Summary Paragraph
Cytochrome c oxidase is a member of the heme copper oxidase superfamily (HCO)1. HCOs function as the terminal enzymes in the respiratory chain of mitochondria and aerobic prokaryotes, coupling molecular oxygen reduction to transmembrane proton pumping. Integral to the enzyme’s function is the transfer of electrons from cytochrome c to the oxidase via a transient association of the two proteins. Electron entry and exit are proposed to occur from the same site on cytochrome c2–4. Here we report the crystal structure of the caa3-type cytochrome oxidase from Thermus thermophilus, which has a covalently tethered cytochrome c domain. Crystals were grown in a bicontinuous mesophase using a synthetic short-chain monoacylglycerol as the hosting lipid. From the electron density map, at 2.36 Å resolution, a novel integral membrane subunit and a native glycoglycerophospholipid embedded in the complex were identified. Contrary to previous electron transfer mechanisms observed for soluble cytochrome c, the structure reveals the architecture of the electron transfer complex for the fused cupredoxin/cytochrome c domain which implicates different sites on cytochrome c for electron entry and exit. Support for an alternative to the classical proton gate characteristic of this HCO class is presented.
HCO members are classified based on sequence signatures and conservation of the D-and K-proton pathways, and fall into three main families5,6: Types A, B and C. Type A HCOs can be further divided into two subclasses, according to the amino acid motif at the hydrophobic end of the D-pathway (Supplementary Fig. 1). Type A1 contains a highly conserved glutamate residue in the sequence motif –XGHPEV–. In Type A2, the gating glutamate has been proposed to be replaced spatially by a consecutive tyrosine and serine in a –YSHPXV– motif5.
The crystal structure of caa3-type cytochrome c oxidase from T. thermophilus is the first for a Type A2 HCO (Fig. 1). The structure was determined to 2.36 Å resolution with crystals (Supplementary Fig. 2) grown by the in meso method using a synthetic, short chain (14 carbon) monoacylglycerol (7.7 MAG) as the hosting lipid7. 7.7 MAG was chosen for the larger water channel and reduced interfacial curvature of the cubic mesophase it forms to more suitably accommodate the enzyme’s extensive intra-and extra-membrane domains7,8. Increasingly, 7.7 and related short chain MAGs are proving to be useful for in meso crystallization8–11. Initial phases were determined by molecular replacement with the mitochondrially encoded SU I, II and III, from bovine heart cytochrome oxidase (PDB ID: 2OCC), identifying two molecules (Molecules 1 and 2) in the asymmetric unit. Consistent with the hypothesis for crystallization in meso12, packing within the crystal was layered or Type I (Supplementary Fig. 3). The cytochrome c domain was built manually after the heme iron was assigned by an anomalous map. The residual Rwork and Rfree of the refined structure were 17.1 % and 21.8 % (Supplementary Table 1, Methods, Supplementary Fig. 4 shows an experimental map of a portion of the oxidase structure) and the model ranked in the 99th percentile in MolProbity geometry analysis13.
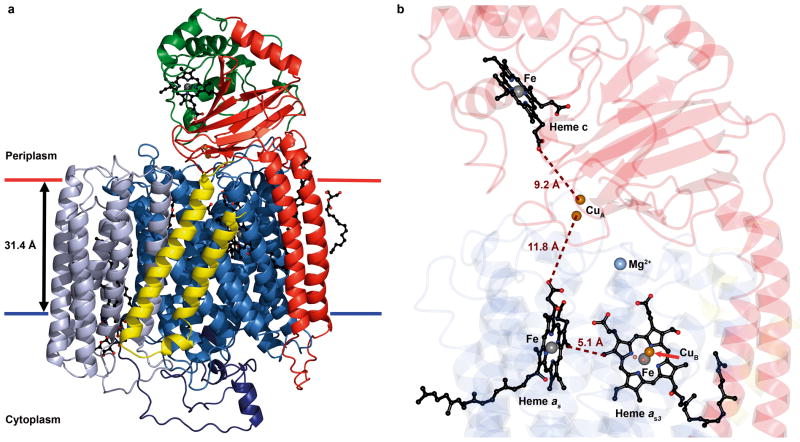
Figure 1. Structure of and cofactor arrangement in caa3-oxidase.
a, Structure (ribbon model). SU I/III is colored to highlight its canonical subunits: SU I, blue; SU III, blue-grey and the fusion linker, dark blue. SU IIc is colored to highlight the classical SU II (red) and the fused cytochrome c domain (green). SU IV is in yellow. The hemes are in ball and stick with the iron and copper metal centers as grey and copper spheres, respectively. Membrane boundaries are based on hydrophobic thickness calculations from the OPM server57. b, Cofactor arrangement. Hemes c, as and as3, and the iron and copper ions are shown as in a. The magnesium ion is represented as a light blue sphere. Distances are shown in brown. SU I/III, IIc and IV are color coded as faded blue, red and yellow ribbons, respectively.
The caa3-oxidase from T. thermophilus has been described as consisting of two subunits, SU I/III and IIc, that contain the metal centers characteristic of cytochrome c oxidases14,15. SU I/III exists as a fusion of the classical SU I and SU III16 while SU IIc is a fusion of a canonical SU II and a cytochrome c domain17. During the course of caa3-oxidase structure determination, electron density for a third subunit, SU IV, consisting of two connected transmembrane (TM) helices located along the hydrophobic surface of SU I/III (Fig. 1a and Supplementary Fig. 5) and a native glycoglycerophospholipid (see Supplementary Discussion and Supplementary Figs 6 and 7) embedded in SU I/III were identified. SU IV was modeled based on a 66 residue, N-terminally formylated protein sequence determined using vapor diffusion grown crystals (see Methods and Supplementary Fig. 5).
