Abstract
One of the primary pathways for removal of DNA damage is nucleotide excision repair (NER). In bacteria, the UvrA protein is the component of NER that locates the lesion. A notable feature of NER is its ability to act on many DNA modifications that vary in chemical structure. So far, the mechanism underlying this broad specificity has been unclear. Here, we report the first crystal structure of a UvrA protein in complex with a chemically modified oligonucleotide. The structure shows that the UvrA dimer does not contact the site of lesion directly, but rather binds the DNA regions on both sides of the modification. The DNA region harboring the modification is deformed, with the double helix bent and unwound. UvrA uses damage-induced deformations of the DNA and a less rigid structure of the modified double helix for indirect readout of the lesion.
DNA damage can occur spontaneously or be caused by mutagenic chemicals and physical factors such as radiation and sunlight. These modifications must be corrected to avoid detrimental effects to the cell. One of the primary pathways of DNA repair is NER, in which a stretch of bases harboring the lesion is removed, and the gap is filled by a DNA polymerase. The unique feature of NER is its ability to detect and correct a wide spectrum of DNA modifications of different sizes and chemical structures. NER can target single base modifications, bulky adducts, backbone modifications and inter- or intrastrand cross-links1. NER was first described in bacteria2, and its key genes were identified and named uvrA, uvrB and uvrC3. These proteins are used only by bacteria and some archaea, but the principles of NER are the same in all three kingdoms of life. In eukaryotes, a larger number of proteins are used, and only a few have similarities with the bacterial system4. In humans, mutations in NER genes lead to several diseases, such as xeroderma pigmentosum with extreme susceptibility to ultraviolet radiation and an increased risk of skin cancer5, Cockayne syndrome with impaired development, premature aging and sunlight sensitivity and trichothiodystrophy with impaired development and mental retardation6.
The first step in NER is damage detection, which is carried out by the UvrA or UvrA–UvrB complex in bacteria1. The first contacts with the DNA occur through the UvrA protein. When the damage is located, the DNA is handed over to the UvrB protein for damage verification. UvrC nuclease is then recruited and cleaves the DNA at the fourth or fifth phosphate 3′ to the lesion and at the eighth phosphate 5′ to the lesion7. UvrD helicase removes the excised oligonucleotide, and polymerase I fills the gap. The repair is completed by DNA ligase I, which seals the nick.
The UvrA, UvrB and UvrC proteins have been extensively characterized biochemically. The crystal structures of UvrB show that the protein adopts a helicase fold similar to that of PcrA8-10. The characteristic feature of the structure is a β-hairpin inserted between the two DNA strands to clamp the substrate11. UvrC has two separate nuclease domains: an N-terminal domain resembling GIY-YIG homing endonuclease12,13 that is responsible for 3′ cleavage and a C-terminal domain that adopts the RNase H fold and executes the 5′ cleavage14.
UvrA is a dimeric protein that belongs to the ATP-binding cassette (ABC) family of ATPases, together with transporters15,16 and MutS DNA repair protein17,18. Crystal structures of Bacillus stearothermophilus UvrA (Bst-UvrA) and Deinococcus radiodurans UvrA2 (Dr-UvrA2) have recently been reported19,20. UvrA contains two ATP-binding domains, I and II. Signature domains I and II are inserted into the corresponding ATP-binding domains. The first signature domain also contains two additional insertions, one responsible for UvrB binding19,21 and the other for DNA binding20. The two composite ATPase active sites are formed between ATP-binding domain I and signature motif II (that is, the proximal site) and between ATP-binding domain II and signature motif I (that is, the distal site). Three structural zinc-binding elements are present in the structure.
UvrA is hypothesized to be the first NER component to detect the DNA lesion, but its mechanism of DNA binding and damage recognition has been unclear. The key unanswered question has been how the markedly wide specificity of UvrA for different types of DNA lesions is achieved. Here we report a crystal structure of Thermotoga maritima UvrA in complex with modified DNA that indicates a mechanism of indirect readout in which UvrA detects DNA modification through deformations of the double helix.
