF-1), antisilencing factor 1 (Asf1), facilitates chromatin transcription (FACT), regulator of Ty1 transposition 106 (Rtt106), chaperone for Htz1/H2A-H2B dimer (Chz1) and nucleosome assembly protein 1 (Nap1) (Das et al., 2010; Hansen et al., 2010). High resolution crystal structures have been obtained for a number of histone chaperones including Asf1 (Daganzo et al., 2003; Mousson et al., 2005), Vps75 (Berndsen et al., 2008; Park and Luger, 2008; Tang et al., 2008) nucleosplasmin (Dutta et al., 2001) and Nap1 (Park and Luger, 2006b). However, only for Asf1 is there detailed information regarding the nature of the interaction between a chaperone and the histones it binds. In this case crystallography and NMR have been used to show that the C-terminal helix of H3 interacts with a concave groove on Asf1 (Mousson et al., 2005) (Henikoff, 2008). Subsequently, Asf1 has been co-crystallised with truncated histones H3 and H4 (English et al., 2006; Natsume et al., 2007). This structure indicates that Asf1 makes contacts with both H3 and H4 and, consistent with previous biochemical investigations (English et al., 2005), these histones are present as a dimer rather than tetramer. Furthermore the interaction of Asf1 with the C-terminus of H3 obscures an interaction surface required for formation of a H3-H4 tetramer. Further support for the splitting of the tetramer has come from pull down experiments suggesting that the Asf1-H3-H4 heterotrimer prevails in complex with CAF-1 and HIRA in mammalian cells (Tagami et al., 2004).
Although these observations all support the splitting of histone tetramers following association with histone chaperones, there is historic literature (reviewed in (Annunziato, 2005), supported by recent studies (Xu et al., 2010) indicating that the integrity of a substantial proportion of histone tetramers is maintained following DNA replication. These observations are difficult to reconcile with the splitting of the (H3-H4)2-tetramer when associated with chaperones as has been proposed to occur during chromatin assembly and disassembly reactions. One possible means of reconciling the persistence of the parental tetramer is that the nature of the interaction between alternate classes of chaperone and their histone cargos differs. Importantly, interactions between Nap1 related chaperones have not been characterised in as much detail as has been possible for Asf1.
S. cerevisiae has at least two Nap proteins, Nap1 and Vps75. Crystal structures of these two chaperones show they exist as tightly bound homodimers adopting the so called ‘headphone’ fold (Berndsen et al., 2008; Park and Luger, 2006b; Park et al., 2008b; Tang et al., 2008; Tsubota et al., 2007). Nap1 and Vps75 bind all four core histones with nanomolar affinity in vitro (Andrews et al., 2008; Berndsen et al., 2008; Park et al., 2008b; Tsubota et al., 2007), with Nap1 being shown to associate with all four core histones in vivo (Krogan et al., 2006). Vps75 is found in complex with the acetyl transferase Rtt109 in vivo where it aids in acetylation of newly synthesised histones (Fillingham et al., 2008; Li et al., 2008; Selth and Svejstrup, 2007; Tsubota et al., 2007) possibly assisting histone deposition during S-phase (Kaplan et al., 2008; Li et al., 2008). Nap1 has roles in histone transport into the nucleus (Chang et al., 1997; Mosammaparast et al., 2001), and acts in conjunction with ATP-dependent chromatin remodeling factors in the assembly and disassembly of nucleosomes in vitro (Lorch et al., 2006; Tyler et al., 1999).
Initially it was reported that a Nap1 dimer binds a single H3-H4 dimer (McBryant et al., 2003; Toth et al., 2005). These calculations were derived from observing complex formation under native polyacrylamide gel electrophoresis, an approach which is often prone to error, especially with a self associating system (Park et al., 2008a; Toth et al., 2005). More recent studies using a fluorescent binding assay to analyse the thermodynamics of histone-Nap1/Vps75 interactions clearly show that two H3-H4 dimers bind a single Nap1/Vps75 dimer (Andrews et al., 2010; Andrews et al., 2008; Park et al., 2008b). However, using these approaches, it was not possible to distinguish whether histones H3 and H4 are bound as two dimers or one tetramer. The presence of an exposed beta sheet face within the earmuff domain of Nap proteins has led some to suggest, guided by homology modelling to Asf1, that binding may occur via splitting of the tetramer, one H3-H4 dimer binding per beta sheet face in a similar fashion to Asf1 (Gill et al., 2009).
Here we apply protein crosslinking and site directed spin labelling (SDSL) with pulsed electron double resonance (PELDOR) to probe the conformation of H3 and H4 when in complex with Nap1, Vps75 and Asf1. Consistent with previous reports we find that association with Asf1 causes dissociation of the (H3-H4)2-tetramer, resulting in binding of a single H3-H4 dimer. In contrast, histones H3 and H4 bound to Nap1 or Vps75 remain associated as a tetramer, showing for the first time that a histone chaperone can bind H3 and H4 in their nucleosomal conformation. Furthermore, we show H3 and H4 remain as a tetramer during deposition onto DNA by Nap1 and that Vps75, but not Asf1, preferentially stimulates Rtt109 directed acetylation of H3 and H4 locked in their tetrameric conformation. The prevalence of a tetramer of H3 and H4 when bound to a histone chaperone potentially provides a molecular explanation as to how the integrity of the histone tetramer is maintained during the course of DNA replication.
Results
The interactions of Asf1 and Nap1 with histones H3 and H4 display contrasting sensitivity to ionic conditions
Within the nucleosome the predominant mode of histone-DNA interaction is electrostatic: histones present a basic ramp around which the acidic DNA molecule wraps. Outside of the nucleosome, for instance during histone shuttling into the nucleus, the basic surfaces of the histones have to be shielded from spurious interactions with other cell components. Histone chaperones therefore have been defined as proteins that prevent such non-specific interactions occurring, promoting proper chromatin assembly (Andrews et al., 2010; Laskey et al., 1978). As a first step towards comparing the mode by which Asf1 and Nap1 interact with H3 and H4 we monitored complex formation during gel filtration chromatography under different ionic conditions.