The final model consists of three subunits, SU I/III, IIc and IV, which collectively form a bundle of 23 TM helices and a bulky extra-membrane cytochrome c/cupredoxin domain (Fig. 1a). SU I/III is comprised of 19 TM helices, which hosts the low spin heme, a high spin heme-CuB binuclear center (Fig 2 a, b)and a conserved magnesium binding site (Fig. 2c). The fusion linker between the SU I and III domains is approximately 70 residues long, largely devoid of secondary structure save for a two-turn helix, and drapes the cytoplasmic surface of SU I/III, possibly conferring stability to the helical bundle. SU IIc consists of a membrane anchored cupredoxin domain, containing the electron accepting homo-binuclear copper (CuA)-centre, with a C-terminal fusion to a cytochrome c domain. The arrangement of the active site and redox centers is shown in Fig. 1b. In the context of number of amino acids and structured waters modeled, Molecule 1 is the most complete and is the focus of the discussion that follows.
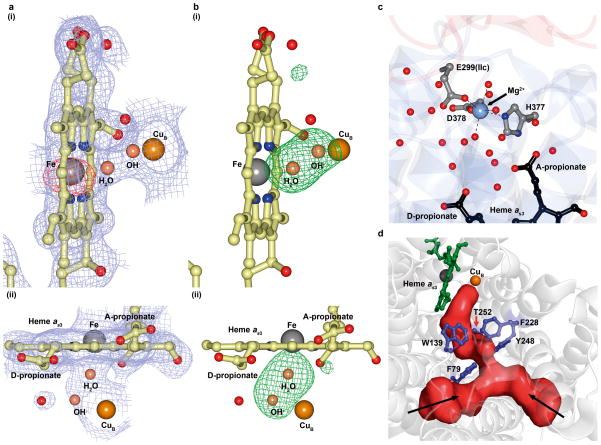
Figure 2. Active site, water pool and oxygen channel in caa3-oxidase.
a, b, The active site of heme as3 viewed from (i) the oxygen entry side and (ii) the periplasm. In a is shown a 2mFo-DFc electron density map contoured to 1σ (blue) and an anomalous difference map contoured to 5σ (red). In b is shown an mFo-DFc electron density difference map calculated without the bridging ligands contoured to 4σ (green). For clarity, the anomalous map is not shown in a(ii). c, Water pool. A water filled cavity centered on a magnesium ion is situated above heme as3. Coordinate bonds are shown as dashed black lines. d, The calculated oxygen channel is Y-shaped and hydrophobic. Two extremities of the channel (red) contact the apolar surface of the protein (black arrows); a third contacts the binuclear centre.
The core subunit, SU I/III, of caa3-oxidase shares a distinct sequence and structure homology with its Type A1 counterparts (Supplementary Fig. 1). Akin to ba3-oxidase from T. thermophilus, the a-type hemes in caa3-oxidase have geranylgeranyl tails and are designated heme as18. Both hemes as and as3 are located in the hydrophobic core of the SU I domain (Fig. 1b) with their metal centers ~11 Å from the periplasmic surface. As in other HCOs, two histidines, His 73 and 387, act as axial ligands to the low spin heme as iron at a distance of 2.0 and 2.1 Å, respectively (Supplementary Fig. 8a). Ligating the high spin heme as3 iron at its proximal side is a highly conserved His 385. The CuB is coordinated by the three canonical histidines, His 250, 299 and 300, and is located 4.9 Å from the heme as3 iron (Supplementary Fig. 8b). The ligands bridging CuB and the high spin iron have been modeled as a hydroxide and water pair 2.3 Å apart, with the hydroxide ion located 2.2 Å from the copper and the water 2.2 Å from the heme iron (Fig. 2a, b). Ligand assignments in the active site and oxygen entry into the enzyme (Fig. 2d and Supplementary Fig. 9) are discussed in the Supplementary Information.
SU I/III contains two proton channels, the D- and K-pathways (Fig. 3a, c). These are conduits for the protons that are 1) pumped across the membrane and 2) required for the chemical reduction of oxygen in the binuclear centre. A fuller description and comparison of the proton channels and exit pathways are included in the Supplementary Information (Supplementary Discussion and Supplementary Figs 10–12). The end of the D-pathway in caa3-oxidase is distinctly different from the proton gating site of Type A1 HCOs (Fig. 3b and Supplementary Fig. 13). The canonical Type A1 gating glutamate, Glu 278 (P. denitrificans numbering), is replaced by Thr 252 in caa3-oxidase while the phenylalanine-glycine pair, a single turn upstream of the Type A1 glutamate position in helix VI, is replaced by a consecutive tyrosine (Tyr 248) and serine (Ser 249) residues (Supplementary Fig. 13a). This YS pair (Supplementary Fig. 13b) offers a functional equivalent to the gating glutamate as determined by single-, double- and triple-mutant studies performed on the P. denitrificans aa3-type oxidase mimicking the Type A2 pump configuration in both T. thermophilus and Rhodothermus marinus19. In caa3-oxidaseTyr 248 is held in position through interactions with the side chain amide of Asn 205 and the hydroxyl of Thr 252, while a hydrogen bond between Tyr 248 and a structured water (Wat 2074)connects the tyrosine to the D-pathway water chain (Fig. 3a, b).
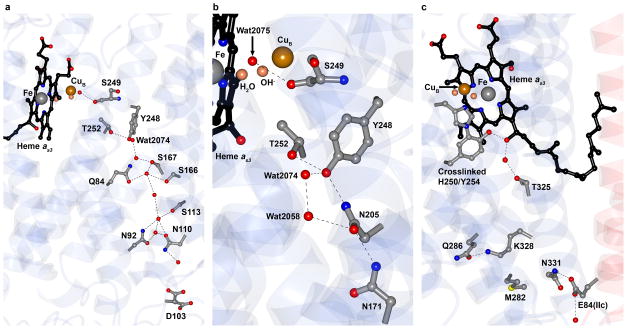
Figure 3. Proton pathways in caa3-oxidase.
a, The D-pathway begins at Asp 103 and leads through a solvent filled cavity to Tyr 248. Alternative conformations for the Asp 103 side chain were modeled into electron density and are shown. The conformer pointing towards Asn 110 represents the conformation observed in other known oxidases. b, Detailed view of the YS gate, the structural equivalent in caa3-oxidase of the classical Type A1 gating glutamate. c, The K-pathway originates at Glu 84 of SU IIc and continues up to the binuclear centre via the cross-linked His 250 - Tyr 254, by way of Lys 328. Hydrogen bonds are shown as dashed black lines in a–c. The known covalent linkage between His 250 and Tyr 254 was modeled based on separate biochemical characterization58.