RESULTS
Protein characterization and crystallization
To solve the crystal structure of the UvrA protein in complex with chemically modified DNA, we cloned and purified T. maritima UvrA (Tm-UvrA). We then used the malachite green colorimetric assay to verify that Tm-UvrA has ATPase activity modulated by DNA binding (Supplementary Table 1). The KM for ATPase activity of Tm-UvrA decreased by 37% upon the addition of a modified 32-mer oligonucleotide and decreased by 27% upon the addition of a corresponding unmodified oligonucleotide. The kcat for Tm-UvrA activity was also slightly reduced in the presence of DNA. For Escherichia coli UvrA, the reported KM was similar to the one we measured for Tm-UvrA, and it was also reduced upon the addition of the DNA22,23. In contrast, the kcat values were lower for Tm-UvrA than for the E. coli enzyme by a factor of 4 to 17 (ref. 23). The difference can be explained by the fact that our assays were carried out at 50 °C to ensure DNA duplex stability, and this temperature may be lower than the optimum temperature for Tm-UvrA activity.
For crystallization of Tm-UvrA in complex with the DNA, we used oligonucleotides that contained a fluorescein modification of a thymine residue (Supplementary Fig. 1). This modification is efficiently repaired by NER24 and has been used in biochemical and structural studies of NER proteins11,25. We used nonpalindromic oligonucleotides that contained the modification in only one strand of double-stranded DNA (dsDNA) and palindromic (self-annealing) oligonucleotides that contained symmetrically positioned fluorescein modifications in both strands of the double helix. The crystal structures of UvrA showed that the core of its dimer has two-fold symmetry19,20. We hypothesized that the palindromic DNA would match this symmetry, which would favor crystallization. Without nucleotides, the palindromic 32-base pair (bp) oligonucleotide was bound by Tm-UvrA with a Kd of 1.9 nM, whereas the corresponding unmodified DNA bound with an affinity reduced by a factor of six (Fig. 1a,b). The Kd for the binding of the fluorescein-modified oligonucleotide is very similar to the values reported for binding of various modified DNAs by other UvrA proteins (reviewed in ref. 26). We also tested whether the palindromic duplex with closely positioned fluorescein modifications in each strand can be processed by the complete NER machinery comprising UvrA, UvrB and UvrC proteins. Such processing should lead to two cleavages: one on the 5′ side of the modification and another one on the 3′ side7. UvrA covers about 30–34 bp on the DNA, so to allow loading of the UvrB onto the duplex, we added 10 bp to the 32-bp palindromic oligonucleotide either on one or on both ends. The oligonucleotides were radiolabeled on either the 5′ or the 3′ end and treated with UvrA, UvrB and UvrC proteins. The 52-bp duplex with an additional 10 bp on both ends (10-32-10) underwent only the 5′ incision (Fig. 1c–e). UvrB molecules could be loaded on this oligonucleotide from both ends, which would cause a steric hindrance inhibiting the 3′ incision. In contrast, the 42-bp oligonucleotide with 10 bp added only on one end (10-32) underwent both the 5′ and the 3′ incision at the expected positions (Fig. 1c–e). The cleavages occurred only in the strand that was extended on its 5′ end by the addition of the 10 bp. Even though the other strand also contained the modification, it was not cleaved, probably because it lacked the additional 5′ sequence. These results are in agreement with previous work showing that the loading of UvrB to form the preincision complex starts on the 5′ side of the lesion27. In conclusion, oligonucleotide with a palindromic segment harboring two fluorescein modifications in both strands can be processed by the NER machinery comprising UvrA, UvrB and UvrC.
Figure 1.