If Nap1 binds H3 and H4 in a similar fashion to Asf1 (i.e. via the β-sheet face within the earmuff domain) we reasoned its interaction should display similar properties. In 0.6 M sodium chloride both Nap1 and Asf1 elute as higher molecular weight complexes consisting of the chaperone, H3 and H4 (Figure 1A top, fractions 5-8 and 10-12, respectively). However, in the high ionic environment of 1 M sodium chloride, H3 and H4 no longer stably associate with Nap1 (Figure 1A bottom, Nap1 fractions 8-11, H3 and H4 fractions 13-15). In contrast, the complex between Asf1 and histones H3 and H4 remains stable. The robustness of the Asf1 histone complex to ionic conditions is likely to result from interactions predominantly with the largely hydrophobic dyad interface of H3 (English et al., 2006; Natsume et al., 2007). The sensitivity of the Nap1 histone complex to salt conditions is more reminiscent of the interaction of histones with DNA and suggests a different mode of contact involving ionic interactions. One means by which this could be achieved is if the hydrophobic surfaces of H3 and H4 are sequestered in their tetrameric conformation (observed within the nucleosome) when they are bound to Nap1. This would leave the positively charged DNA binding surfaces available for interactions with Nap1. This led us to hypothesise that the sensitivity of the Nap1-H3-H4 complex to ionic strength, over the Asf1-H3-H4 complex allows for the tetramerisation of H3 and H4.
Figure 1.
Histone binding to Nap1, but not Asf1, shows sensitivity to ionic strength. (A) The elution profiles of Nap1-H3-H4 from gel filtration shown as consecutive fractions separated by SDS-PAGE. Complex formation under 0.6 M NaCl and 1.0 M NaCl was monitored. Fraction numbers are denoted at the bottom of the gel and the elution points of molecular weight markers shown above. (B) The same experiment carried out for Asf1 in complex with H3 and H4.
Chemical crosslinking of H3 suggests Nap1 binds a constitutive (H3-H4)2-tetramer
To test the hypothesis that Nap1 can bind H3 and H4 in their tetrameric conformation we developed a cysteine crosslinking approach to report on the tetramerisation of H3 and H4. Using the 1.9 Å nucleosome structure as a guide (Richmond and Davey, 2003), we scouted for residues on H3 that are within a few Angstroms of each other yet not involved in the H3-H3’ interaction. We identified H3 lysine 115 as a potential crosslinking site and engineered a cysteine at this location (Figure 2A). The Cβ atoms of this residue are within 9 Angstroms of each other, therefore formation of a tetramer should result in crosslinking at this position in the presence of a homobifunctional thiol reactive crosslinker. This crosslinked, H3-H3’ species can then be easily resolved from non crosslinked H3 by SDS-PAGE. A methanethiosulfonate crosslinker with a 3 carbon spacer (termed MTS-3-MTS, Figure 2A) was found to be most efficient (data not shown). Using a 1:1 ratio of tetramer to MTS-3-MTS, crosslinking occurred rapidly and was all but complete by one minute, resulting in 56% of total H3 crosslinked at 1 μM complex (Supplemental Figure S1 A, B). Importantly, the gel filtration profile of crosslinked tetramer was unchanged indicating that H3 crosslinks occurred specifically within the tetramer rather than between different H3-H4 tetramers (Figure S1C).
Figure 2.
Directed sulfhydryl reactive crosslinking suggests Nap1 binds H3 and H4 in their tetrameric conformation. (A) The structure of the (H3-H4)2-tetramer from the nucleosome crystal structure showing the proximity of H3 K115 residues (green spheres) (image created using Pymol, http://www.pymol.org). Chemical structure of the compound MTS-3-MTS used to crosslink cysteines engineered at position 115 is shown alongside. (B) The extent of crosslinking under increasing concentrations of cysteine free Nap1 or Asf1 shown by separation of the reaction components by SDS-PAGE. Concentration of chaperone from left to right was 0.25, 0.5, 1.0, 2.0 μM monomer. (C) Gel filtration analysis of the H3 K115C crosslinked tetramer in the presence of Nap1 showing they remain stably associated under the conditions used. (D) Globular Asf1 was mixed with non-crosslinked and crosslinked tetramer and the complexes separated by gel filtration chromatography. The majority of Asf1 does not form a complex with crosslinked tetramers.
Using this crosslinking approach we probed the conformation of H3 and H4 when bound to Asf1 and Nap1. Asf1 has been shown to disrupt the histone tetramer by binding to the C-terminus of H3, obscuring the H3-H3’ interface (English et al., 2006; English et al., 2005; Natsume et al., 2007). Therefore, one would expect that Asf1 would inhibit crosslinking in our assay. Indeed this is exactly what we see (Figure 2B). Titrating Asf1 against the (H3-H4)2-tetramer prior to crosslinking, we see a gradual disappearance of the H3-H3’ crosslinked species and an increase in non-crosslinked H3. In contrast to this, when a cysteine free form of Nap1 (Andrews et al., 2008) is titrated into the reaction prior to crosslinking the H3-H3’ crosslinked species persists (Figure 2B), strongly suggesting that the H3-H3’ interface is retained when bound to these chaperones. The final titration point for Nap1 was subject to gel filtration chromatography (Figure 2C) showing that crosslinked H3 remains stably associated with Nap1. Furthermore, pre-crosslinked tetramers formed complexes with Nap1 that were indistinguishable from those formed in the absence of crosslinking (compare Figure S2 and Figure 1A). This indicates that crosslinking does not affect the Nap1-(H3-H4) interaction. In contrast, the ability of Asf1 to bind histones H3 and H4 was impaired if they were first crosslinked (figure 2D). As Asf1, Asf1-H3-H4 and the (H3-H4)2-tetramer all elute in similar fractions following gel filtration chromatography, a globular form of Asf1 (Asf1g)(English et al., 2005) comprising the histone binding domain was used to aid in the separation of complexes as it elutes in later fractions (Figure 2D, fractions 19-21). In the absence of crosslinker Asf1g migrates with the histones in a heterotrimeric complex (Figure 2D, fractions 14-16). In contrast to this, in the presence of crosslinked tetramer Asf1g’s ability to bind histones is severely impaired. The vast majority of Asf1g elutes as a monomer (Figure 2D, fractions 19-21) with a small amount eluting with residual non-crosslinked tetramer (fractions 14-16) and a smaller fraction eluting with crosslinked tetramer (fractions 12-13). This is consistent with the crosslink acting to stabilise H3 and H4 in a nucleosomal conformation disfavoured during Asf1 binding. Note that this stabilisation is not necessarily absolute. Some reorganisation of tetramer structure may still be possible, despite the presence of the crosslink. This could account for the residual binding of Asf1 to crosslinked H3. Overall these observations demonstrate that our crosslinking approach is both an excellent reporter on tetramer structure and an effective method of stabilising the (H3-H4)2-tetramer.