Molecular dynamics simulations (MDS)19 that accompanied the above mutagenesis work, predicted a chain of four water molecules linking the engineered tyrosine to the active site. The water chain was anchored by the Nε2 of His 326, the hydroxyl of Tyr 274 and the carbonyl oxygen of Ser 275 (P. denitrificans numbering). The side chain of Ser 275, however, did not interact with the water chain. Similarly, in caa3-oxidase, Wat 2075 hydrogen bonds to the carbonyl of Ser 249, whose side chain points away from the active site (Fig. 3b). Given that Thr 252 is not conserved in Type A2 oxidases, we speculate that it functions as part of a protonic chain from Tyr 248 to the active site replacing the water that hydrogen bonds to the tyrosine hydroxyl in the aforementioned MDS studies. These observations lend support to the presence of a similar water chain in caa3-oxidase which is only partially resolved in the current structure (Fig. 3b). MDS, carried out on the model reported here, identify transient water chains in the YS gate region that provide clear protonic connectivity to the binuclear centre and to the D-propionate of heme as3 (Supplementary Fig. 11a,b).
In caa3-oxidase from Rh. marinus, the tyrosine of the YS gate has been reported to deprotonate during the gating event as evidenced by FTIR difference spectroscopy20. This observation is counter to earlier pKa calculations, which indicate that the tyrosine remains protonated21. The pKa of a tyrosine hydroxyl in aqueous solution is approximately 10. Given the hydrophobic environment surrounding Tyr 248 in the caa3-oxidase of T. thermophilus, the effective pKa of its hydroxyl is expected to be above 10. This has been confirmed by pKa calculations performed on the caa3 oxidase model reported here (Supplementary Table 2). The higher pKa favors protonation possibly limiting the role of Tyr 248 to one of stabilizing the water network in the proton pathway.
Subunit IIc is a fusion between the classical SU II and a cytochrome c (Fig. 1a). The SU II domain consists of two N-terminal TM helices anchoring a cupredoxin-like domain composed of a 10-stranded β-barrel that hosts the CuA homobinuclear centre, the primary acceptor of electrons from cytochrome c (Fig. 4a). The asymmetric ligands for the CuA centre in SU IIc are highly conserved among Type A and B cytochrome c oxidases. The cytochrome c domain of SU IIc consists of four α-helices connected by random coil segments (Fig. 4a, Supplementary Discussion and Supplementary Fig. 14). The heme c resides within the hydrophobic core of the domain and is held in place by the classical axial iron ligands, His 251 and Met 303, and by two thioether linkages to Cys 247 and Cys 250.
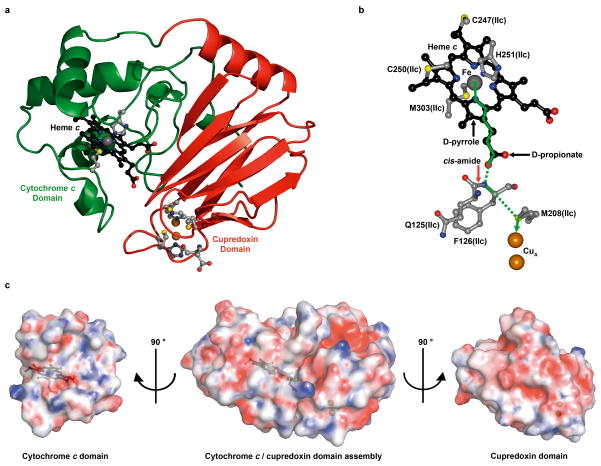
Figure 4. Cytochrome c/cupredoxin complex and electron transfer pathway in caa3-oxidase.
a, Structure of the cytochrome c (green) and cupredoxin (red) domains of SU IIc. b, The optimum electron transfer pathway between the heme c iron and the CuA center was calculated with HARLEM. Through bond and through space tunneling are shown in solid and dashed green lines, respectively. c, “Open book” view of surface complementarities between the cytochrome c and cupredoxin domains in caa3-oxidase. Electrostatic potentials are viewed at ±10 kbT. The linker (residues 217-235) between the two domains has been removed for clarity. Surfaces are at 25 % transparency to reveal the positions of heme c and the CuA centre shown in black ball and stick and copper spheres, respectively.
Cytochrome c is oriented at the face of the cupredoxin domain such that the D-propionate of heme c is pointing towards the CuA centre (Fig. 4a, b). This arrangement is consistent with speculation regarding the assembly of caa3-oxidase in Rh. marinus22,23. It is, however, counter to that described on the basis of MDS and nuclear magnetic resonance studies of the interaction between cytochrome c and the cupredoxin domains of ba3- and aa3-oxidase, respectively, in solution2,24,25. Here, contact was proposed by way of the exposed heme cleft of the type reported for the cytochrome c/bc1 complex from Saccharomyces cerevisiae26. Interestingly, the assembly of the cytochrome c/cupredoxin domain in caa3-oxidase is quite distinct from the arrangement of the mono- and di-heme domains of cbb3-oxidase (Supplementary Fig. 15).
The cytochrome c (SU IIc residues 236-325) and cupredoxin (SU IIc residues 117-216) domains share an interface with a calculated buried surface area of 934 Å2/domain (www.ebi.ac.uk/msd-srv/prot_int/pistart.html). Interaction between the domains is by surface and electrostatic complementarity (Fig. 4c). The contact surface of the cupredoxin domain is anionic and complementary to the cationic contact surface of the cytochrome c domain created by Lys 298 and Arg 272 in the vicinity of the heme propionates (Fig. 4c). Interdomain contacts are tabulated in Supplementary Table 3.