Binding and processing of the palindromic DNA substrate. (a) DNA binding by Tm-UvrA. Binding curves were determined in filter binding assays. Values are mean ± s.e.m. of four independent measurements. Modified DNA was the palindromic 32-mer with fluorescein modification in position 14 used for crystallization (see Online Methods for sequence). The unmodified oligonucleotide had the same sequence and length but did not contain the fluorescein moiety. WT, wild type. (b) Kd values determined in the filter binding assays. (c,d) Incision assay. The oligonucleotides were radiolabeled on 5′ end (c) or 3′ end (d). Oligonucleotides added to each reaction are specified for 10-32, the labeled strand is indicated. UvrA, UvrB and/or UvrC were added to the reaction as indicated and the reaction products were resolved on the TBE-urea denaturing PAGE. DNA was visualized by phosphorimaging. Positions of DNA size markers are at right of each gel (nt, nucleotides). (e) Sequences of oligonucleotides used in incision assays. Sequences added to the 32-mer palindromic duplex are underlined. Fluorescein-modified thymines are boxed. Arrows indicate observed cleavage sites.
In supporting of our initial hypothesis, our extensive crystallization trials only yielded crystals of protein–nucleic acid complex with the self-annealing (palindromic) 32-bp oligonucleotide. After optimization of its length and the position of the modification, we produced crystals that diffracted X-rays to 2.9 Å. The structure was solved using molecular replacement and refined (Fig. 2 and Table 1). Samples of electron density maps are shown in Supplementary Figure 2. The asymmetric unit contains a protein monomer and one strand of DNA. The UvrA dimer and double-stranded nucleic acid are generated by the crystal symmetry. The structure of the Tm-UvrA protein closely resembles previously reported structures of Bst-UvrA and Dr-UvrA2 (refs. 19, 20) (both solved with bound ADP). No nucleotides are present in our structure, but we observed strong oblong electron densities at both active sites, which were refined as pyrophosphate ions. These ions are probably impurities of the orthophosphate present at 0.2 M in our crystallization conditions. At this concentration, even trace amounts of pyrophosphate are stoichiometric to UvrA.
Figure 2.
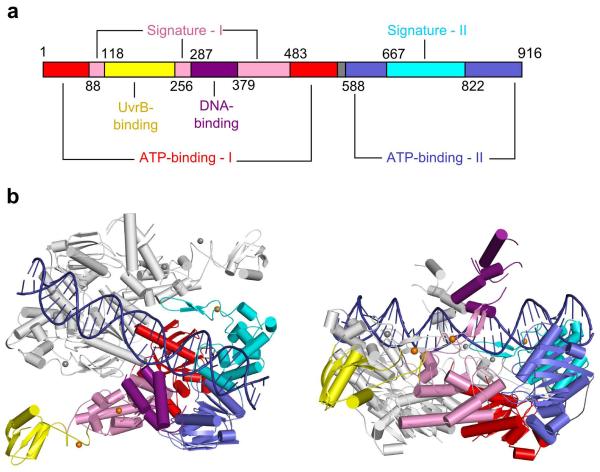
Structure of complex. (a) Domain organization of UvrA protein. The domain names are the same as in reference 19. Numbers indicate Tm-UvrA residues at the domain boundaries. (b) Two views of the structure of the Tm-UvrA–DNA complex. Protein is in ribbon representation, and domains of one subunit of the dimer are colored as in a. DNA, blue. Structural zinc ions, orange and gray spheres.
Table 1. Data collection and refinement statistics.
| Tm-UvrA–DNA | |
|---|---|
| Data collection | |
| Space group | P42 |
| Cell dimensions | |
| a, b, c (Å) | 107.5, 107.5, 108.3 |
| Resolution (Å) | 50–2.9 (2.95–2.9)* |
| R merge | 0.06 (0.41) |
| I / σI | 27.4 (2.1) |
| Completeness (%) | 96.0 (62.7) |
| Redundancy | 5.4 (2.4) |
| Refinement | |
| Resolution (Å) | 2.9 |
| No. reflections | 25,333 |
| Rwork / Rfree | 19.8 / 26.5 |
| No. atoms | 7,093 |
| Protein | 6,395 |
| Ligand/ion | 674 |
| Water | 24 |
| B-factors | 65.2 |
| Protein | 64.2 |
| Ligand/ion | 75.5 |
| Water | 54.6 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.05 |
Values in parentheses are for highest-resolution shell.
