SUMMARY
Azaphilones are a class of fungal metabolites characterized by a highly oxygenated pyrano-quinone bicyclic core and exhibits a broad range of bioactivities. While widespread among various fungi, their biosynthesis has not been thoroughly elucidated. By activation of a silent (aza) gene cluster in Aspergillus niger ATCC 1015, we have discovered six new azaphilone compounds, azanigerones A-F (1, 3-7). Transcriptional analysis and deletion of a key polyketide synthase (PKS) gene further confirmed the involvement of the aza gene cluster. The biosynthetic pathway was shown to involve the convergent actions of a highly-reducing and a non-reducing PKSs. Most significantly, in vitro reaction of a key flavin-dependent monooxygenase encoded in the cluster with an early benzaldehyde intermediate revealed its roles in hydroxylation and pyran-ring formation to afford the characteristic bicylic core shared by azaphilones.
INTRODUCTION
Filamentous fungi are known to be prolific producers of secondary metabolites, such as the penicillin, lovastatin and cyclosporine, and are an important resource for discovering small molecules of pharmaceutical and industrial value (Keller, et al., 2005). In the last decade, whole genome sequencing of various fungi has revealed that these microorganisms have immense biosynthetic potential that far surpasses the chemical diversity that we observe in laboratory culture (Sanchez, et al., 2012). For example, the genome of many aspergilli are found to encode for a combined 30 to 80 polyketide synthases (PKSs), nonribosomal peptide synthetases (NRPSs) and PKS-NRPS hybrids, which far exceeds the total number of known polyketides and nonribosomal peptides (Sanchez, et al., 2012). Of these, the fungal PKSs are of considerable interest due to their interesting enzymology and the polyketide structural diversity.
Fungal type I PKSs contain multiple catalytic domains and resemble the animal fatty acid synthases, where a single set of catalytic domains is used iteratively. The chain extension by decarboxylative condensation of malonyl-CoA units is catalyzed by the minimal PKS domains, including ketosynthase (KS), malonyl-CoA:ACP transacylase (AT) and acyl carrier protein (ACP) (Cox, 2007). Non-reducing PKSs (NR-PKSs) synthesize a poly-β-ketone backbone which is cyclized by a product template (PT) domain to yield aromatic compounds such as orsellinic acid and norsolorinic acid (Crawford, et al., 2009). In contrast, highly-reducing PKSs (HR-PKSs) utilize different combinations of ketoreductase (KR), dehydratase (DH), and enoyl reductase (ER) domains following each chain extension to reduce the β-keto positions in different extent, and produces reduced polyketides such as lovastatin and fumonisin (Cox, 2007). Together with tailoring enzymes that are typically clustered in a biosynthetic pathway at the genetic level, the different fungal PKSs produce a large array of polyketides (Keller, et al., 2005).
Bioinformatic analyses of different fungal genomes have revealed that it is common for two PKSs to be located in the same gene cluster (Sanchez, et al., 2012). The polyketide products of several of these dual PKS-containing gene clusters are known, including hypothemycin (Reeves, et al., 2008; Zhou, et al., 2010), asperfuranone (Chiang, et al., 2009) and lovastatin (Kennedy, et al., 1999; Ma, et al., 2009). The two PKSs can either work in sequence or in convergence to synthesize the polyketide product. When the two PKSs function sequentially, the polyketide chain formed by the first PKS is transferred to the second PKS to continue the chain extension process. This has been demonstrated in the biosynthesis of the resorcylic acid lactones and asperfuranone, in which the upstream HR-PKS produces a partially reduced polyketide chain that is transferred to the downstream NR-PKS to be further elongated (Chiang, et al., 2009; Zhou, et al., 2010). In the convergent model, the two PKSs can function independently in parallel, and the two polyketide products are ultimately connected via accessory enzymes. An example is the biosynthesis of the lovastatin, in which the nonaketide and a diketide chains produced by two different HR-PKSs are combined via the action of the acyltransferase LovD (Xie, et al., 2009). With a limited number of dual-PKS systems characterized so far, it is currently not possible to predict which mode of crosstalk (sequential or convergent) between the two PKSs will take place through bioinformatic means alone. Therefore, characterization of additional dual PKS-containing pathways will facilitate our understanding of the molecular and genetic basis that underlie the differences between the PKS-PKS partnerships, and enable better prediction of the gene cluster products.
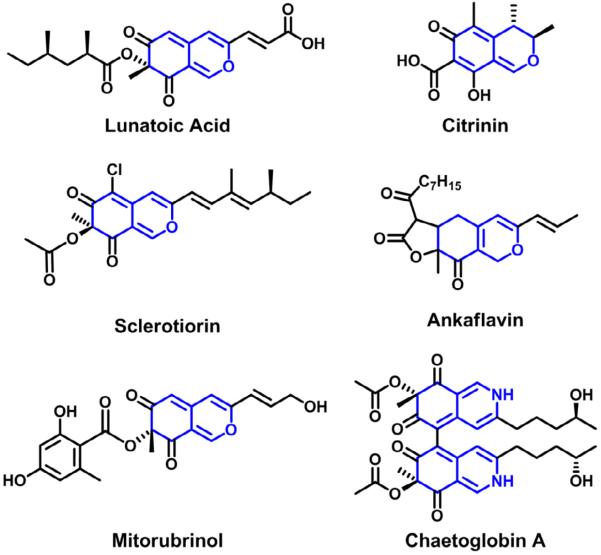
Aspergillus niger and closely related black aspergilli are known to produce a large number of secondary metabolites, with up to 145 compounds catalogued (Nielsen, et al., 2009). Annotation of the sequenced A. niger genomes unveiled an impressive number of PKS genes, including 34 PKSs and 7 PKS-NRPS hybrids (Andersen, et al., 2011; Pel, et al., 2007). In this work, we mined the genome of A. niger ATCC 1015 and identified a polyketide gene cluster that contains a pair of HR-PKS and NR-PKS. Overexpression of a pathway-specific transcriptional regulator found in the gene cluster led to the overproduction of a number of previously unknown azaphilone natural products. Azaphilones possess a signature bicyclic chromophore and is known to react readily with amines to produce the vinylogous γ-pyridones (Osmanova, et al., 2010). This family is structurally diverse and includes bioactive compounds such as the antifungal lunatoic acid (Nukina and Marumo, 1977), the anti-inflammatory agent monascorubrin (Yasukawa, et al., 1994), the lipoxygenase inhibitor sclerotiorin (Chidananda and Sattur, 2007), the food dye ankaflavin (Manchand, et al., 1973) as well as the nephrotoxic mycotoxin citrinin (Endo and Kuroda, 1976; Sakai, et al., 2008) (Figure 1). We show here that the two partnering PKSs function in a convergent manner toward azaphilone biosynthesis.
Figure 1.
Examples of azaphilone compounds isolated from fungi. The conserved bicyclic core (highlighted in gray) is the hallmark of these compounds.