The optimal electron tunneling pathway between heme c and CuA in caa3-oxidase was calculated using the program HARLEM (www.kurnikov.org) (Fig. 4b). It proceeds through the D-pyrrole and D-propionate of heme c, the backbone cis-amide nitrogen and α-carbon of Phe 126(IIc) and the sulfur of Met 208(IIc). The effective tunneling length of the pathway is 18.8 Å with a direct metal-to-metal separation of 17.2 Å. By contrast, the closest ‘edge-to-edge’ distance between the heme c D-propionate donor and the CuA centre acceptor moieties is 9.2 Å which too can be bridged efficiently by a tunneling electron27. Such a process would involve two through-space jumps; the first from the D-propionate of heme c to the phenyl ring of Phe 126(IIc)(separation, 4.4 Å) and a second to either the sulfur of Cys 201(IIc) (separation, 3.7 Å) or directly to the CuA centre (separation, 5.1 Å). Interestingly, mutating Trp 121 of the P. denitrificans aa3-oxidase, a structural homolog of Phe 126(IIc), to a glutamine inactivates the enzyme28–30. Furthermore, changing the homologous Phe 88 in ba3-oxidase to leucine resulted in a drop to 68 % in electron transfer efficiency2. Together, these data suggest that the bulky hydrophobic side chain of Phe 126(IIc) is required to stabilize the conserved cis-amide for efficient ET. A mechanism for electron entry into the cytochrome c domain is proposed (Supplementary Information).
The structure of caa3-oxidase as described here offers the first view of a Type A2 HCO consisting of a pair of fused canonical subunits and a previously unknown SU IV. It shows the cytochrome c/oxidase complex where the D-propionate of heme c is the most probable conduit for electron loading into CuA through the cis-amide of an aromatic residue conserved across Type A and B cytochrome c oxidases. Electrons are proposed to enter the cytochrome c domain by way of its exposed heme edge. MDS calculations on the Type A2 pump gating site lend support to protonic water chains linking Tyr 248 to both the binuclear centre and the D-propionate of heme as3. Further, interaction between Lys 328 and Gln 286 is proposed to stabilize the canonical lysine in its protonated state in the K-pathway, replacing the functionally equivalent serine-water-lysine motif in Type A1 HCOs.
The caa3-oxidase structure provides a framework for examining how this integral membrane complex couples energy transduction to the complete reduction of oxygen. The manner in which Type A2 oxidases direct protons across the membrane and electrons and protons to the active site of the complex can now be studied in detail as a result of having available the 3-D arrangement of key residues, metal ions and structured waters. Crystal structures of representatives from all HCO families have now been solved and the stage is set for a more complete understanding of how these intricate proteinaceous machines, with diverse oxygen affinities, have evolved.
ONLINE METHODS
Protein production and purification
Biomass of the extreme thermophilic bacterium T. thermophilus HB8 was produced by fermentation at the Helmholtz Centre for Infection Research (Braunschweig, Germany), as described previously31. The protocol used for isolating and purifying caa3-oxidase from the biomass is similar to that used for the ba3-oxidase with slight modifications31. Briefly, 100 g of T. thermophilus biomass was suspended in 0.5 L 0.1 MTris-HCl pH 7.6 buffer (Buffer A) containing 0.2 M KCl and homogenized using a blender. Lysozyme was added to a final concentration of 0.6 μM and the suspension was stirred for 3 h at 4 °C. After centrifugation at 53,936g for 45 min at 4°C the pellet was resuspended in 0.5 L Buffer A, homogenized and centrifuged again at 53,936g for 30 min at 4 °C. This washing step was repeated three times. The final pellet was resuspended in 0.5 L Buffer A and incubated with stirring for 3 h at 4°C in the presence of 5%(v/v) Triton X-100 (Sigma Aldrich) to solubilize the membranes. After centrifugation at 53,936g for 1 h at 4°C, the supernatant was collected and diluted with MilliQ water at 4 °C to a final volume of 5 L. It was subsequently loaded at 4°C on DEAE-Biogel agarose anion exchange resin (Biorad) packed in a 30 cm × 10 cm column equilibrated with 0.01 M Tris-HCl pH 7.6, 0.1%(v/v) Triton X-100(Buffer B). After washing with 2 L Buffer B, the respiratory complexes were eluted with 4 L of a 0 to 0.2 M linear gradient of NaCl in Buffer B.
The caa3-oxidase containing fractions, identified by UV/visible spectroscopy, from the anion exchange column were pooled (ca. 0.8 L) and dialyzed against 10 L Buffer B for 5 h at 4 °C. The dialyzed material was loaded on an XK 26/20 column packed with 30 mL Fractogel EMD TMAE anion exchange resin (Merck) pre-equilibrated with Buffer B. With the protein bound to the resin, the Triton X-100 detergent was exchanged for 0.05 %(w/v) n-dodecyl-β-D-maltoside (DDM). The protein was eluted with a 0 to 0.3M linear gradient of NaCl in 0.01 M Tris-HCl pH 7.6, 0.05 %(w/v) DDM (Buffer C) for 1 h at 22° C and at a flow rate of 4 mL/min. The caa3-oxidase containing fractions (ca. 0.16 L) were pooled and concentrated to 8 mL using a centrifugal filter (Centricon YM100, Millipore) and applied to a Highload XK 16/60 Superdex 200 gel filtration column (GE-Healthcare) previously equilibrated with 0.05 M Tris-HCl pH 7.6, 0.05%(w/v) DDM. Fractions across the caa3-oxidase peak were pooled (ca. 90 mL) and diluted 10-fold with 0.01M sodium phosphate pH 6.8 containing 0.05 %(w/v) DDM, and applied to an XK 16/20 column packed with 20 mL hydroxylapatite ‘high resolution’ ion exchange resin (Fluka, BioChemika) pre-equilibrated with the same buffer. The caa3-oxidase was eluted with a 0.01 to 0.04 M linear gradient of sodium phosphate pH 6.8 containing 0.05 %(w/v) DDM within 0.5 h at a flow rate 2 mL/min at 22° C. The pooled caa3-oxidase fractions (ca. 50 mL) were diluted 5-fold with Buffer C reducing the sample conductivity to < 2 mS/cm. The previous anion exchange and concentration steps using Buffer C was repeated once. The detergent was exchanged to 0.2 %(w/v) n-decyl-β-D-maltoside (DM) in 0.01 M Tris-HCl pH 7.6 and 150 mM NaCl during a final Highload XK 16/60 Superdex 200 gel filtration step and resulted in ca. 10 mg of purified caa3-oxidase. The protein was concentrated to 10 mg/mL (Centricon YM100, Millipore), flash frozen in liquid nitrogen and stored in aliquots of 20 μL at −80 °C. The protein was used in the ‘as-prepared’ oxidized state for crystallization studies.