DNA binding
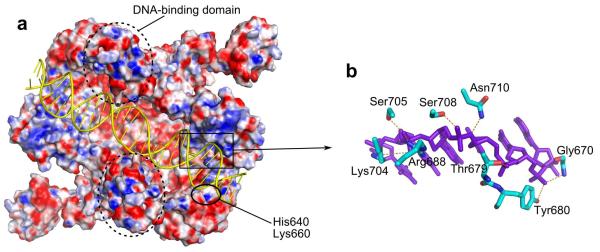
In the Tm-UvrA-DNA complex, the nucleic acid binds in a cleft running across the protein dimer (Figs. 2 and 3a), in a region accurately predicted from the structure of the Dr-UvrA2–ADP complex20. Tm-UvrA interacts only with the DNA backbone, leading to non-sequence-specific binding. The protein forms the most extensive interactions with the terminal regions of the oligonucleotide (Fig. 3a). The most important DNA contact seems to be a patch comprising eight residues, Gly670, Thr679, Tyr680, Arg688, Lys704, Ser705, Ser708 and Asn710, which interact with four consecutive nucleotides of the DNA (Fig. 3b and Supplementary Fig. 2a). Most of these residues are strongly conserved in evolution (Supplementary Fig. 3). To verify the importance of this patch, we prepared expression constructs for three Tm-UvrA proteins with substitutions in this region, but only the TY679AA variant could be expressed in E. coli. The affinity of TY679AA protein for fluorescein-modified DNA was reduced by a factor of nine compared with wild-type protein, and its affinity for the unmodified duplex was even lower, confirming the role of the patch in nucleic acid binding (Fig. 1a,b). An equivalent of Arg688 was substituted in Bst-UvrA together with three other basic residues, and this altered protein also had defective DNA binding19. In addition to the patch, another potential protein-DNA contact is formed by His640 and Lys660 from ATP-binding domain II. Indeed, substitutions of the equivalent of Lys660 in Bacillus caldotenax UvrA (Bca-UvrA) decreased the affinity for DNA28. In contrast, H640A substitution in Tm-UvrA did not affect DNA binding (data not shown).
Figure 3.
DNA binding. (a) Surface representation of Tm-UvrA with surface potential in red (negative) and blue (positive) (±20 kT e−1). DNA is yellow. (b) Details of key protein-DNA interactions mediated by signature domain II. DNA, purple; protein residues, cyan.
The DNA-binding insertion domain is located close to the midpoint of the DNA substrate and seems to clamp it from the sides. The insertion domain is involved in DNA binding: its deletion in the UvrA2 protein leads to a loss of damage-specific binding20. We do not observe any strong and specific contacts between the DNA-binding domain and the nucleic acid. In our structure, this domain forms crystal contacts with the UvrB-binding domain of a symmetry-related molecule. We cannot exclude the possibility that the crystal contacts affect the positioning of the DNA-binding domain and its interactions with the DNA. On the other hand, the sequence conservation on the side of this domain, which faces the DNA, is rather low, and the domain may form only nonspecific interactions with the nucleic acid.