RESULTS
Bioinformatic analysis of A. niger ATCC 1015 genome reveals a gene cluster with two PKS genes
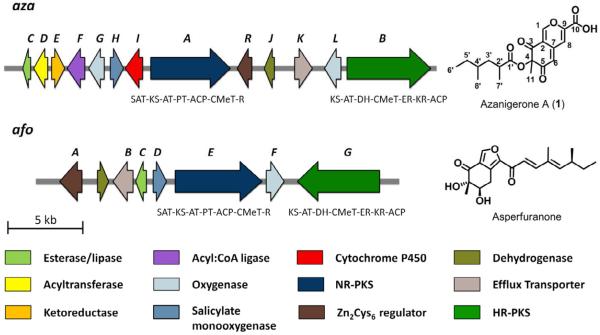
We scanned the sequenced genome of A. niger ATCC 1015 for dual PKS gene clusters and found a single gene cluster containing an HR- and an NR-PKS. The NR-PKS gene ASPNIDRAFT_56946 and the HR-PKS gene ASPNIDRAFT_188817 (herein referred to as azaA and azaB, respectively) appear to be homologs of afoE and afoG (44% and 43% protein identity) in the A. nidulans afo cluster, which is responsible for the synthesis of asperfuranone (Chiang, et al., 2009) (Figure 2 and Table 1). The azaA gene encodes for a Clade III NR-PKS according to a previous fungal PKS phylogenetic classification scheme (Kroken, et al., 2003). In addition to the minimal PKS domains, it also includes an N-terminus starter unit: acyl-CoA transacylase (SAT) and a C-terminus didomain of C-methyltransferase (CMeT) and reductase (R). On the other hand, AzaB is a typical HR-PKS consisting of the minimal PKS domains, the β-keto processing domains (DH, ER and KR) and CMeT domain. The domain organizations of AzaA and AzaB are parallel to that of AfoE and AfoG, respectively.
Figure 2.
Comparison of the A. niger aza cluster and the A. nidulans afo cluster and their respective products. PKS domain abbreviations: SAT, starter unit:ACP transacylase; KS, ketosynthase; AT, malonyl-CoA:ACP transacylase; PT, product template; ACP, acyl carrier protein; CMeT, C-methyltransferase; DH, dehydratase; KR, ketoreductase; ER, enoyl reductase; R, reductase. Structure of 1 is confirmed by NMR (see Table S1 and Figure S5) and MS/MS analysis (Figure S2).
Table 1.
Putative functions and homologs of the genes in the aza cluster.
| Locus Tag (ASPNIDRAFT_) |
Gene Name |
Putative function | Characterized Homolog |
Protein Identity / Similarity (%) |
|---|---|---|---|---|
| 54405 | n/a | hydrolase | - | - |
| 127101 | n/a | thioesterase | - | - |
| 50208 | azaC | esterase/lipase | AN1032.3, afoC | 44/56 |
| 189181 | azaD | acyltransferase | F. sporotrichioides Tri101 | 33/49 |
| 212676 | azaE | ketoreductase | S. cerevisiae GRE2 | 35/50 |
| 188806 | azaF | acyl:CoA ligase |
Physcomitrella patens 4CL2 |
35/53 |
| 189194 | azaG | FAD-dependent oxygenase |
AN1035.3, afoF | 37/64 |
| 188800 | azaH | salicylate monooxygenase |
AN1033.3, afoD | 41/58 |
| 43449 | azaI | cytochrome P450 | A. flavus CypX | 30/49 |
| 56946 | azaA | NR-PKS | AN1034.3, afoE | 44/60 |
| 132962 | azaR | Zn2Cys6 regulator | AN1029.3, afoA | 34/48 |
| 43447 | azaJ | dehydrogenase | AN1030.3 | 40/53 |
| 188912 | azaK | efflux transporter | - | - |
| 132654 | azaL | FAD-dependent oxygenase |
AN1035.3, afoF | 33/55 |
| 188817 | azaB | HR-PKS | AN1036.3, afoG | 43/60 |
| 188504 | n/a | cytochrome P450 | - | - |
| 50199 | n/a | oxidoreductase | - | - |
Note: Assignment of genes to aza cluster is based on transcriptional analysis on A. niger T1 (Figure 3A); n/a, not assigned (upstream and downstream genes).
Analysis of nearby genes in the dual-PKS cluster reveals the presence of ASPNIDRAFT_132962 (azaR) encoding for a Zn(II)2Cys6 zinc finger transcription factor, which could regulate the coordinated expression of the genes in a cluster (Keller, et al., 2005). Upstream and downstream of the PKSs, we find several tailoring enzymes that may modify the polyketide product. Interestingly, most of these genes have a corresponding homolog in the asperfuranone gene cluster, including azaC (afoC homolog), azaG (afoF homolog), azaH (afoD homolog), azaJ (AN1030.3 homolog), azaL (afoF homolog), each with > 30% identity to their afo counterparts (Table 1). However, four new genes are also found in the cluster including an acyltransferase (azaD), a ketoreductase (azaE), an AMP-dependent CoA ligase (azaF) and a P450 monooxygenase (azaI) (Table 1). The extensive overlap between the aza and afo gene clusters suggests that the aza cluster may encode for the production of an asperfuranone-like compound.
Overexpression of azaR activates the aza pathway
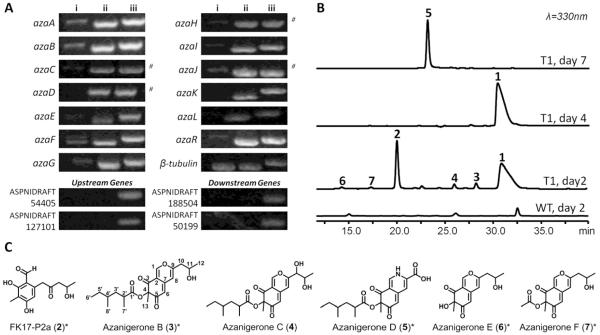
Among the polyketide metabolites catalogued for A. niger (Nielsen, et al., 2009), none appears to be a likely candidate of the aza cluster. Reverse transcription PCR (RT-PCR) analysis of azaA and azaB showed that both PKS genes were weakly transcribed under our laboratory culturing conditions (Figure 3A). Ectopic integration and overexpression of pathway-specific transcriptional activator has been used as a strategy to activate the expression of silent gene cluster for production of cryptic metabolites (Bergmann, et al., 2007). In order to interrogate the aza cluster, we constructed the overexpression plasmid pAZ44 by cloning azaR into pBARGPE1 (obtained from FGSC), which contains a PgpdA for gene expression in fungi and a bar gene that confers resistance to glufosinate. Upon PEG-mediated protoplast transformation and selection, several transformants emerged and produced a yellow pigmentation not observed in the wild type strain. The genomic DNA from one of the transformants (T1) was extracted and integration of the vector was confirmed via amplification of PgpdA::azaR present in the pAZ44 plasmid.
Figure 3.
Activation of the aza pathway via overexpression of azaR. (A) Transcriptional analysis of the aza genes in (i) A. niger WT and (ii) the activated T1 strain by RT-PCR. (ii) PCR from genomic DNA are shown for comparison. # indicates intron-less genes. (B) Time course of A. niger T1 metabolites. Metabolic profiles of the activated A. niger T1 strain on day 2, 4 and 7; and WT profile at day 2 are shown (C) Compounds observed in culture of A. niger T1 strain (see Figure S1). Structures indicated with * are confirmed by NMR (see Tables S1-3 and Figures S6-10). MS/MS analyses of 3 and 4 are in Figures S3-4.
In order to determine which genes are transcriptionally activated by the regulator, RT-PCR analysis was performed on the wild type (WT) and the A. niger T1 strains under identical conditions. We amplified a ~600 bp segment in each gene, particularly in those regions with introns in order to distinguish between the products from spliced cDNA and potential genomic DNA carryover during RNA extraction. In addition to azaA, azaB and azaR, 10 other genes were significantly upregulated (Figure 3A). The genes immediately upstream of azaC and downstream of azaB showed no increase in transcription levels compared to the WT (Figure 3A). We therefore hypothesize that the 13 transcribed genes comprise the aza cluster.
LC-MS analysis of the metabolic profile of the activated A. niger T1 reveals the presence of several new compounds not found in the WT strain (Figure 3B). The culture was scaled up to 2 L to obtain sufficient compound for structure elucidation. In addition, a time course of the metabolites produced by T1 was performed in order to identify possible biosynthetic intermediates of the aza pathway (Figure 3B).