Crystallization
7.7 MAG was synthesized according to Caffrey et al8. The protein-laden mesophase was prepared by homogenizing 7.7 MAG and a 10 mg/mL protein solution, supplemented with sodium ascorbate to a final concentration of 0.2 μM, in a 1:1 ratio by weight using a dual syringe mixing device at 20 °C32. Crystallization trials were carried out at 20 °C in 96-well glass sandwich plates with 50 nL mesophase and 0.8 μL precipitant solution using an in meso robot33. Precipitant solutions consisted of 14–21 %(v/v) PEG 400, 0.1 M NaCl, 0–0.1 M Li2SO4 and 0.1 M sodium citrate pH 4.5–5.0. Triangular plate-like crystals grew to a maximum size of 100 × 60 × 5 μm3 in 7 to 10 days (Supplementary Fig. 2). Wells were opened using a tungsten-carbide glass cutter and the crystals were harvested using 100 μm micromounts (MiTeGen). Crystals were cryo-cooled directly in liquid nitrogen.
Subunit IV identification and sequencing
The caa3-oxidase complex, pre-purified by vapor diffusion crystallization using a precipitant of 10 mM bis-Tris pH 6.0, 6 %(w/v) PEG 2000 and 100 mM NaCl, was separated into its individual subunits on a reversed phase Synchropak C4 column (250 mm × 4.6 mm; Supelco) using a Hewlett-Packard (HP) 1050 HPLC system at 40 °C. The solvents used were (A) 5% formic acid, (B) 95% formic acid, (C) n-propanol, and (D) acetonitrile. Gradients were formed with a microprocessor-controlled quaternary pump (HP) by low-pressure mixing of the four solvents. Two solvent gradients were used. Gradient 1 employed (A) 40–5 %, (B) constant at 60 %, (C) 0–10 %, and (D) 0–25 % in the 0 – 30 min run time. Gradient 2 employed (A) constant at 5 %, (B) constant at 60 %, (C) 10–25 %, and (D) 25–10 % in the 30–60 min run time. Separated subunits were identified by N-terminal sequencing after deformylation as appropriate. Parenthetically, we note that vapor diffusion grown crystals, referred to above, diffracted to no better than 3.8 Å and were unsuitable for structure determination.
The full length SU IV was characterized by N- (residues 1-41) and C-terminal (residues 60-66) sequencing and mass spectrometry. Sequencing was performed on peptide fragments prepared using cyanogen bromide, as reported for SU IIa in ba3-oxidase34. The peptide fragments were separated by HPLC at 45 °C, as described above. The solvents used were (A) 70 % formic acid and (B) 70 % formic acid, 15 % n-propanol and 11 % acetonitrile with a linear gradient of 100 %A to 100 %B. Sequencing results are shown in Supplementary Fig. 5.
Data collection and processing
X-ray diffraction data were collected onthe 23-ID-D beamline of the General Medicine and Cancer Institutes Collaborative Access Team (GM/CA-CAT) at the Advanced Photon Source (APS), Argonne, Illinois, USA, the I24 beamline at the Diamond Light Source (DLS), Didcot, Oxford, UK and the PX1 beamline at the Swiss Light Source (SLS), Villigen, Switzerland. Data were acquired using a 10 μm collimated minibeam at GM/CA-CAT35, while a 10 μm and a 20 μm microfocus beam was used at DLS and SLS, respectively. Attenuated (10X) images were used to locate and to centre on highly ordered regions of the crystal36. Oscillation data were measured in 1.0 ° frames with 1–2 s exposures using a 1X or 10X attenuated beam. All data were initially reduced in xia237 using XDS38, XSCALE and SCALA39. Optimum data wedges were identified by data quality and isomorphic unit cell parameters. These data were rescaled in XSCALE and merged in SCALA39 (Supplementary Table 1). The data reduction strategy involved combining a complete low resolution (3.5 Å) data set recorded from a single crystal with twenty high resolution (up to 1.9 Å) 10–20° wedges of data collected from multiple crystals.
Structure solution and refinement
Molecular replacement search models were prepared from SU I, SU II and SU III of the bovine heart cytochrome c oxidase complex (PDB ID: 2OCC) pruned of all non-protein atoms using Chainsaw in CCP440. Initial phases were obtained by molecular replacement as performed by the program Phaser41, identifying two molecules (Molecules 1 and 2) in the asymmetric unit. Initial stages of structure refinement and model building were performed after simulated annealing in CNS42 with subsequent rounds of refinement carried out in PHENIX43. NCS restraints were used throughout and the inclusion of TLS refinement (20 groups over Molecules 1 and 2) was implemented in the latter stages of refinement. The structural model was revised in real space with the program COOT44 using sigma-A weighted 2Fo−Fc and Fo−Fc electron density maps. The geometric quality of the model was assessed with MolProbity13. Structures of cofactors, lipids and water molecules were determined and refined as described below.