Long range distance measurements between histones indicate that H3 and H4 exist as a tetramer when bound to Nap1
Our cysteine reactive crosslinking approach demonstrated that position K115 on H3 and H3’ are in close proximity when bound to Nap1 and Vps75. Although this strongly suggests that this is the result of (H3-H4)2-tetramer formation, the possibility still exists that H3 and H4 may be bound as two separate dimers in an orientation where K115 of H3 and H3’ are within crosslinking distance. Thus, to further test our hypothesis that H3 and H4 are in their tetrameric conformation we used SDSL and PELDOR to extract distance measurements from H3 and H4 when in complex with the chaperones Nap1 and Asf1. This involves attaching nitroxide reporters via engineered cysteine residues and extracting distance information from spin-spin dipole interactions observed following pulsed microwave radiation. This approach was previously used to characterise the structure of the (H3-H4)2-tetramer and identified labelling sites on both H3 and H4 that give discrete distance distributions in close agreement with distances calculated from the nucleosome crystal structure (Bowman et al., 2010). These sites, H4 R45R1 and H3 Q125R1 (Figure 3A), were used to probe the structure of the (H3-H4)2-tetramer under conditions established to result in stable complexes with Nap1 and Asf1.
Figure 3.
Extraction of long-range distances between spin labelled H3-H4 dimers agrees with a tetrameric conformation when bound to Nap1. (A) Positions of spin labels on H4 R45 and H3 Q125 (green spheres) are shown on the structure of the tetramer taken from the nucleosome (PDB code 1KX5, images created by Pymol, http://www.pymol.org). (B) The background corrected dipolar evolution and the distance distribution for each labelling site in complex with Nap1 and Asf1 is shown (red trace). For comparison the tetramer value extracted from the tetramer alone is shown alongside (black trace).
In these experiments three possible outcomes were anticipated. Firstly, if a (H3-H4)2-tetramer is bound then the distance distributions from a complex of proteins should be comparable to that of free tetramer. Secondly, if two H3-H4 dimers are bound in a non-tetrameric conformation an alternative distance distribution should be observed. Thirdly, if the tetramer is split with just a single H3-H4 dimer being bound, as has been suggested (McBryant et al., 2003; Toth et al., 2005), there would be no spin-spin dipolar coupling, and thus no distance.
It can be seen for both labelling sites, H4 R45R1 and H3 Q125R1, that the distance distribution corresponding to a tetramer of H3 and H4 persists when the histones are in complex with Nap1 (Figure 3B). It is also notable that minor distance distributions increase in the presence of histone chaperones. The conversion of PELDOR data to distance distributions by Tikhonov regularisation is prone to background derived errors. This is often observed as minor apparent distance distributions at longer distances (Bowman et al., 2010). Adjustment of background correction parameters results in apparent movement of artefactual distance distributions but does not have a large effect on real distance distributions. This is illustrated in Figure S3. For this reason, we believe that the major distance distributions are significant whereas the minor ones are not. In contrast, formation of Asf1-H3-H4 complexes completely ablates the dipolar coupling, and this is observed in the time trace as an unmodulated exponential decay. Thus, in addition to the short range interaction between H3 K115C identified by crosslinking, longer range distance measurements between H4 R45R1 and H3 Q125R1 indicate a tetrameric configuration predominates in Nap1-H3-H4 complexes. We conclude from this that H3 and H4 are bound to Nap1 in a tetrameric conformation very similar to that observed within nucleosomes (Bowman et al., 2010).
The Nap1 related chaperone Vps75 also binds H3 and H4 as a tetramer
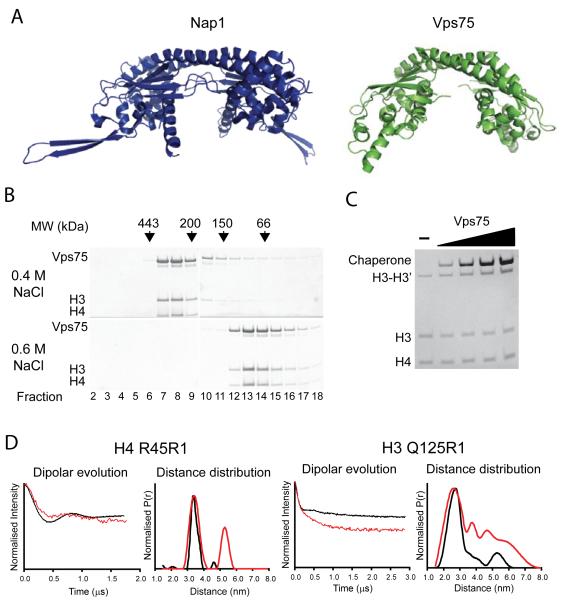
Differences in the structures of Nap1 and Asf1 are likely to confer the different modes of histone interaction we have detected for these two proteins. Sequence alignments indicate that Nap1 related proteins are broadly conserved through evolution and that many eukaryotes encode multiple Nap1 related proteins (Park and Luger, 2006a). For example, budding yeast encode the Nap1 related protein Vps75. The structure of Vps75 is closely related to Nap1 (Figure 4A). This raises the possibility that the interaction with tetrameric H3-H4 may be a general feature of Nap1 related histone chaperones. To investigate this further we set out to characterise the interaction of histones H3 and H4 with Vps75.
Figure 4.
Vps75 binds H3 and H4 in their tetrameric conformation. (A) Nap1 (PDB code: 2ZR2) and Vps75 (PDB code: 3C9D) both adopt the headphone fold. Structures shown for comparison (images created by Pymol, http://www.pymol.org). (B) H3 and H4 binding by Vps75 displays sensitivity to ionic strength. Fractions from gel filtration chromatography are shown under 0.4 M and 0.6 M sodium chloride. Vps75-H3-H4 form a complex at 0.4 M sodium chloride (fractions 7-9, top panel), but elute in their separate fractions at 0.6 M (note, both free Vps75 dimer and H3-H4 tetramer elute in fractions 13-15) (C) Histone tetramer specific crosslinking of H3 K115C by MTS-3-MTS persists in the presence of cysteine free Vps75. Concentration of Vps75 from left to right was 0.25, 0.5, 1.0, 2.0 μM monomer. (D) Long range distance from histone tetramer spin labelled at position H4 R45R1 and H3 Q125R1 complexed with Vps75 were extracted using the PELDOR experiment. Background correct dipolar evolutions and distance distributions for each are shown (red trace). For comparison, distances measured for the tetramer alone are shown alongside (black trace).
Just as we had previously observed for Nap1, Vps75 formed complexes with histones H3 and H4 that were sensitive to ionic conditions, though in this case 0.6 M sodium chloride was sufficient to cause dissociation (Figure 4B) with a stable association occurring at 0.4 M sodium chloride. Crosslinking at H3 K115C was not reduced when Vps75 was bound to tetramers (Figure 4C) and crosslinking was retained in the complexes when assessed by gel filtration (not shown). Finally PELDOR measurements between H4 R45R1 and H3 Q125R1 indicate that a tetrameric configuration is retained within these complexes (Figure 4D). From these observations we conclude that Vps75, like Nap1 binds to histones H3 and H4 in a tetrameric configuration, suggesting that tetramer binding may be a general property of Nap proteins.