From the second and fourth day cultures of A. niger T1, compound 1 (m/z 363 [M+H]+, λmax 330 nm) was the most abundant product (Figure 3B and S1A); it was purified as a yellow gum from the 4-day culture at an approximate yield of 60 mg/L. High-resolution mass of 1 is consistent with the molecular formula of C19H22O7 (observed m/z 363.1465 [M+H]+, calculated 363.1444). The pure compound was characterized by 1H and 13C NMR spectroscopy, which showed all of the 19 carbon and 22 proton signals (Table S1 and Figure S5). From the 2D HSQC-INEPT135 and HMBC correlations (Figure S5), it is apparent that there are two substructures in the molecule – a fully-saturated aliphatic chain and a conjugated cyclic moiety. The highly-reduced chain is consistent with a 2,4-dimethyl hexanoyl structure characterized by 3 methyls, 2 methylenes and 2 methines. We did not find any HMBC correlation beyond the carbonyl carbon with a chemical shift of 176.86 ppm (Figure S5D). Meanwhile, we assigned the rest of the signals to a bicyclic ring system consisting of three aromatic protons [δH 7.93 ppm (s), 7.31 ppm (s), 5.88 ppm (s)] with a carboxylic acid moiety, which corresponded to a low-field proton signal [δH 10.2 ppm (br s)]. We identified that the only available position on the bicyclic ring system for the linkage to the 2,4-dimethyl hexanoyl moiety is the oxygen atom bonded to the methylated carbon (δC 84.06 ppm). We therefore propose the structure of 1 as shown in Figure 2, with the two substructures connected by an ester bond at C1’ and C4. MS/MS analysis supports this linkage, which showed a fragment consistent with the mass of the bicyclic core (m/z 237 [M+H]+) (Figure S2). Thus, 1 was determined to be a new member of azaphilones, which bears the typical pyrano-quinone bicyclic core, and was named azanigerone A. The configurations of the chiral centers in 1 are yet to be determined. The structure of 1 resembles lunatoic acid, a previously characterized azaphilone from the plant pathogen Cochliobolus lunatus (Nukina and Marumo, 1977) (Figure 1). The aliphatic acyl chains of the two molecules are identical as evidenced by comparison to published values for lunatoic acid (Nukina and Marumo, 1977). The main structural difference is the side chain on C9, where a longer acrylic acid is in place in lunatoic acid instead of the C1 carboxylic acid in 1.
In addition to 1, several other compounds were observed in the culture at different time points. Compound 2 (m/z 251 [M+H]+, λmax 300 nm) is produced on day 2 but disappeared at day 4 and beyond (Figure 3B and S1B). This compound was purified and its structure was determined to be the tetra-substituted benzaldehyde FK17-P2a (Figures 3C and Table S1) that was previously isolated from Aspergillus versicolor and Pseudobotrytis terrestris (Arai T, 1994; Yamaguchi, et al., 2004). FK17-P2a should be a polyketide that originates from a methylated hexaketide that can be synthesized by a NR-PKS, which undergoes C2-C7 intramolecular cyclization and reductive release to yield a 1,3-diketo benzaldehyde (Bailey, et al., 2007). Further ketoreduction of the terminal ketone affords 2.
Compound 3 (m/z 377 [M+H]+, λmax 330 nm) was produced at a much lower yield during the earlier culture periods (Figure 3B and S1C). The bicyclic core structure was resolved to be similar to 1, but instead of the carboxylic acid moiety three new proton signals were detected: a terminal methyl doublet (δH 1.32 ppm), a methylene doublet (δH 2.54 ppm) and a methane multiplet (δH 4.16 ppm) (Table S2 and Figure S7). These signals are consistent with a 2-hydroxypropyl substitution at the C9, which is consistent with the m/z 251 [M+H]+ fragment from MS/MS analysis (Figure S3). The 2-hydroxypropyl moiety is reminiscent of the terminal portion of the 2-oxo-4-hydroxypentyl substituent in 2, which suggests that 2 may be an intermediate in the synthesis of 3 (Figure 3C). Meanwhile, 4 (m/z 393 [M+H]+, λmax 330 nm) was produced at an even lower titer and its structure was not fully elucidated by NMR due to insufficient material obtained (Figure 3B and S1D). Through comparison of MS/MS peaks of 3 and 4, as well as the molecular formula of 4 obtained through high resolution MS (C21H28O7, observed m/z 393.1908 [M+H]+,calculated 393.1913) (Figure S3 and S4), we propose that 4 could be a 10-hydroxy derivative of 3 (Figure 3C), and may also be an intermediate toward the biosynthesis of 1.
After seven days of culturing, a new compound 5 (m/z 362 [M+H]+, λmax 357 nm) replaces 1 as the dominant product in the A. niger T1 culture (Figure 3B and S1E). 5 was purified from a 7-day culture and its structure was confirmed via NMR to be the vinylogous γ-pyridone derivative of 1 (Figures 3C and S8, Table S2). This compound is likely originated from 1 via a characteristic azaphilone reaction with amines present in the culture medium. Indeed, a compound with the identical retention time, mass and λmax was obtained by reacting purified 1 with NH4OH (data not shown).
Knockout of azaB demonstrates a convergent synthesis of 1 by AzaA and AzaB
Inspection of the structure of 1 suggests that the molecule is assembled convergently from two discrete polyketide chains. We propose that the dimethylhexanoate moiety is produced by the HR-PKS AzaB, while the bicyclic core is cyclized from an unreduced polyketide backbone synthesized by the NR-PKS AzaA. This is in stark contrast to the sequential mode of collaboration between AfoE and AfoG in the highly parallel asperfuranone gene cluster (Chiang, et al., 2009), and underscores the difficulties in accurately predicting product structures among fungal PKSs.
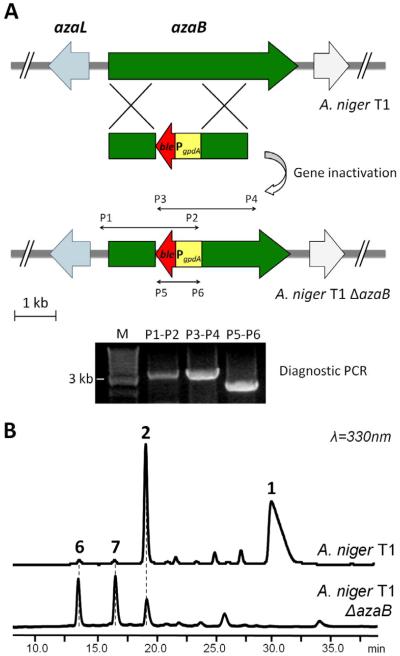
Therefore in the convergent synthesis model, disruption of azaB in T1 should not affect the function of AzaA, and should lead to accumulation of the pyrano-quinone. To test this model, genetic disruption of azaB in A. niger T1 was performed using a double-crossover recombination with the zeocin-resistant gene ble (Figure 4A). Two desired azaB transformants were identified via diagnostic PCR (Figure 4A). After culturing and metabolite analysis, the production of 1 was confirmed to be abolished in the azaB strains. Instead, two new compounds (6 and 7) that have UV spectra characteristic of pyrano-quinone (identical to that of 1) emerged (Figure 4B). Compound 6 and 7 have molecular masses of 250 and 292, respectively (Figure S1F and S1G). Both compounds were purified and their structures were elucidated by 1D and 2D NMR (Table S3 and Figure S9-10). 6 contains the same bicyclic core and the 2-hydroxypropyl side chain as 3, but lacks the 2,4-dimethylhexanoyl ester as a result of azaB disruption (Figure 3C). Compound 7 is the O4 acetylated version of 6 (Figure 3C). In addition, the benzaldehyde 2 observed in the A. niger T1 strain was present in the early cultures of the azaB strains (Figure 4B). Upon closer examination, trace amounts of 6 and 7 were also found in A. niger T1 culture on Day 2 (Figure 3B); the non-acetylated 6 is likely an intermediate in the biosynthesis of 1.
Figure 4.