Cofactors
The locations of two hemes as, heme c, the binuclear CuA, CuB and magnesium in each molecule in the asymmetric unit were determined using the 2Fo−Fc electron density map. Iron atoms of the three hemes were located using an anomalous difference Fourier map from data collected at the iron K-edge (1.7397 Å) at the SLS.
Lipids and unassigned density
Three types of lipid molecule were identified in a composite omit 2Fo−Fc map. The first, a native Thermus lipid, was found buried within the protein at the interface between the SU I and SU III domains of the fused SU I/III. The second includes the 1- and 2- MAG isoforms of 7.7 MAG, where 1- and 2- refers to the position of the acyl chain on the glycerol headgroup. The third was modeled as a diacylglycerol with two fatty acyl chains. The diacylglycerol may derive from the hosting 7.7 MAG by way of transesterification or from a more complex native lipid. Electron density in the vicinity of the headgroup region of the diacylglycerol was ill-defined. Tentatively modeled lipid and solvent molecules were removed if no clear hydrogen bonding was present and/or the model was not justified by the electron density.
Water molecules
Both surface associated and integral water molecules were assigned based on sigma-A weighted 2Fo−Fc electron density maps contoured at 1σ using standard geometrical and chemical restraints. Molecules 1 and 2 have 247 and 145 waters, respectively, for a total of 392 water molecules per asymmetric unit.
Electrostatic surfaces
The electrostatic potential was calculated separately on the cupredoxin domain (residues 117-216) and the cytochrome c domain (residues 236-325) of SU IIc using the Poisson-Boltzmann method as implemented in the Potential module from the package MEAD45. The ionic strength in the calculations was 0.1 M. A grid of 141 points with 1 nm spacing was used to enclose the protein. The solvent probe radius was set to 1.4 Å and the ion exclusion layer thickness was 2.0 Å. An external dielectric constant (ε) of 80 was used for the solvent and the internal ε was set to 4. PARSE atomic charges and radii were used. The reduced heme c iron was assigned a charge of +1.4 with the remaining +0.6 charge assigned equally amongst its axial ligands. The oxidized CuA site was assigned a mixed valence of +1.0 on each copper ion with the remaining +1.0 charge assigned proportionally based on coordination to its axial ligands.
MD simulations
An all atom simulation system included caa3-oxidase embedded into a pre-equilibrated POPE lipid bilayer solvated in 0.15 M NaCl. All crystallographic water molecules were kept and additional water molecules were modeled in the protein internal hydrophilic cavities using Dowser46. The total simulation system size was ~159,000 atoms. MDS were performed using NAMD47 with the CHARMM force field for proteins (C22 with the CMAP correction)48,49, lipids (C36)50 and water (the TIP3P water model). A lipid molecule identified in SU I/III (Supplementary Fig. 6) was parameterized using the CHARMM General Force Field51. All protein residues were modeled in their standard protonation states and metal centers were modeled as in the reduced state. Production runs (two independent simulations) were performed in the NPT ensemble at a constant temperature (310 K) and pressure (1 atm) for the combined simulation time of ~300 ns. Coordinates were saved every 2 ps. VMD52 was used for trajectory analysis.
pKa calculations
Electrostatic calculations were performed using MEAD45 with the following options: protein atomic coordinates were taken from the X-ray structure; hydrogen atoms were added and minimized. MEAD requires that the dielectric constant used for the protein is also used for the membrane. Accordingly the pKa calculations were carried out using ε = 4 and 10 for the protein and membrane regions and ε = 80 for solvent. Calculations were performed, and results are reported, with and without explicit water molecules.
Electron transfer
The electron transfer (ET) pathway calculation between the heme c iron of the cytochrome c domain and homobinuclear CuA centre, with both sites selected interactively, was performed using the pathways module as implemented in HARLEM. The non-bonding and hydrogen bonding decay was 1.7 Å−1 and the covalent bond decay was 0.6 Å−1. The choice of the best ET pathway was based on couplings.
Oxygen channels
Oxygen channel cavities were calculated with HOLLOW53 using a grid spacing of 0.25 Å and a probe radius of 1.4 Å.
Structure based sequence alignment
Coordinates for the Type A cytochrome c oxidases present in the PDB (PDB IDs: 3HB3, 2GSM, 1V54) were aligned with the caa3-oxidase structure, presented here (PDB ID: 2YEV), using the secondary structure matching algorithm (SSM)54 as implemented in COOT44. This initial alignment was further refined with Modeller55 with a threshold of 3.5 Å between Cα atoms for inter-protein residue matching. Figures for the resulting structure based sequence alignments were produced with ALINE56. Percentage sequence identity was calculated from the number of identical residues/minimum length.
Protein figures were generated in CCP4mg. Figures describing oxygen cavities and electrostatic surfaces were generated in PyMOL.
Supplementary Material
Acknowledgments
We acknowledge support from Science Foundation Ireland Grants 07/IN.1/B1836 (M.C)and BICF685 (T.S), the National Institutes of Health Grants GM75915, P50GM073210, U54GM094599 (M.C), and FP7 COST CM0902 (M.C) and Marie Curie Actions PIEF-GA-2009-235612 (D.A). We thank David Hart, Jean Lee, and Timothy Smyth for help with 7.7 MAG synthesis, Manfred Dewor for amino acid sequencing, Cláudio Soares for discussions regarding surface potential calculations, and Graeme Winter for help with data reduction from multiple crystals. The assistance and support of beamline scientists at the Advanced Photon Source (23-ID), Diamond Light Source (I24) and Swiss Light Source (PX1) are gratefully acknowledged.
Footnotes
Supplementary information is linked to the online version of the paper at www.nature.com/nature
Author Contributions
J.A.L. synthesized 7.7 MAG, optimized crystallization conditions, collected and processed data, solved, refined and analyzed the structure, wrote manuscript.