In vivo evidence for the association of histone chaperones with histones in a tetrameric conformation
Asf1 and Vps75 are known chaperones for H3 and H4, however, Nap1 is typically regarded as an H2A-H2B chaperone, despite its high affinity of H3 and H4 in vitro (Andrews et al., 2008). To investigate whether Nap1 also associates with histones H3 and H4 in vivo, we isolated Nap1 by Tandem Affinity Purification (TAP) (Puig et al., 2001) and then immunoblotted for histones. In addition to H2A we could detect H4 associated with Nap1 (Figure S4A), suggesting that Nap1 is likely to form complexes with all histones in vivo. We next generated a strain in which the sole source of H3 was myc tagged and included the H3K115C mutation we had used in vitro. This strain was viable with no growth defects detected (data not shown). TAP tagging the chaperones Asf1, Nap1 and Vps75 in this background provided a means of recovering the H3 associated with each chaperone. We next sought to monitor the conformation of histones associating with these different chaperones by crosslinking.
Initial experiments using the chemical crosslinkers such as MTS-3-MTS and BMOE resulted in low levels of H3-H3 crosslinking and the appearance of additional high molecular weight crosslinked species. When using these compounds with recombinant proteins we had observed that the stoichiometry of crosslinker to cysteines was critical and likely to be more difficult to control using native proteins. Instead, we pursued an alternative crosslinking approach whereby disulphide bond formation between juxtaposed cysteine residues is catalysed by copper(II) chelated with 1,10-phenanthroline (CuP) (Kobashi, 1968). Crosslinking using CuP gave similar results to chemical crosslinkers in vitro (Figure S4C), but did not display the same sensitivity to stoichiometry, with crosslinking efficiency increasing with CuP concentration, as previously reported (Kobashi, 1968).
Using this approach, crosslinking of H3K115C-myc associated with Nap1 and Vps75 was found to be considerably more efficient than H3 bound by Asf1 (Figure 5A). Indeed, 78% of Nap1 and 86% of Vps75 associated H3K115C-myc was crosslinked at 1mM CuP, whereas only 13% of Asf1 associated H3K115C-myc was crosslinked at the same CuP concentration. The minor crosslinked fraction in the Asf1 pull down may result from an equilibrium between (H3-H4)2 and Asf1-H3-H4 at the low concentrations used in the crosslinking experiment or the ability of some Asf1 to associate with H3 despite the presence of the crosslink as observed in vitro (Figure 2D). To verify the size of the crosslinked H3 recombinant H3 was run alongside (Figure 5A). The lack of the myc tag means that the recombinant H3 and crosslinked H3 migrate slightly faster than there tagged counterparts (Figure 5A).
Figure 5.
Nap1 and Vps75 co-purify with tetrameric H3-H4 and can use H3 and H4 trapped in their tetrameric conformation for their biological functions in vitro. (A)TAP-tagged Asf1, Nap1 and Vps75 were purified from strains expressing H3K115C-myc as the sole source of histone H3. Complexes were subject to crosslinking in the presence of 0.064, 0.25 or 1.0 mM CuP (Asf1, lanes 2-4; Nap1, lanes 6-8; Vps75, lanes 10-12), separated by SDS-PAGE and immuno-blotted within an anti-myc antibody. The intensity of the anti-myc signal was quantified and the proportion of crosslinked H3 of the total displayed at the bottom of each lane. Lane 13 contains recombinant partially cross-linked (non-tagged) H3 developed with a H3 antibody as a size standard (B) (H3-H4)2-tetramer deposition by Nap1. Left panel: crosslinked tetramer input stained with coomassie. This indicates that the majority of histone H3 present is crosslinked. Centre panel: monotetrasomes were separated from free DNA by native gel electrophoresis. Lane 1, without Nap1; lane 2, 3, 4 and 5: 0.2, 0.5, 1.0 and 2.0 μM Nap1, respectively. Right panel: the mono-tetrasomal bands were excised from the native gel and subject to SDS-PAGE and probed by western blotting for H3. Lanes 6-10 correspond to lanes 1-5 from the centre panel. This shows that crosslinked tetramers are present in tetrasomes assembled by Nap1. (C) Vps75 preferentially promotes Rtt109 acetylation of crosslinked tetramer, whereas Asf1 preferentially directs Rtt109 acetylation in favour of non-crosslinked tetramer. Left panel: equimolar amounts of crosslinked to non-crosslinked H3 K115C was used as an input. Right panel: lane 1, Rtt109 alone (140 nM). Lanes 2-6: 60, 80, 100, 120 and 140 nM each of Asf1 and Rtt109, respectively. Lanes 7-11: 10, 15, 20, 25 and 30 nM of (Vps75)2-Rtt109 complex, respectively.
Nap1 facilitates the loading of intact H3-H4 tetramers onto DNA
Having established that Nap1 and Vps75 bind H3 and H4 in their tetrameric conformation, we wanted to see if these chaperones could utilise H3-H4 trapped in their tetrameric conformation by crosslinking at H3K115C performing functional assays. The observation that Asf1 causes a structural rearrangement within H4 has been used to propose a model in which tetramer splitting is an obligate step in the action of Asf1 in the assembly and disassembly of nucleosomes (English et al., 2006). To investigate whether Nap1 is able to deposit tetrameric H3-H4 complexes onto DNA we prepared tetramers that were cross-linked at H3 K115C to an efficiency of 80% (Figure 5B). This improved crosslinking efficiency was achieved by crosslinking at 10 μm rather than 1 μm. Deposition of H3 and H4 onto DNA to form a tetrasome is the first step in nucleosome assembly. To isolate this first stage of nucleosome assembly we used a 91 bp DNA fragment (termed −28W-28) derived from the well defined 601 positioning sequence (Thastrom et al., 1999). This size fragment allows only a single tetramer to be assembled onto it, migrating as a single super-shifted band upon native gel electrophoresis, and thus allowing for unambiguous assignment. Assembly of crosslinked tetramers onto the 91 bp DNA fragment was inefficient in the absence of Nap1 (Figure 5B, lane 1), but enhanced in the presence of Nap1 (Figure 5B, lane 2-5). In order to investigate whether the tetramers transferred onto DNA by Nap1 retained a tetrameric conformation, and were not derived from the uncrosslinked population, we excised the tetrasomal band, resolved the histones by SDS-PAGE and assessed the crosslinking status of H3 following western blotting. Figure 5B, lanes 6-10, confirms that the majority of the H3 assembled onto DNA was crosslinked, suggesting that dissociation of the histone tetramer is not an obligate step in nucleosome assembly directed by Nap1.