Deletion of azaB confirms the involvement of the cluster in biosynthesis of 1. (A) Knockout strategy employed for azaB. Correct integration is verified by using 3 sets of primers shown. (B) HPLC traces demonstrating the abolishment of 1 production in the azaB strain and the accumulation of 6 and 7 (See Table 3 and Figures S9-10).
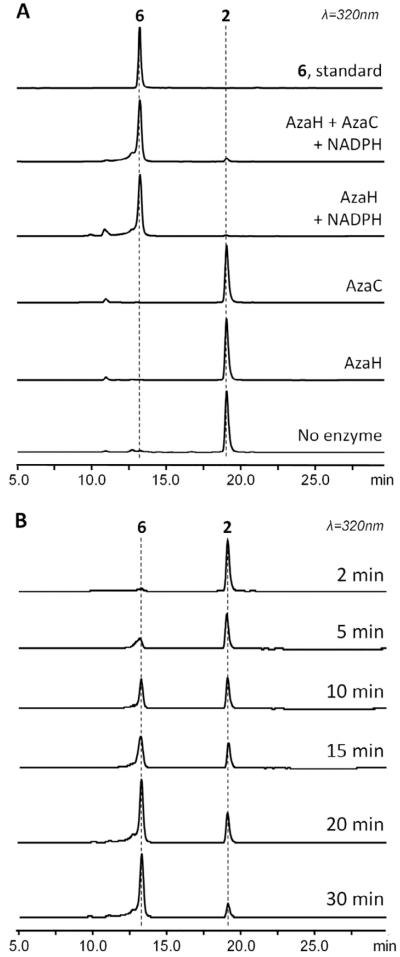
In vitro reaction of AzaH with the benzaldehyde intermediate confirms its roles in hydroxylation and pyran-ring formation
The formation of the bicyclic core of azaphilones has not been elucidated to date. It has been suggested that it proceeds from a benzaldehyde precursor in asperfuranone biosynthesis (Chiang, et al., 2009), which is structurally similar to 2 isolated from A. niger T1 in early culturing times. Since 2 is isolated in stable form, the route to the formation of the pyran ring in the biosynthesis of 1 must be enzyme-catalyzed.
We initially proposed that AzaC is involved in the pyran-ring cyclization. NCBI Conserved Domain Search results indicate that both AzaC and its homologs (CtnB from the citrinin pathway, and AfoD) belong to the esterase-lipase family of serine hydrolases and are not oxidoreductases as assigned previously for CtnB and AfoD (Chiang, et al., 2009; Sakai, et al., 2008). Since 1, citrinin and asperfuranone all contain a fused heterocyclic ring system, AzaC, CtnB and AfoB may be responsible for their corresponding heterocycle formation; a similar hypothesis has been proposed in a recent study as well (Davison, et al., 2012). Accordingly, the recombinant AzaC protein (30 kDa) was purified and assayed with 2. However, no conversion of the substrate 2 was observed in the in vitro reaction (Figure 5A). We therefore reasoned that AzaC might act on a different substrate, possibly the C4-hydroxylated derivative of 2.
Figure 5.
In vitro assays demonstrate AzaH-mediated pyran-cyclization. (A) HPLC analysis of the in vitro assays of AzaH and/or AzaC (10 μM) with 2 (50 μM), and comparison with the standard 6. Conversion of 2 to 6 was observed in the presence of AzaH and NADPH (0.5 mM). (B) Time course of AzaH-catalyzed formation of 6 from 2. 0.5 M AzaH was assayed with 50 M substrate 2 and 0.5mM NADPH. Reactions were quenched at specified time with 100 L of acetonitrile.
We then searched for genes in the aza cluster that may hydroxylate the C4 position in 2. AzaH, an FAD-dependent monooxygenase, was found to be homologous to a recently characterized monooxygenase TropB (43% identity, 61% similarity), which was recently shown to hydroxylate the corresponding carbon of 3-methylorcinaldehyde during tropolone biosynthesis (Davison, et al., 2012). To examine its role in the synthesis of 1, AzaH was expressed and purified from E. coli BL21(DE3) and a series of in vitro assays including AzaH and AzaC/AzaH in the presence of 2 was performed. Surprisingly, incubation of both purified AzaH or AzaC/AzaH with 2 and NADPH yielded a new product that has an identical UV spectrum, retention time and mass as 6 (Figure 5A). This indicates that AzaH alone is capable of converting 2 to 6. However, in the absence of NADPH, which is needed for the regeneration of the reduced flavin cofactor, no substrate conversion by AzaH was observed. Therefore, the flavin-dependent hydroxylation of C-4 is likely a prerequisite for the subsequent pyran formation. Further LC/MS time-course analysis of the reaction of AzaH with 2 showed that the conversion proceeds rapidly with no detectable uncyclized C4-hydroxylated intermediate (Figure 5B). These results confirmed the critical role of AzaH in morphing the benzaldehyde intermediate into the bicyclic, pyran-containing azaphilone chromophore.
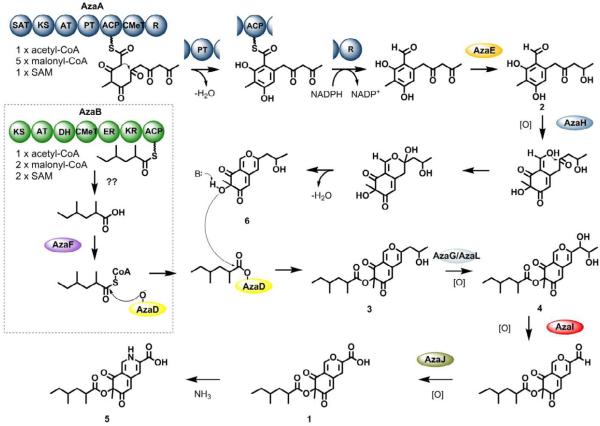
Proposed pathway for biosynthesis of the azanigerones
The RT-PCR transcriptional analysis indicates that there are a total of 12 genes in the aza cluster, which were activated upon overexpression of the pathway-specific regulator azaR (Figure 3A). Combining the structural characterization of the compounds from the activated A. niger T1 culture and T1 ΔazaB, as well as in vitro confirmation of the role of AzaH, a biosynthetic pathway leading to the production of 1 can be proposed (Figure 6). Based on the time course studies of the metabolic profile of A. niger T1 (Figure 3B), 1 is most likely to be the end product of the pathway, as it is the dominant compound in the culture after 4 days. We propose that the biosynthesis of 1 begins with the polyketide assembly by AzaA, which is a typical Clade III NR-PKS with a domain organization of SAT-KS-AT-PT-ACP-CMeT-R from the N- to C-terminal. AzaA forms the hexaketide precursor from successive condensations of five malonyl-CoA units presumably with a simple acetyl-CoA starter unit. The reactive polyketide chain then undergoes a PT-mediated C2-C7 cyclization to afford the aromatic ring and is eventually released as an aldehyde through the R-domain, in a manner similar to the previously characterized 3-methylorcinaldehyde synthase in Acremonium strictum (Bailey, et al., 2007). The putative ketoreductase, AzaE, a homolog of the methylglyoxal reductase in Saccharomyces cerevisiae (35% identity), is proposed to catalyze the reduction of the terminal ketone resulting in the early culture product 2.
Figure 6.
Proposed biosynthetic pathway for production of azanigerones from the aza cluster.