D.A. harvested crystals, collected and processed data, solved, refined and analyzed the structure, wrote manuscript.
O.S. performed initial protein production, purification, crystallization and data collection.
A.V.P carried out molecular dynamics simulations and pKa calculations.
T.S. initiated the project, produced, purified and characterized protein, helped with crystallization, data collection and processing and structure analysis, wrote manuscript.
M.C. responsible for the overall project strategy and management and oversaw manuscript preparation.
Author Information
Coordinates and structure factors for caa3-oxidase are deposited in the Protein Data Bank (accession code 2YEV). Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Ferguson-Miller S, Babcock GT. Heme/copper terminal oxidases. Chem Rev. 1996;96:2889–2908. doi: 10.1021/cr950051s. [DOI] [PubMed] [Google Scholar]
- 2.Muresanu L, et al. The electron transfer complex between cytochrome c552 and the CuA domain of the Thermus thermophilus ba3 oxidase. A combined NMR and computational approach. J Biol Chem. 2006;281:14503–14513. doi: 10.1074/jbc.M601108200. [DOI] [PubMed] [Google Scholar]
- 3.Maneg O, Ludwig B, Malatesta F. Different interaction modes of two cytochrome-c oxidase soluble CuA fragments with their substrates. J Biol Chem. 2003;278:46734–46740. doi: 10.1074/jbc.M307594200. [DOI] [PubMed] [Google Scholar]
- 4.Maneg O, Malatesta F, Ludwig B, Drosou V. Interaction of cytochrome c with cytochrome oxidase: two different docking scenarios. Biochim Biophys Acta. 2004;1655:274–281. doi: 10.1016/j.bbabio.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 5.Pereira MM, Santana M, Teixeira M. A novel scenario for the evolution of haem-copper oxygen reductases. Biochim Biophys Acta. 2001;1505:185–208. doi: 10.1016/s0005-2728(01)00169-4. [DOI] [PubMed] [Google Scholar]
- 6.Pereira MM, Sousa FL, Verissimo AF, Teixeira M. Looking for the minimum common denominator in haem-copper oxygen reductases: towards a unified catalytic mechanism. Biochim Biophys Acta. 2008;1777:929–934. doi: 10.1016/j.bbabio.2008.05.441. [DOI] [PubMed] [Google Scholar]
- 7.Misquitta LV, et al. Membrane protein crystallization in lipidic mesophases with tailored bilayers. Structure. 2004;12:2113–2124. doi: 10.1016/j.str.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 8.Caffrey M, Lyons J, Smyth T, Hart DJ. In: Current Topics in Membranes. Ch 4. DeLucas LJ, editor. Vol. 63. Academic Press; 2009. pp. 83–108. [Google Scholar]
- 9.Höfer N, Aragão D, Lyons JA, Caffrey M. Membrane protein crystallization in lipidic mesophases. Hosting lipid effects on the crystallization and structure of a transmembrane peptide. Cryst Growth Des. 2011;11:1182–1192. doi: 10.1021/cg101384p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li D, Lee J, Caffrey M. Crystallizing membrane proteins in lipidic mesophases. A host lipid screen. Cryst Growth Des. 2011;11:530–537. doi: 10.1021/cg101378s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rasmussen SG, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caffrey M. Crystallizing membrane proteins for structure determination: use of lipidic mesophases. Annu Rev Biophys. 2009;38:29–51. doi: 10.1146/annurev.biophys.050708.133655. [DOI] [PubMed] [Google Scholar]
- 13.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fee JA, Choc MG, Findling KL, Lorence R, Yoshida T. Properties of a copper-containing cytochrome c1aa3 complex: a terminal oxidase of the extreme thermophile Thermus thermophilus HB8. Proc Natl Acad Sci USA. 1980;77:147–151. doi: 10.1073/pnas.77.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steffens GC, Biewald R, Buse G. Cytochrome c oxidase is a three-copper, two-heme-A protein. Eur J Biochem. 1987;164:295–300. doi: 10.1111/j.1432-1033.1987.tb11057.x. [DOI] [PubMed] [Google Scholar]
- 16.Mather MW, Springer P, Hensel S, Buse G, Fee JA. Cytochrome oxidase genes from Thermus thermophilusNucleotide sequence of the fused gene and analysis of the deduced primary structures for subunits I and III of cytochrome caa3. J Biol Chem. 1993;268:5395–5408. [PubMed] [Google Scholar]
- 17.Mather MW, Springer P, Fee JA. Cytochrome oxidase genes from Thermus thermophilusNucleotide sequence and analysis of the deduced primary structure of subunit IIc of cytochrome caa3. J Biol Chem. 1991;266:5025–5035. [PubMed] [Google Scholar]
- 18.Lubben M, Morand K. Novel prenylated hemes as cofactors of cytochrome oxidases. Archaea have modified hemes A and O. J Biol Chem. 1994;269:21473–21479. [PubMed] [Google Scholar]
- 19.Backgren C, Hummer G, Wikstrom M, Puustinen A. Proton translocation by cytochrome c oxidase can take place without the conserved glutamic acid in subunit I. Biochemistry. 2000;39:7863–7867. doi: 10.1021/bi000806b. [DOI] [PubMed] [Google Scholar]
- 20.Pereira MM, Sousa FL, Teixeira M, Nyquist RM, Heberle J. A tyrosine residue deprotonates during oxygen reduction by the caa3 reductase from Rhodothermus marinus. FEBS Lett. 2006;580:1350–1354. doi: 10.1016/j.febslet.2006.01.055. [DOI] [PubMed] [Google Scholar]
- 21.Soares CM, Baptista AM, Pereira MM, Teixeira M. Investigation of protonatable residues in Rhodothermus marinus caa3 haem-copper oxygen reductase: comparison with Paracoccus denitrificansaa3 haem-copper oxygen reductase. J Biol Inorg Chem. 2004;9:124–134. doi: 10.1007/s00775-003-0509-9. [DOI] [PubMed] [Google Scholar]