Vps75 presents a tetramer of H3-H4 to Rtt109 for acetylation
The majority of Vps75 in vivo is found in complex with the acetyl transferase Rtt109, where it is proposed to enhance its acetyl transferase activity (Fillingham et al., 2008; Selth and Svejstrup, 2007; Tsubota et al., 2007). Asf1 also regulates the activity of Rtt109 (Chen et al., 2008; Fillingham et al., 2008; Li et al., 2008; Tsubota et al., 2007) and both enzymes have been shown to stimulate Rtt109’s activity in vitro (Berndsen et al., 2008; Tsubota et al., 2007). Thus, we reasoned that if Vps75 binds a tetramer of H3 and H4 the Vps75-Rtt109 complex should acetylate crosslinked tetramer as efficiently as non-crosslinked tetramer. On the contrary, one would expect acetylation of H3 by an Asf1-Rtt109 complex to be diminished when using crosslinked tetramer as a substrate. To test this we used a competition assay where half of the H3 was in a crosslinked tetramer and half was in a non-crosslinked tetramer. Rtt109 on it own was very poor at acetylating H3 (probed by an antibody to acetylated H3 K56) (Figure 5C, lane 1). Addition of Asf1 increased the efficiency of acetylation by Rtt109 largely on non-crosslinked H3, as expected (Figure 5C, lanes 2-6). In contrast to this, acetylation of H3 by Vps75-Rtt109 was directed predominantly towards crosslinked (H3-H4)2-tetramer (Figure 5C, lanes 7-11). This suggests that Vps75 not only tolerates a tetramer of H3 and H4, but its chaperoning activity is actually enhanced by the stabilisation of the tetramer. Indeed, at physiological ionic strength, in which the acetylations were carried out, the interaction between H3-H3’ is weakened, resulting in an equilibrium between H3-H4 dimers and (H3-H4)2-tetramers (Baxevanis et al., 1991). Therefore, the preference we see towards acetylation of H3 crosslinked at K115C may be due to the stabilising effect that this crosslink has on the tetramer at these salt concentrations.
Discussion
Histone chaperones have been defined based on their ability to bind histones and affect chromatin metabolism without the requirement for ATP hydroylsis. Although they all share this function, crystallographic analysis shows histone chaperones adopt diverse tertiary structures, suggesting they may harbour alternate modes of histone binding. Understanding these interactions at a structural and biochemical level is likely to give key insights into their biological function.
Both site directed crosslinking and PELDOR measurements indicate that histones H3 and H4 adopt a tetrasomal configuration related to that observed in the nucleosome when associated with the histone chaperones Nap1 and Vps75. However, we cannot rule out the possibility that under some circumstances, for example in the presence of limiting histones that dimeric H3-H4 could be bound. Nonetheless, we have found that these Nap proteins retain H3-H4 in their tetrameric conformation when purified from yeast, and can utilise H3 and H4 locked in their tetrameric conformation as substrates in assays related to their biological functions: Vps75 preferentially enhances the activity of Rtt109 for tetrameric H3-H4, whereas Nap1 is able to deliver a crosslinked histone tetramer to DNA. These key steps in chromatin assembly therefore, do not necessarily involve splitting apart tetramers of H3 and H4. This is consistent with both historical and recent observations indicating that following DNA replication the majority of maternal H3 and H4 dimers do not mix with nascent histones (Jackson, 1990; Prior et al., 1980; Xu et al., 2010).
As well as being able to bind H3 and H4 in their tetrameric conformation both Nap proteins analysed in this study show a strong dependency on the ionic environment for their interaction, that has also been observed for Nap1’s interaction with H2A-H2B dimers (Park and Luger, 2006b). Thus, in some respects Nap1 and Vps75 seem to act as a functional DNA mimic. All Nap proteins crystalised to date have shown a distinct charge distribution with the cleft formed between the two earmuff domains being highly acidic. This could be a possible interaction site for the basic ramp of the histone tetramer, and indeed after removing the H3 αN helix, which is likely to be flexible in solution (Bowman et al., 2010), the basic geometry would allow binding in this manner. In this way, Nap1 may function to drive nucleosome assembly by presenting a tetramer of H3 and H4 to DNA in a manner that is conducive to nucleosome formation.
The binding specificity of Nap proteins for histone tetramers may be related to their existence as obligate dimers. Interestingly, homo-dimerisation doesn’t appear to be ubiquitous amongst histone chaperones characterized to date. Indeed, only one other chaperone, nuclear autoantigenic sperm protein (Nasp), prevalent in metazons, has been reported to exist as a homodimer (Finn et al., 2008). Additionally, Nasp has been shown to interact with all four core histones and the linker histone H1 in vitro and in vivo (Finn et al., 2008; Richardson et al., 2000; Tagami et al., 2004; Wang et al., 2008), a characteristic shared by Nap1. The conformation of histones H3 and H4 when bound to Nasp has yet to be determined. The quantities of material required for pulsed EPR limit this technique to chaperones that can be over-expressed recombinantly. However, our crosslinking approach is limited only to protein dectection by immuno-blotting, thus providing an avenue for determining the conformation of H3 and H4 when in complex with larger chaperone complexes and proteins that are not amenable to recombinant expression, such as the CAF1 and HIR complexes.
Our observations indicate that Nap1 related histone chaperones have the properties required to shuttle histone tetramers off and on DNA without a requirement for separating H3-H4 dimers. In contrast, it has previously been shown that Asf1, interacts with histones H3 and H4 in a dimeric conformation. This may represent a normal step in the delivery of newly synthesised histones into chromatin assembly pathways and also contribute to replication independent chromatin assembly where tetramer splitting has been observed to be more frequent (Xu et al., 2010). The existence of distinct pathways by which different classes of histone chaperone act during chromatin assembly is anticipated to influence the way in which histone modifications and variants are transmitted. Investigating the nature of the interaction between histone chaperones and their cargos is likely to provide further insight into this process.
Experimental procedures
Protein expression and purification
A detailed description of protein expression and purification procedures can be found in the supplementary experimental procedures. Histones were essentially expressed as described previously (Luger et al., 1997). Cysteine free version of Asf1, Nap1 and Vps75 were generated by site-directed mutagenesis, expressed from a pET15b vector (Novagen) in E. coli and purified by cobalt affinity, anion exchange and gel filtration chromatography. Rtt109 was expressed from a pET28a vector and purified by cobalt affinity, cation exchange and gel filtration chromatography. Cysteine to alanine mutations were found not to affect interactions with histones. 6His-tagged Vps75 (pET15b vector) and GST tagged Rtt109 (pGEX 6P1 vector, GE Healthcare) were co-expressed in E. coli and purified by sequential cobalt/glutathione affinity and cation exchange chromatography. Asf1g, consisting of residues 1-164, was PCR amplified, clone into pET15b and expressed and purified the same as the full length.