The monooxygenase AzaH was demonstrated to be the only enzyme required to convert 2 to 6 in vitro. We propose that AzaH first hydroxylates the benzaldehyde intermediate 2 at C4, which triggers the formation of the pyran-ring to afford 6 (Figure 6). In parallel, the 2,4-dimethylhexanoyl chain is synthesized by the HR-PKS AzaB, and is proposed to be transferred to the C4-hydroxyl of 6 by the acyltransferase AzaD directly from the ACP domain of AzaB via a similar mechanism to the LovD acyltransferase (Xie, et al., 2009). Alternatively, the 2,4-dimethylhexanoyl chain may be offloaded from the HR-PKS as a carboxylic acid and converted to an acyl-CoA by AzaF, which shares 35% identity to the 4-coumarate:CoA ligase (Silber, et al., 2008). The resulting acyl-CoA molecule could then be taken up as a substrate by AzaD to form 3 (Figure 6). Like the homolog acetyltransferase Tri101 in trichothecene biosynthesis (33% identity to AzaD) (Garvey, et al., 2008), AzaD belongs to the BAHD family of acyltransferases that catalyze transfer of various acyl-CoA substrates (D’Auria, 2006). It is interesting to note that the C4-O-acetylated 7 is also observed in the activated A. niger T1 and azaB cultures (Figure 4B). This suggests that AzaD may have a relaxed specificity toward acyl-CoA substrates, although it is entirely possible that other endogenous acyltransferases might be involved in this reaction.
In order to yield the carboxylic acid substituent in 1, the hydroxypropyl side chain of 3 would need to undergo a C-C oxidative cleavage. We propose that this C-C bond cleavage is catalyzed by a cytochrome P450 AzaI encoded in the cluster. Similar reactions have been observed in other P450s, with the CYP11A involved in the decomposition of cholesterol into pregnenolone and 4-methylpentanal as the archetypal enzyme (Burstein and Gut, 1971). Such P450 is proposed to act on a vicinal diol that leads to a C-C bond scission either through an alkoxyradical intermediate or a peroxy complex (Ortiz de Montellano, 2005). In the biosynthesis of 1, we propose that 3 first undergoes hydroxylation at C10, possibly catalyzed by one of the two FAD-dependent monooxygenases encoded in the cluster, AzaG or AzaL, resulting in the vicinal diol (Figure 6). A small amount of this putative intermediate 4, with mass and fragmentation pattern consistent with such a diol, was isolated from the 2-day culture of A. niger T1 (Figure 3B and S4). Oxidative cleavage 4 by AzaI would yield the corresponding aldehyde derivative of 1. A homolog of AzaI, CypX, also exists in the aflatoxin biosynthesis in A. parasiticus (35% identity) and is found to convert averufin to hydroxyversicolorone via a still unknown oxidative cleavage and ring rearrangement step (Wen, et al., 2005). Finally, the dehydrogenase AzaJ (homologous to the AN1030.3, 40% identity) is proposed to convert the aldehyde functional group into the carboxylic acid, completing the conversion from 3 to 1. Alternatively, the oxidation of aldehyde to carboxylic acid may be catalyzed by the same P450 enzyme AzaI via consecutive oxidation or by endogenous alcohol dehydrogenase. Interestingly, the proposed oxidative chain-shortening does not occur in the absence of the 2,4-dimethylhexanoate ester. Both 5 and 6 contain the hydroxypropyl chain and are not converted to the corresponding acids in azaB culture.
DISCUSSION
Genome mining, in particular activation of silent secondary metabolic pathways by overexpression of pathway-specific regulator, is becoming a useful strategy for natural product discovery in fungi (Brakhage, et al., 2008; Winter and Tang, 2012). From overexpression of the cluster-specific regulator, we have uncovered novel compounds that belong to the family of azaphilones. The hallmark feature of azaphilones is the presence of a highly oxygenated pyrano-quinone bicyclic core of polyketide origin (Osmanova, et al., 2010). The name derives from their tendency to react with primary amines to yield γ-pyridones that result from the exchange of the pyrane oxygen with nitrogen. Over 170 different azaphilone compounds have been identified from hundreds of fungal species across 23 genera from 13 families, and many of the azaphilones exhibit diverse biological activities, including antibacterial, antifungal and antitumor (Osmanova, et al., 2010). Several common modifications that contribute to the structural diversification of the common bicyclic chromophore include but are not limited to a) aliphatic side chains at C9; b) aliphatic esters at O4; c) halogen substitution on the bicyclic core, as in sclerotiorin (Chidananda and Sattur, 2007); d) additional fused rings, as in ankaflavin (Manchand, et al., 1973); e) aromatic esters at O4, as in mitorubrinol (Quang, et al., 2005); and f) molecular dimers, as in chaetoglobin A (Ge, et al., 2008) (Figure 1).
Despite their apparent ubiquity among fungi, the genetic and molecular basis for the biosynthesis of these azaphilones has not been thoroughly explored or understood. Besides labeling studies suggesting a polyketide origin for the bicyclic core (Ogihara, et al., 2000), only the azaphilone-like compound asperfuranone and citrinin have been linked to their corresponding biosynthetic gene clusters (Chiang, et al., 2009; Sakai, et al., 2008). In this study, we have discovered a silent gene cluster in A. niger responsible for producing a series of azaphilones. Characterization of this aza gene cluster has provided important insights on the biosynthesis of this family of compounds.
In total, six new azaphilones, azanigerone A-F, were isolated and all share a substituted bicyclic core indicative of a common biosynthetic origin. We also observed their signature reaction with amine groups in the conversion of the major product 1 into the vinylogous γ-pyridone 5. All six compounds contain a C4 hydroxyl group, which is a post-PKS tailoring modification observed in a multitude of other azaphilones. With the exception of 6, the tertiary alcohol at C4 of azanigerones is acylated with an aliphatic acyl or an acetyl group, which is another common feature among many azaphilones. The isolation of pyran intermediates at early stages of the A. niger T1 culture underscores the convergent actions of the two PKSs in the production of these molecules (Figure 3B).
A key discovery in this study is the enzyme responsible for the formation of the characteristic pyrano-quinone core common to all azaphiliones. It has been proposed that the biosynthesis of azaphilones proceeds from a benzaldehyde intermediate (Chiang, et al., 2009), but the enzymology of the pyran-ring formation step has not been elucidated. We have discovered that the cyclization of the benzaldehyde is mediated by the C4-hydroxylation catalyzed by the FAD-dependent monooxygenase AzaH. We propose that the hydroxylation at C4 by AzaH dearomatizes the ring and causes keto-enol tautomerization at the C1 aldehyde leading to the condensation between the C1 enol and the C9 carbonyl to yield the pyran ring (Figure 6). Our finding that the C4-hydroxylation and pyran-ring cyclization occur very rapidly suggests that these two features are coupled. This is supported by the fact that with a few exceptions, majority of the azaphilones contain such geminal methyl, hydroxyl (or ester) substitution at the pyrano-quinone core (Figure 1) (Osmanova, et al., 2010). For the few azaphilones that do not have this feature, such as in the case of citrinin, a different mechanism of cyclization might take place. Coincidentally, the citrinin gene cluster lacks the azaH homolog (Sakai, et al., 2008).
It is curious to note that with the exception of a few genes, the aza and the afo clusters are highly similar, with multiple gene homologs sharing >30% protein sequence identity (Figure 2 and Table 1). However, the structures of the two compounds are considerably different. A striking disparity between the two biosynthetic pathways is on how the dual PKS systems partner to form their respective products. In the case of the afo cluster, the HR-PKS AfoG synthesizes the tetraketide intermediate, which is passed on to the SAT domain of the NR-PKS AfoE. Such sequential mode of collaboration occurs in other HR/NR-PKS tandem systems, such as in the biosynthesis of resorcylic acid lactones (Zhou et al. 2010). We were expecting the two PKSs in aza cluster, AzaA and AzaB, to follow the same tandem mode of PKS-PKS collaboration. To our surprise, the two PKSs acted independently of each other in the convergent synthesis of the two substructures of 1. Whereas the afoG culture completely disrupted the production of asperfuranone or any appreciable intermediate, compounds 6 and 7 lacking the reduced triketide chain were isolated in the azaB culture. Previous studies have pointed to the N-terminus SAT as a specificity-conferring domain that selects the starter unit (Crawford, et al., 2008). By multiple protein sequence alignment, it was noticed that the SAT domains of AzaB and AfoG have a lower identity (35% vs. 43%) compared to the rest of the protein, although both contain the GXCXG motif found in most SATs. It is likely that the selectivity of the SAT and its ability to interact with HR-PKS hold the key for which mode of collaboration will take place between a pair of HR- and NR-PKSs. The azaA-azaB dual PKS system in A. niger is the first example of a convergent mode of collaboration between an HR and an NR-PKS; elucidation of the basis for the intriguing difference between aza and afo pathways could facilitate better prediction of polyketide products from such pathways.