- 22.Santana M, Pereira MM, Elias NP, Soares CM, Teixeira M. Gene cluster of Rhodothermus marinus high-potential iron-sulfur protein: oxygen oxidoreductase, a caa(3)-type oxidase belonging to the superfamily of heme-copper oxidases. J Bacteriol. 2001;183:687–699. doi: 10.1128/JB.183.2.687-699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srinivasan V, et al. Structure at 1.3 Å resolution of Rhodothermus marinuscaa(3) cytochrome c domain. J Mol Biol. 2005;345:1047–1057. doi: 10.1016/j.jmb.2004.10.069. [DOI] [PubMed] [Google Scholar]
- 24.Wienk H, et al. Interaction of cytochrome c with cytochrome c oxidase: an NMR study on two soluble fragments derived from Paracoccus denitrificans. Biochemistry. 2003;42:6005–6012. doi: 10.1021/bi027198f. [DOI] [PubMed] [Google Scholar]
- 25.Roberts VA, Pique ME. Definition of the interaction domain for cytochrome c on cytochrome c oxidase. J Biol Chem. 1999;274:38051–38060. doi: 10.1074/jbc.274.53.38051. [DOI] [PubMed] [Google Scholar]
- 26.Lange C, Hunte C. Crystal structure of the yeast cytochrome bc1 complex with its bound substrate cytochrome c. Proc Natl Acad Sci USA. 2002;99:2800–2805. doi: 10.1073/pnas.052704699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Page CC, Moser CC, Dutton PL. Mechanism for electron transfer within and between proteins. Curr Opin Chem Biol. 2003;7:551–556. doi: 10.1016/j.cbpa.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 28.Drosou V, Malatesta F, Ludwig B. Mutations in the docking site for cytochrome c on the Paracoccus heme aa3 oxidase. Electron entry and kinetic phases of the reaction. Eur J Biochem. 2002;269:2980–2988. doi: 10.1046/j.1432-1033.2002.02979.x. [DOI] [PubMed] [Google Scholar]
- 29.Drosou V, Reincke B, Schneider M, Ludwig B. Specificity of the interaction between the Paracoccus denitrificans oxidase and its substrate cytochrome c: comparing the mitochondrial to the homologous bacterial cytochrome c(552), and its truncated and site-directed mutants. Biochemistry. 2002;41:10629–10634. doi: 10.1021/bi020094k. [DOI] [PubMed] [Google Scholar]
- 30.Witt H, Malatesta F, Nicoletti F, Brunori M, Ludwig B. Tryptophan 121 of subunit II is the electron entry site to cytochrome-c oxidase in Paracoccus denitrificans Involvement of a hydrophobic patch in the docking reaction. J Biol Chem. 1998;273:5132–5136. doi: 10.1074/jbc.273.9.5132. [DOI] [PubMed] [Google Scholar]
- 31.Soulimane T, Kiefersauer R, Than ME. In: Membrane Protein Purification and Crystallization. 2. Hunte C, von Jagow G, Schägger H, editors. Ch 14. Academic Press; 2003. pp. 229–251. [Google Scholar]
- 32.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat Protoc. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cherezov V, Peddi A, Muthusubramaniam L, Zheng YF, Caffrey M. A robotic system for crystallizing membrane and soluble proteins in lipidic mesophases. Acta Crystallogr D. 2004;60:1795–1807. doi: 10.1107/S0907444904019109. [DOI] [PubMed] [Google Scholar]
- 34.Soulimane T, Than ME, Dewor M, Huber R, Buse G. Primary structure of a novel subunit in ba3-cytochrome oxidase from Thermus thermophilus. Protein Sci. 2000;9:2068–2073. doi: 10.1110/ps.9.11.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischetti RF, et al. Mini-beam collimator enables microcrystallography experiments on standard beamlines. J Synchrotron Radiat. 2009;16:217–225. doi: 10.1107/S0909049508040612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cherezov V, et al. Rastering strategy for screening and centring of microcrystal samples of human membrane proteins with a sub-10 micron size X-ray synchrotron beam. J R Soc Interface. 2009;6:S587–597. doi: 10.1098/rsif.2009.0142.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winter G. xia2: an expert system for macromolecular crystallography data reduction. J Appl Cryst. 2010;43:186–190. [Google Scholar]
- 38.Kabsch W. XDS. Acta Crystallogr D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evans P. Scaling and assessment of data quality. Acta Crystallogr D. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 40.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 41.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 43.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Bashford D, Gerwert K. Electrostatic calculations of the pKa values of ionizable groups in bacteriorhodopsin. J Mol Biol. 1992;224:473–486. doi: 10.1016/0022-2836(92)91009-e. [DOI] [PubMed] [Google Scholar]
- 46.Zhang L, Hermans J. Hydrophilicity of cavities in proteins. Proteins. 1996;24:433–438. doi: 10.1002/(SICI)1097-0134(199604)24:4<433::AID-PROT3>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 47.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MacKerell AD, et al. All atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 49.MacKerell AD, Feig M, Brooks CL. Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 50.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vanommeslaeghe K, et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 53.Ho BK, Gruswitz F. HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct Biol. 2008;8:49. doi: 10.1186/1472-6807-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 55.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 56.Bond CS, Schuttelkopf AW. ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr D. 2009;65:510–512. doi: 10.1107/S0907444909007835. [DOI] [PubMed] [Google Scholar]
- 57.Lomize AL, Pogozheva ID, Lomize MA, Mosberg HI. Positioning of proteins in membranes: a computational approach. ProteinSci. 2006;15:1318–1333. doi: 10.1110/ps.062126106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buse G, Soulimane T, Dewor M, Meyer HE, Bluggel M. Evidence for a copper-coordinated histidine-tyrosine cross-link in the active site of cytochrome oxidase. Protein Sci. 1999;8:985–990. doi: 10.1110/ps.8.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.