Analytical gel filtration
Complexes were analysed by micro gel filtration chromatography using a Superdex S200 PC 3.2/30 column (GE Healthcare) under the indicated sodium chloride concentrations in 20 mM HEPES-KOH pH 7.5, 1 mM EDTA, 30 % glycerol. 20 μL of 25 μM complex was loaded on to the column and thirty fractions collected spanning the void to bed volume of the column and analysed by SDS-PAGE.
PELDOR experiments
Spin labelled histone-chaperone complexes were prepared as described in the supplementary methods. PELDOR experiments were executed using a Bruker ELEXSYS E580 spectrometer operating at X-band with a dielectric ring resonator and a Bruker 400U second microwave source unit. All measurements were made at 50 K with an overcoupled resonator giving a Q factor of approximately 100. The video bandwidth was set to 20 MHz. The four pulse, dead-time free, PELDOR sequence, was used, with the pump pulse frequency positioned at the center of the nitroxide spectrum; the frequency of the observer pulses was increased by 80 MHz. The observer sequence used a 32 ns π- pulse; the pump π- pulse was in a range of 12-18 ns. The experiment repetition time was 4.08 ms, and the number of scans was sufficient to obtain a suitable signal to noise with 50 shots at each time point, e.g. sample Vps75(C>A)-H4R45R1 was run for 904 scans with 159 time points. PELDOR data analysis was carried out as described in the supplementary methods.
Sulfhydryl reactive crosslinking
For probing the H3-H4 conformation when in complex with Asf1, Vps75 and Nap1, H3 K115C tetramer-chaperone complexes (cysteine null chaperones) were made up in 20 mM HEPES-KOH pH 7.5, 0.2 M sodium chloride, 1 mM EDTA at a concentration of 1 μM histone tetramer. 1,3-Propanediyl bismethanethiosulfonate (MTS-3-MTS) (Toronto Research Chemicals) was made up in dimethyl sulfoxide (DMSO) to a concentration of 25 μM. 48 μL of 1 μM complex was added to 2 μL of MTS-3-MTS to achieve a 1:1 ratio of crosslinker to histone tetramer, and the crosslinking reaction allowed to proceed for 3 minutes, at which point MTSL was added to a concentration of 200 μM to quench any unreacted cysteine residues. Quenching proceeded for one minute before samples were subjected to non-reducing SDS-PAGE.
To look at binding of Asf1 to the crosslinked tetramer, Asf1 was mixed at 20 μM with an equal stoichiometry of pre-crosslinked tetramer (10 μM) in a total volume of 30 μL. Binding was allowed to take place for 1 hour at 25 °C and complexes separated by filtration chromatography. Fractions were analysed by SDS-PAGE and bands quantified by the software AIDA (Raytest).
For crosslinking of endogenous histones, purified complexes were reduced with 20 mM DTT before microdialysis for 12-14 hours against E-buffer (20 mM HEPES-KOH pH 7.5, 150 mM NaCl, 10 % glycerol, 0.05 % Tween20). The reduced samples (normalized to total H3K115C-myc as probed by immuno-blotting) were diluted to 40 μL in E-buffer and subjected to crosslinking for two hours at 26°C by the addition of 0.064, 0.25 or 1.0 mM Cu(II):1,10-phenanthroline3 (Melford Laboratories Ltd) in 50 % dimethylsulfoxide. The reactions were quenched by the addition of 200 μM MTSL and 10 mM EDTA before separation by SDS-PAGE and visualization by immuno-blotting using and anti-myc antibody (Upstate).
Additional methods for crosslinking related to tetrasome assembly and acetyltrasferase assays are included in the supplementary methods.
Strain construction and TAP-tag purifications
A cysteine at H3K115 was introduced using PCR based quick-change method for crosslinking studies. Plasmid pNOY439 (CEN6 ARS4 TRP1 MYC-HHT2-HHF2) (Schermer et al., 2005) was used as a template to generate pNOY439-H3K115C. To generate the HHT2-Myc tag strain, WZY42 strain (Zhang et al., 1998) (Δ hht1-hhf1, Δ hht2-hhf2, and Ycp50 carrying HHT1-HHF1) was transformed with the plasmid pNOY439-H3K115C, driving the expression of Myc-Hht2K115C and untagged Hhf2 (H4) from their endogenous promoter. Subsequently, loss of Ycp50 was selected for on 5-FOA resulting in ectopic myc tagged H3K115C being the only source of histone H3. At this stage TAP tag was introduced at the C-terminus of the genomic copies of three histone chaperones (ASF1, NAP1 and VPS75) by transforming PCR products with the tag. The tagging methodology has been detailed elsewhere (Longtine et al., 1998). The primers used in the construction of the strains and quick-change to introduce K115C mutation in HHT2 (pNOY439) will be provided on request.
For TAP-tag purifications from strains containing myc-tagged H3K115C, cells were grown at 30 °C in 3xYPAD media until mid-log phase was reached. Cells were resuspended in E-buffer, frozen in liquid nitrogen and lysed in a planetary grinder. TAP-tagged complexes were isolated on IgG sepharose beads (GE Healthcare), washed extensively in E-buffer, and released by cleavage of the Protein A tag. TAP-tag purifications from strains carrying endogenous histones was essentially the same, except a second purification step using calmodulin affinity resin (Stratagene) was performed. Histone H2A and H4 were probed with antibodies ab13923 and ab10158 (Abcam), respectively.
Tetrasome assembly assay
A DNA fragment derived from the 601 sequence (Thastrom et al., 1999) comprising 45 bp each side of the dyad nucleotide was generated by PCR amplification resulting in a 91 bp fragment and purified by anion exchange chromatography. 20 μL reactions containing 1 μM crosslinked tetramer, 1 μM −28W-28, 10 mM dithiothreitol, 0.2 M NaCl, 20 mM HEPES-KOH pH 7.5 and 1 mM EDTA were made. To these reactions Nap1 was added to a final concentration of 0, 0.1, 0.25, 0.5 and 1 μM dimer. Assembly was allowed to proceed for 2 hours at 25 °C at which point glycerol was added to a final concentration of 5 %. Tetrasomes were resolved from free DNA on a 6.5 % polyacrylamide gel in 0.5x TBE. After staining with ethidium bromide the tetrasome bands were excised, soaked in 50 mM β-mercaptoethanol, 0.1 % sodium dodecyl sulphate for 10 minutes and cast within an SDS-PAGE gel. This step insured that the crosslinked histone tetramer was derived from only the BMOE crosslinked fraction, and not from H3 K115C that had disulphide bonded during native gel electrophoresis. Crosslinked tetramer was separated from non crosslinked tetramer and probed by western blotting with an anti-H3 antibody (ab1791, Abcam).