Another notable difference between the structures of azanigerones and asperfuranone is the formation of a furan in the latter pathway instead of a pyran ring that is common in azaphilones. Both pathways are proposed to go through a benzaldehyde intermediate. However, in the case of asperfuranone, it is likely that the benzylic carbon hydroxylation precedes the dearomatizing hydroxylation. It was previously proposed that AfoD catalyzes this benzylic carbon hydroxylation, which is the key branching point in the biosynthesis of asperfuranone and the azaphilone sclerotiorin (Davison, et al., 2012; Somoza, et al., 2012). However, it is more likely that AfoD has a similar function as the homolog AzaH characterized in this study, i.e. the formation of the tertiary alcohol. Thus, AfoF, another FAD-dependent monooxygenase, may catalyze this benzylic carbon hydroxylation step in the asperfuranone pathway. Two AfoF homologs, AzaG and AzaL, are also found in the A. niger aza cluster (Table 1). We proposed that one of them may act later in the pathway in the hydroxylation of C10 to form the pre-cleavage vicinal diol.
Among the aza genes that share a homolog to the afo cluster, the function of AzaC remains enigmatic. The possibility of its involvement in the heterocycle formation has been excluded by the results in this study. In line with the NCBI Conserved Domain Search results that suggest an esterase-lipase function for this enzyme, we propose that AzaC might participate in the offloading of the 2,4-dimethylhexanoyl chain from the ACP of AzaB (Figure 6). The role of the homologs AfoC and CtnB in the biosynthesis of asperfuranone and citrinin respectively is also unclear and should be the subject of further investigation.
SIGNIFICANCE
Activation of the silent aza cluster has led to the discovery of new compounds named azanigerones, which have not been isolated from A. niger before. Characterization of the pathway has shed light to the biosynthesis of the azaphilone group of compounds, which are structurally diverse and exhibit a wide range of bioactivities. The aza pathway represents the first example of a convergent mode of collaboration between an HR- and an NR-PKS, as opposed to the sequential mode of collaboration commonly observed in other dual PKS systems. More importantly, the enzymatic basis for the formation of the pyrano-quinone core common to azaphilones has been revealed for the first time, and the mechanism can likely be generalized to biosynthesis of other azaphilones. Thus, this study provides an important basis for further molecular genetics and biochemical investigations of azaphilone pathways. The sequence information will aid in identification of the gene clusters encode for known bioactive azaphilones and discovery of new azaphilones by genome mining. The many structural diversifications on a common bicyclic core observed among this family of fungal natural products also offer a unique opportunity for combinatorial biosynthesis to expand the library of azaphilones and screen for enhanced bioactivities.
EXPERIMENTAL PROCEDURES
Strain and culture conditions
A. niger ATCC 1015 was obtained from Agricultural Research Service (ARS) culture collection (as NRRL 328) and cultured at 28 °C in potato dextrose agar medium. The activated A. niger T1 strain and the azaB mutant strains were maintained in glucose minimal medium (GMM) containing 10 mM ammonium tartrate as nitrogen source (Chooi, et al., 2010; Li, et al., 2011) or yeast glucose medium (YG) (Szewczyk, et al., 2006). E. coli strains XL1 (Stratagene) and TOPO10 (Invitrogen) were used for routine cloning. Saccharomyces cerevisiae BJ5464 was used for in vivo yeast DNA recombination cloning. E. coli BL21(DE3) was used for protein expression.
Molecular genetic manipulations
Polymerase chain reactions were performed using Phusion high-fidelity DNA polymerase (New England Biolabs), Platinum Pfx DNA polymerase (Invitrogen) and GoTaq Green Master Mix (Promega). PCR products were cloned into a PCR-Blunt vector (Invitrogen) for DNA sequencing and subcloning. Restriction enzymes (New England Biolabs) and T4 ligase (Invitrogen) were used respectively for the digestion and ligation of DNA fragments. Primers used for amplification were synthesized by Integrated DNA Technologies and are listed in Table S4.
Genomic DNA of A. niger ATCC 1015 was extracted using the ZYMO ZR fungal/bacterial DNA kit according to manufacturer’s protocols. The azaR gene (including introns) was amplified from the genomic DNA and inserted into the fungal shuttle vector pBARGPE1 (digested with BglII and EcoRI) to yield pAZ44. The pBARGPE1 vector was obtained from Fungal Genetic Stock Center (FGSC) (Pall and Brunelli, 1993). The intronless azaH and azaC were also amplified from genomic DNA and inserted into the p8HIS expression vector (Jez, et al., 2000) digested with EcoRI and NotI to yield pAZ81 and pAZ83, respectively.
The knockout cassette for azaB was assembled in yeast using the yeast recombination method (Colot, et al., 2006). The two homologous regions were amplified from A. niger genomic DNA using primers containing overlapping regions with the yeast vector and the zeocin-resistance cassette. The zeocin-resistance cassette containing the ble gene under the gpdA promoter was amplified from pAN8-1 (Punt, et al., 1987) using gpdAfor and blerev. The three DNA fragments were co-transformed with the linearized vector backbone derived from YEplac195 (Gietz and Sugino, 1988) into the S. cerevisiae BJ5464 (Jones, 1991) using S. c. EasyComp™ Transformation Kit (Invitrogen) and selected on uracil-dropout semisynthetic media. The resulting transformants were screened by colony-direct PCR and the plasmid in the correct transformant was rescued using the Zymoprep Yeast Plasmid Miniprep Kit (Zymo Research) and transformed into E. coli for propagation and sequencing verification. The resulting plasmid was designated AZ61. This plasmid was used as template to obtain the linear azaB knockout cassette for transformation of A. niger T1.
Fungal transformation
The preparation of A. niger protoplasts and the PEG-mediated transformations were carried out as previously described (Li, et al., 2011). The A. niger transformed with pAZ44 for azaR overexpression was selected on approximately 8 mg/mL of glufosinate prepared as described previously (Chooi, et al., 2010). For genetic manipulation in the activated A. niger strain T1, a modified protocol was used derived from the one developed for Aspergillus nidulans (Szewczyk, et al., 2006). The following modifications were implemented: a) the 2x protoplasting solution was prepared with 3g of VinoTaste Pro; b) the PEG solution used comprised of 25% PEG 4000, 10mM CaCl2 and 10mM Tris-HCl; c) the transforming protoplasts were first plated on non-selective media for five hours before overlaying with soft agar (8g/L) with 200 μg/mL of Zeocin; and d) the protoplasts were stabilized with 1.2M sorbitol.
Expression analysis by reverse transcription polymerase chain reaction (RT-PCR)
The total RNA of A. niger mutant and wild-type strains were extracted using the Ambion RNA extraction kit. The first strand cDNA was synthesized using the Oligo-dT primer and Improm-II reverse transcription system (Promega) according to the manufacturer’s instructions. cDNA was then amplified with GoTaq Green Master Mix using gene-specific primers (Table S4).