Chaperone mediated acetyltransferase activity assay
BMOE crosslinked tetramer on H3 K115C was analysed by SDS Page and coomassie staining. Acetyltransferase reactions were performed in 50 mM Tris HCl pH 7.5 and 100 mM NaCl using 1 mM acetyl CoA. The 10 μL reactions contained 0.5 μM BMOE treated tetramer and enzymes as stated and were incubated for 2 h at 37°C. The histone acetyltransferase activity of Rtt109 was analysed by immunblotting using an anti-acetyl-Histone H3 (Lys56) antibody (Upstate).
Supplementary Material
Highlights.
Site specific crosslinking of H3 shows Nap proteins bind a tetramer of H3 and H4.
Tetrameric conformation confirmed by long distance EPR measurements and in vivo crosslinking.
Nap1 can deposit a whole (H3-H4)2-tetramer onto DNA.
Vps75:Rtt109 preferentially acetylates H3 and H4 locked in their tetrameric conformation.
Acknowledgements
We would like to thank Karolin Luger, John Rouse and Paul Kaufman for protein expression constructs, Philip Korber and Sharon Dent for yeast strains. A. B. is support by a Wellcome Trust PhD Fellowship. T. O. H. and N.W. are supported by Wellcome Trust Senior Research Fellowship number 064414. R.W., D.G.N. and H. E-M were supported by Biotechnology and Biological Sciences Research Council (BB/E022286/1) and Engineering and Physical Sciences Research Council (EP/F0390341). Author contributions: A. B. expressed and purified recombinant proteins, carried out the gel filtration, crosslinking and nucleosome assembly experiments and prepared spin-labelled complexes. R. W., D. G. N. and H. E. carried out PELDOR experiments and associated data analysis. N. W. carried out the histone acetyl transferase assays. V. S. and N. W. created yeast strains and A. B & N. W. carried out TAP-tag purifications and crosslinking. The manuscript was prepared by A. B. and T. O. H. in consultation with all authors.
References
- Andrews AJ, Chen X, Zevin A, Stargell LA, Luger K. The Histone Chaperone Nap1 Promotes Nucleosome Assembly by Eliminating Nonnucleosomal Histone DNA Interactions. Molecular Cell. 2010;37:834–842. doi: 10.1016/j.molcel.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews AJ, Downing G, Brown K, Park YJ, Luger K. A thermodynamic model for Nap1-histone interactions. Journal of Biological Chemistry. 2008;283:32412–32418. doi: 10.1074/jbc.M805918200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziato AT. Split decision: What happens to nucleosomes during DNA replication? Journal of Biological Chemistry. 2005;280:12065–12068. doi: 10.1074/jbc.R400039200. [DOI] [PubMed] [Google Scholar]
- Baxevanis AD, Godfrey JE, Moudrianakis EN. Associative behavior of the histone (H3-H4)2 Tetramer: Dependence on ionic environment. Biochemistry. 1991;30:8817–8823. doi: 10.1021/bi00100a013. [DOI] [PubMed] [Google Scholar]
- Berndsen CE, Tsubota T, Lindner SE, Lee S, Holton JM, Kaufman PD, Keck JL, Denu JM. Molecular functions of the histone acetyltransferase chaperone complex Rtt109-Vps75. Nature Structural and Molecular Biology. 2008;15:948–956. doi: 10.1038/nsmb.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman A, Ward R, El-Mkami H, Owen-Hughes T, Norman DG. Probing the (H3-H4)2 histone tetramer structure using pulsed EPR spectroscopy combined with site-directed spin labelling. Nucleic Acids Research. 2010;38:695–707. doi: 10.1093/nar/gkp1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Loranger SS, Mizzen C, Ernst SG, Allis CD, Annunziato AT. Histones in transit: Cytosolic histone complexes and diacetylation of H4 during nucleosome assembly in human cells. Biochemistry. 1997;36:469–480. doi: 10.1021/bi962069i. [DOI] [PubMed] [Google Scholar]
- Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK. Acetylated Lysine 56 on Histone H3 Drives Chromatin Assembly after Repair and Signals for the Completion of Repair. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daganzo SM, Erzberger JP, Lam WM, Skordalakes E, Zhang R, Franco AA, Brill SJ, Adams PD, Berger JM, Kaufman PD. Structure and Function of the Conserved Core of Histone Deposition Protein Asf1. Current Biology. 2003;13:2148–2158. doi: 10.1016/j.cub.2003.11.027. [DOI] [PubMed] [Google Scholar]
- Das C, Tyler JK, Churchill MEA. The histone shuffle: histone chaperones in an energetic dance. Trends in Biochemical Sciences. 2010 doi: 10.1016/j.tibs.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S, Akey IV, Dingwall C, Hartman KL, Laue T, Nolte RT, Head JF, Akey CW. The crystal structure of nucleoplasmin-core: Implications for histone binding and nucleosome assembly. Molecular Cell. 2001;8:841–853. doi: 10.1016/s1097-2765(01)00354-9. [DOI] [PubMed] [Google Scholar]
- English CM, Adkins MW, Carson JJ, Churchill MEA, Tyler JK. Structural Basis for the Histone Chaperone Activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English CM, Maluf NK, Tripet B, Churchill MEA, Tyler JK. ASF1 binds to a heterodimer of histones H3 and H4: A two-step mechanism for the assembly of the H3-H4 heterotetramer on DNA. Biochemistry. 2005;44:13673–13682. doi: 10.1021/bi051333h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingham J, Recht J, Silva AC, Suter B, Emili A, Stagljar I, Krogan NJ, Allis CD, Keogh MC, Greenblatt JF. Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Molecular and Cellular Biology. 2008;28:4342–4353. doi: 10.1128/MCB.00182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RM, Browne K, Hodgson KC, Ausio J. sNASP, a histone H1-specific eukaryotic chaperone dimer that facilitates chromatin assembly. Biophys J. 2008;95:1314–1325. doi: 10.1529/biophysj.108.130021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill J, Yogavel M, Kumar A, Belrhali H, Jain SK, Rug M, Brown M, Maier AG, Sharma A. Crystal structure of malaria parasite nucleosome assembly protein: Distinct modes of protein localization and histone recognition. Journal of Biological Chemistry. 2009;284:10076–10087. doi: 10.1074/jbc.M808633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JC, Nyborg JK, Luger K, Stargell LA. Histone chaperones, histone acetylation, and the fluidity of the chromogenome. J Cell Physiol. 2010;224:289–299. doi: 10.1002/jcp.22150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nature Reviews Genetics. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- Jackson V. In vivo studies on the dynamics of histone-DNA interaction: evidence for nucleosome dissolution during replication and transcription and a low level of dissolution independent of both. Biochemistry. 1990;29:719–731. doi: 10.1021/bi00455a019. [DOI] [PubMed] [Google Scholar]
- Kaplan T, Liu CL, Erkmann JA, Holik J, Grunstein M, Kaufman PD, Friedman N, Rando OJ. Cell cycle- and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genetics. 2008;4 doi: 10.1371/journal.pgen.1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobashi K. Catalytic oxidation of sulfhydryl groups by o-phenanthroline copper complex. Biochim Biophys Acta. 1968;158:239–245. doi: 10.1016/0304-4165(68)90136-0. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Laskey RA, Honda BM, Mills AD, Finch JT. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978;275:416–420. doi: 10.1038/275416a0. [DOI] [PubMed] [Google Scholar]
- Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. Acetylation of Histone H3 Lysine 56 Regulates Replication-Coupled Nucleosome Assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lorch Y, Maier-Davis B, Kornberg RD. Chromatin remodeling by nucleosome disassembly in vitro. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3090–3093. doi: 10.1073/pnas.0511050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Flaus AJ, Waye MMY, Richmond TJ. Characterization of nucleosome core particles containing histone proteins made in bacteria. Journal of Molecular Biology. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- McBryant SJ, Park YJ, Abernathy SM, Laybourn PJ, Nyborg JK, Luger K. Preferential Binding of the Histone (H3-H4)2 Tetramer by NAP1 Is Mediated by the Amino-terminal Histone Tails. Journal of Biological Chemistry. 2003;278:44574–44583. doi: 10.1074/jbc.M305636200. [DOI] [PubMed] [Google Scholar]
- Mosammaparast N, Jackson KR, Guo Y, Brame CJ, Shabanowitz J, Hunt DF, Pemberton LF. Nuclear import of histone H2A and H2B is mediated by a network of karyopherins. Journal of Cell Biology. 2001;153:251–262. doi: 10.1083/jcb.153.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousson F, Lautrette A, Thuret JY, Agez M, Courbeyrette R, Amigues B, Becker E, Neumann JM, Guerois R, Mann C, Ochsenbein F. Structural basis for the interaction of Asf1 with histone H3 and its functional implications. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:5975–5980. doi: 10.1073/pnas.0500149102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume R, Eitoku M, Akai Y, Sano N, Horikoshi M, Senda T. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- Park YJ, Luger K. Structure and function of nucleosome assembly proteins. Biochemistry and Cell Biology. 2006a;84:549–558. doi: 10.1139/o06-088. [DOI] [PubMed] [Google Scholar]
- Park YJ, Luger K. The structure of nucleosome assembly protein 1. Proceedings of the National Academy of Sciences of the United States of America. 2006b;103:1248–1253. doi: 10.1073/pnas.0508002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, Luger K. Histone chaperones in nucleosome eviction and histone exchange. Current Opinion in Structural Biology. 2008;18:282–289. doi: 10.1016/j.sbi.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, McBryant SJ, Luger K. A beta-hairpin Comprising the Nuclear Localization Sequence Sustains the Self-associated States of Nucleosome Assembly Protein 1. Journal of Molecular Biology. 2008a;375:1076–1085. doi: 10.1016/j.jmb.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, Sudhoff KB, Andrews AJ, Stargell LA, Luger K. Histone chaperone specificity in Rtt109 activation. Nature Structural and Molecular Biology. 2008b;15:957–964. doi: 10.1038/nsmb.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior CP, Cantor CR, Johnson EM, Allfrey VG. Incorporation of exogenous pyrene-labeled histone into Physarum chromatin: a system for studying changes in nucleosomes assembled in vivo. Cell. 1980;20:597–608. doi: 10.1016/0092-8674(80)90306-2. [DOI] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Richardson RT, Batova IN, Widgren EE, Zheng LX, Whitfield M, Marzluff WF, O’Rand MG. Characterization of the histone H1-binding protein, NASP, as a cell cycle-regulated somatic protein. J Biol Chem. 2000;275:30378–30386. doi: 10.1074/jbc.M003781200. [DOI] [PubMed] [Google Scholar]
- Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- Schermer UJ, Korber P, Horz W. Histones are incorporated in trans during reassembly of the yeast PHO5 promoter. Mol Cell. 2005;19:279–285. doi: 10.1016/j.molcel.2005.05.028. [DOI] [PubMed] [Google Scholar]
- Selth L, Svejstrup JQ. Vps75, a new yeast member of the NAP histone chaperone. Journal of Biological Chemistry. 2007;282:12358–12362. doi: 10.1074/jbc.C700012200. [DOI] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 Complexes Mediate Nucleosome Assembly Pathways Dependent or Independent of DNA Synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- Tang Y, Meeth K, Jiang E, Luo C, Marmorstein R. Structure of Vps75 and implications for histone chaperone function. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12206–12211. doi: 10.1073/pnas.0802393105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thastrom A, Lowary PT, Widlund HR, Cao H, Kubista M, Widom J. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J Mol Biol. 1999;288:213–229. doi: 10.1006/jmbi.1999.2686. [DOI] [PubMed] [Google Scholar]
- Toth KF, Mazurkiewicz J, Rippe K. Association states of nucleosome assembly protein 1 and its complexes with histones. Journal of Biological Chemistry. 2005;280:15690–15699. doi: 10.1074/jbc.M413329200. [DOI] [PubMed] [Google Scholar]
- Tsubota T, Berndsen CE, Erkmann JA, Smith CL, Yang L, Freitas MA, Denu JM, Kaufman PD. Histone H3-K56 Acetylation Is Catalyzed by Histone Chaperone-Dependent Complexes. Molecular Cell. 2007;25:703–712. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- Wang H, Walsh ST, Parthun MR. Expanded binding specificity of the human histone chaperone NASP. Nucleic Acids Res. 2008;36:5763–5772. doi: 10.1093/nar/gkn574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Long C, Chen X, Huang C, Chen S, Zhu B. Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science. 2010;328:94–98. doi: 10.1126/science.1178994. [DOI] [PubMed] [Google Scholar]
- Zhang W, Bone JR, Edmondson DG, Turner BM, Roth SY. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 1998;17:3155–3167. doi: 10.1093/emboj/17.11.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.