Expression and purification of AzaC and AzaH
pAZ81 and pAZ83 were used to transform the BL21(DE3) cells for expression of AzaC and AzaH, respectively. The cells were cultured at 37 °C and 250 rpm in 500 mL of LB medium supplemented with 35 g/mL kanamycin to a final OD600 of 0.4-0.6 followed by addition of 0.1 M isopropylthio-β-D-galactoside (IPTG) to induce protein expression. The proteins were purified by Ni2+ affinity chromatography as described previously (Li, et al., 2011). Purified AzaC and AzaH was concentrated and exchanged into PBS (50mM, pH 7.4) with the centriprep filters (Amicon). Protein concentration was determined with the Bradford assay using bovine serum albumin as a standard.
In vitro assays with AzaC and AzaH
The reactions were performed at 50 L scale containing 100 mM phosphate buffer (pH 7.4) in the presence of 50 μM of 2, 0.5 mM NADPH and 10 μM of AzaH and/or AzaC. After 2 hours, the reactions were quenched by precipitating the enzyme/s with 100 μL of acetonitrile. The acetonitrile/aqueous solution was concentrated 5-fold and directly analyzed on the LC-MS (Figure 5A). The time course analysis was performed with the same assay condition except that 0.5 M AzaH was used and the reactions were quenched at different time points (Figure 5B).
LC/MS analysis
For small-scale analysis, the A. niger wild-type and transformants were grown in 10 mL GMMT under shaking conditions at 28 °C. The cult ures were extracted with equal volume of ethyl acetate (EA) with 0.1% trifluoroacetic acid (TFA) and evaporated to dryness. The dried extracts were dissolved in methanol for LC/MS analysis using a Shimadzu 2010 EV Liquid Chromatography Mass Spectrometer with positive and negative electrospray ionization and Phenomenex Luna 5μL 2.0×10mm C18 reverse phase column. The samples were resolved on a linear gradient from 5 to 95% with CH3CN/H2O + 0.05% formic acid solvent system.
Isolation of compounds
Most compounds were purified from shake flask cultures, except for 2, which was obtained from 2-day static culturing. 1, 3 and 4 were extracted with EA after four days of culturing, 5 after seven days, and the azaB knockout compounds, 6 and 7, after two days. The following scheme was generally used to purify each compound from the crude extract: a) chloroform-water partitioning; b) hexane - 9:1 methanol:water partitioning; c) Sephadex LH-20 (GE) chromatography; and d) preparative high pressure liquid chromatography with Phenomenex Luna 5μL 250×1000mm C18 reverse phase column using CH3CN/H2O + 0.1% TFA or +0.2% formic acid solvent system. DRX500 or ARX500 instruments were used to perform the NMR spectroscopy of the different compounds.
Supplementary Material
HIGHLIGHTS.
Discovery of an azaphilone pathway in Aspergillus niger
Six new azaphilone compounds were isolated and characterized
The biosynthesis involves convergent actions of an HR-PKS and an NR-PKS
Hydroxylation by a monooxygenase promotes pyran-ring formation
ACKNOWLEDGEMENT
We would like to thank Prof. Neil Garg and Dr. Jaclyn Winter for their useful suggestions. This work is supported by NIH Grants 1R01GM085128 to Y.T. and the NIH Biotechnology Training Grant 5T32GM067555 to A.Z. The NMR instrument in The Magnetic Resonance Facility at UCLA used in this study is based upon work supported by the NSF under equipment Grant no. CHE-1048804. The MS instrument used in the UCLA Mass Spectrometry Laboratory was supported by the National Center for Research Resources Grant no. S10RR025631.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Andersen MR, Salazar MP, Schaap PJ, van de Vondervoort PJ, Culley D, Thykaer J, Frisvad JC, Nielsen KF, Albang R, Albermann K, et al. Comparative genomics of citric-acid-producing Aspergillus niger ATCC 1015 versus enzyme-producing CBS 513.88. Genome Res. 2011;21:885–897. doi: 10.1101/gr.112169.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai T, S H. Novel UV-absorbing compounds FK17-P2a, FK17-P2b1, FK17-P2b2, and FK17-P3 and manufacture of the compounds with Aspergillus sp. (in Japanese) Jpn. Kokai Tokkyo Koho JP. 1994:93–121677. [Google Scholar]
- Bailey AM, Cox RJ, Harley K, Lazarus CM, Simpson TJ, Skellam E. Characterisation of 3-methylorcinaldehyde synthase (MOS) in Acremonium strictum: first observation of a reductive release mechanism during polyketide biosynthesis. Chem. Commun. (Camb) 2007:4053–4055. doi: 10.1039/b708614h. [DOI] [PubMed] [Google Scholar]
- Bergmann S, Schumann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat. Chem. Biol. 2007;3:213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- Brakhage AA, Schuemann J, Bergmann S, Scherlach K, Schroeckh V, Hertweck C. Activation of fungal silent gene clusters: a new avenue to drug discovery. Prog. Drug. Res. 2008;66(1):3–12. doi: 10.1007/978-3-7643-8595-8_1. [DOI] [PubMed] [Google Scholar]
- Burstein S, Gut M. Biosynthesis of pregnenolone. Recent Prog. Horm. Res. 1971;27:303–349. doi: 10.1016/b978-0-12-571127-2.50032-8. [DOI] [PubMed] [Google Scholar]
- Chiang YM, Szewczyk E, Davidson AD, Keller N, Oakley BR, Wang CC. A gene cluster containing two fungal polyketide synthases encodes the biosynthetic pathway for a polyketide, asperfuranone, in Aspergillus nidulans. J. Am. Chem. Soc. 2009;131:2965–2970. doi: 10.1021/ja8088185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidananda C, Sattur AP. Sclerotiorin, a novel inhibitor of lipoxygenase from Penicillium frequentans. J. Agric. Food. Chem. 2007;55:2879–2883. doi: 10.1021/jf062032x. [DOI] [PubMed] [Google Scholar]
- Chooi YH, Cacho R, Tang Y. Identification of the viridicatumtoxin and griseofulvin gene clusters from Penicillium aethiopicum. Chem Biol. 2010;17:483–494. doi: 10.1016/j.chembiol.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chooi YH, Cacho R, Tang Y. Identification of the viridicatumtoxin and griseofulvin gene clusters from Penicillium aethiopicum. Chem. Biol. 2010;17:483–494. doi: 10.1016/j.chembiol.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA. 2006;103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RJ. Polyketides, proteins and genes in fungi: programmed nano-machines begin to reveal their secrets. Org. Biomol. Chem. 2007;5:2010–2026. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]
- Crawford JM, Korman TP, Labonte JW, Vagstad AL, Hill EA, Kamari-Bidkorpeh O, Tsai SC, Townsend CA. Structural basis for biosynthetic programming of fungal aromatic polyketide cyclization. Nature. 2009;461:1139–1143. doi: 10.1038/nature08475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JM, Vagstad AL, Ehrlich KC, Townsend CA. Starter unit specificity directs genome mining of polyketide synthase pathways in fungi. Bioorg. Chem. 2008;36:16–22. doi: 10.1016/j.bioorg.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Auria JC. Acyltransferases in plants: a good time to be BAHD. Curr. Opin. Plant Biol. 2006;9:331–340. doi: 10.1016/j.pbi.2006.03.016. [DOI] [PubMed] [Google Scholar]
- Davison J, Al Fahad A, Cai M, Song Z, Yehia SY, Lazarus CM, Bailey AM, Simpson TJ, Cox RJ. Genetic, molecular, and biochemical basis of fungal tropolone biosynthesis. Proc. Natl. Acad. Sci. USA. 2012 doi: 10.1073/pnas.1201469109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo A, Kuroda M. Citrinin, an inhibitor of cholesterol synthesis. J Antibiot (Tokyo) 1976;29:841–843. doi: 10.7164/antibiotics.29.841. [DOI] [PubMed] [Google Scholar]
- Garvey GS, McCormick SP, Rayment I. Structural and functional characterization of the TRI101 trichothecene 3-O-acetyltransferase from Fusarium sporotrichioides and Fusarium graminearum: kinetic insights to combating Fusarium head blight. J. Biol. Chem. 2008;283:1660–1669. doi: 10.1074/jbc.M705752200. [DOI] [PubMed] [Google Scholar]
- Ge HM, Zhang WY, Ding G, Saparpakorn P, Song YC, Hannongbua S, Tan RX. Chaetoglobins A and B, two unusual alkaloids from endophytic Chaetomium globosum culture. Chem. Commun. 2008:5978–5980. doi: 10.1039/b812144c. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Jez JM, Ferrer JL, Bowman ME, Dixon RA, Noel JP. Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry. 2000;39:890–902. doi: 10.1021/bi991489f. [DOI] [PubMed] [Google Scholar]
- Jones EW. Tackling the protease problem in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:428–453. doi: 10.1016/0076-6879(91)94034-a. [DOI] [PubMed] [Google Scholar]
- Keller NP, Turner G, Bennett JW. Fungal secondary metabolism - from biochemistry to genomics. Nat. Rev. Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- Kennedy J, Auclair K, Kendrew SG, Park C, Vederas JC, Hutchinson CR. Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science. 1999;284:1368–1372. doi: 10.1126/science.284.5418.1368. [DOI] [PubMed] [Google Scholar]
- Kroken S, Glass NL, Taylor JW, Yoder OC, Turgeon BG. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc. Natl. Acad. Sci. USA. 2003;100:15670–15675. doi: 10.1073/pnas.2532165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chooi YH, Sheng Y, Valentine JS, Tang Y. Comparative characterization of fungal anthracenone and naphthacenedione biosynthetic pathways reveals an alpha-hydroxylation-dependent Claisen-like cyclization catalyzed by a dimanganese thioesterase. J Am Chem Soc. 2011;133:15773–15785. doi: 10.1021/ja206906d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chooi YH, Sheng Y, Valentine JS, Tang Y. Comparative characterization of fungal anthracenone and naphthacenedione biosynthetic pathways reveals an alpha-hydroxylation-dependent Claisen-like cyclization catalyzed by a dimanganese thioesterase. J. Am. Chem. Soc. 2011;133:15773–15785. doi: 10.1021/ja206906d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma SM, Li JW, Choi JW, Zhou H, Lee KK, Moorthie VA, Xie X, Kealey JT, Da Silva NA, Vederas JC, et al. Complete reconstitution of a highly reducing iterative polyketide synthase. Science. 2009;326:589–592. doi: 10.1126/science.1175602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchand PS, Whalley WB, Chen FC. Isolation and structure of ankaflavin - new pigment from Monascus Anka. Phytochemistry. 1973;12:2531–2532. [Google Scholar]
- Nielsen KF, Mogensen JM, Johansen M, Larsen TO, Frisvad JC. Review of secondary metabolites and mycotoxins from the Aspergillus niger group. Analytical and Bioanalytical Chemistry. 2009;395:1225–1242. doi: 10.1007/s00216-009-3081-5. [DOI] [PubMed] [Google Scholar]
- Nukina M, Marumo S. Lunatoic acid A and acid B aversion factor and its related metabolite of Cochliobolus lunata. Tetrahedron Letters. 1977:2603–2606. [Google Scholar]
- Ogihara J, Kato J, Oishi K, Fujimoto Y. Biosynthesis of PP-V, a monascorubramine homologue, by Penicillium sp AZ. J. Biosci. Bioeng. 2000;90:678–680. doi: 10.1263/jbb.90.678. [DOI] [PubMed] [Google Scholar]
- Ortiz de Montellano PR. Cytochrome P450: structure, mechanism, and biochemistry. Kluwer Academic/Plenum Publishers; New York: 2005. pp. 211–213. [Google Scholar]
- Osmanova N, Schultze W, Ayoub N. Azaphilones: a class of fungal metabolites with diverse biological activities. Phytochemistry Reviews. 2010;9:315–342. [Google Scholar]
- Pall M, Brunelli J. A series of six compact fungal transformation vectors containing polylinkers with multiple unique restriction sites. Fungal Genet. Newslett. 1993:59–62. [Google Scholar]
- Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 2007;25:221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- Punt PJ, Oliver RP, Dingemanse MA, Pouwels PH, van den Hondel CA. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene. 1987;56:117–124. doi: 10.1016/0378-1119(87)90164-8. [DOI] [PubMed] [Google Scholar]
- Quang DN, Hashimoto T, Stadler M, Radulovic N, Asakawa Y. Antimicrobial azaphilones from the fungus Hypoxylon multiforme. Planta Med. 2005;71:1058–1062. doi: 10.1055/s-2005-873129. [DOI] [PubMed] [Google Scholar]
- Reeves CD, Hu Z, Reid R, Kealey JT. Genes for the biosynthesis of the fungal polyketides hypothemycin from Hypomyces subiculosus and radicicol from Pochonia chlamydosporia. Appl. Environ. Microbiol. 2008;74:5121–5129. doi: 10.1128/AEM.00478-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai K, Kinoshita H, Shimizu T, Nihira T. Construction of a citrinin gene cluster expression system in heterologous Aspergillus oryzae. J. Biosci. Bioeng. 2008;106:466–472. doi: 10.1263/jbb.106.466. [DOI] [PubMed] [Google Scholar]
- Sanchez JF, Somoza AD, Keller NP, Wang CC. Advances in Aspergillus secondary metabolite research in the post-genomic era. Nat. Prod. Rep. 2012 doi: 10.1039/c2np00084a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber MV, Meimberg H, Ebel J. Identification of a 4-coumarate:CoA ligase gene family in the moss, Physcomitrella patens. Phytochemistry. 2008;69:2449–2456. doi: 10.1016/j.phytochem.2008.06.014. [DOI] [PubMed] [Google Scholar]
- Somoza AD, Lee KH, Chiang YM, Oakley BR, Wang CC. Reengineering an azaphilone biosynthesis pathway in Aspergillus nidulans to create lipoxygenase inhibitors. Org. Lett. 2012;14:972–975. doi: 10.1021/ol203094k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR. Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- Wen Y, Hatabayashi H, Arai H, Kitamoto HK, Yabe K. Function of the cypX and moxY genes in aflatoxin biosynthesis in Aspergillus parasiticus. Appl. Environ. Microbiol. 2005;71:3192–3198. doi: 10.1128/AEM.71.6.3192-3198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter JM, Tang Y. Synthetic biological approaches to natural product biosynthesis. Curr. Opin. Biotechnol. 2012 doi: 10.1016/j.copbio.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Meehan MJ, Xu W, Dorrestein PC, Tang Y. Acyltransferase mediated polyketide release from a fungal megasynthase. J. Am. Chem. Soc. 2009;131:8388–8389. doi: 10.1021/ja903203g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Masuma R, Kim Y-P, Uchida R, Tomoda H, Omura S. Taxonomy and secondary metabolites of Pseudobotrytis sp. FKA-25. Mycoscience. 2004;45:9–16. [Google Scholar]
- Yasukawa K, Takahashi M, Natori S, Kawai K, Yamazaki M, Takeuchi M, Takido M. Azaphilones inhibit tumor promotion by 12-O-tetradecanoylphorbol-13-acetate in two-stage carcinogenesis in mice. Oncology. 1994;51:108–112. doi: 10.1159/000227320. [DOI] [PubMed] [Google Scholar]
- Zhou H, Qiao K, Gao Z, Meehan MJ, Li JW, Zhao X, Dorrestein PC, Vederas JC, Tang Y. Enzymatic synthesis of resorcylic acid lactones by cooperation of fungal iterative polyketide synthases involved in hypothemycin biosynthesis. J. Am. Chem. Soc. 2010;132:4530–4531. doi: 10.1021/ja100060k. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.