DNA conformation and damage detection
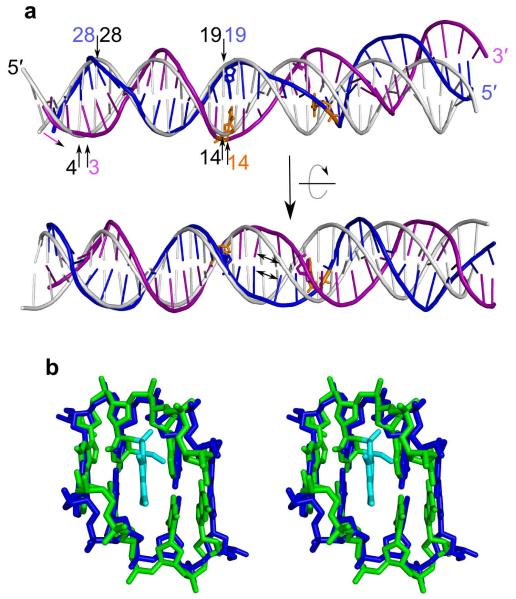
The substrate used for crystallization is self-annealing. Therefore, its structure is symmetrical, with a two-fold axis perpendicular to the double helix going through its midpoint. Each of the symmetrically positioned fluorescein-modified thymine residues (position 14) are 2 bp away from this midpoint (Fig. 4a). The modified bases are slightly pushed out from the double helix, leading to higher stretch (distance between bases) and shear (shift of the bases in the plane) values. This weakening of base-pairing of the modified residues may lead to the general destabilization of the double helix between the two fluorescein-dT moieties. In our structure, this region of the DNA has higher B-factors and less well defined electron densities of the phosphodiester backbone, suggesting mobility.
Figure 4.
Deformation of the DNA. (a) Two views of the DNA from structure of the Tm-UvrA–DNA complex (purple and blue) superimposed on the model of ideal B-form DNA (white) of identical length and sequence. Superimposition was carried out using the positions of the phosphate groups of residues 20–29 from the strand in blue. Fluorescein-modified bases, orange sticks. Numbers of selected residues in both models (blue, purple and orange for DNA from the Tm-UvrA–DNA complex and black for the ideal DNA model) indicate the unwinding of the strand shown in purple. Trailing of the DNA, purple arrow. Unstacking of central bases in DNA from the Tm-UvrA–DNA complex, double-headed arrows. (b) Stereoview showing midpoint of DNA from Tm-UvrA–DNA complex superimposed on NMR solution structure of a psoralen monoadduct (DNA, green; modification, cyan; PDB 203D)30. DNA from the Tm-UvrA–DNA structure is blue.
The DNA is bent in the middle by ~15° (Fig. 4a and Supplementary Movies 1 and 2). The values of the bend angles induced by various lesions have been compiled26,29. They depend greatly on the type of lesion and vary from 0° (straight helix) up to 50°. The bend observed in the Tm-UvrA–DNA structure is smaller than for most modified DNAs in free form. The other evident DNA distortion is a stretching of the helix and an unstacking observed between two nucleotides in the center (residues 16 and 17). The rise for this step is ~6.5 Å, which is almost double the value for regular dsDNA. We observed an electron density between the unstacked bases that corresponded with a flat molecule intercalated into the DNA (Supplementary Fig. 2b). No such flat molecules were present in the crystallization solution, and this density could be interpreted as the triple aromatic ring of fluorescein covalently tethered to T14. Fluorescein intercalation into single-stranded DNA has been shown in the structure of UvrB in complex with DNA carrying the same dT-fluorescein modification11. The flat electron density in our structure, however, is not very well defined and the single aromatic ring of fluorescein and the flexible linker are not visible in the electron density maps, probably owing to their disorder. Therefore, although the density could correspond to the fluorescein molecule, we chose not to build a model into it.
The midpoint of the DNA in the structure of the Tm-UvrA–DNA complex is similar to NMR solution structures of DNA adducts of psoralen30 (Fig. 4b) and some polycyclic aromatic hydrocarbons (PAHs)31. The flat rings of psoralen or PAHs intercalate into the DNA, causing stretching and unstacking of the DNA in a manner similar to that in our structure. Both psoralen modification and covalently tethered fluorescein are efficiently recognized by UvrA and repaired by bacterial NER, and both have been extensively used as model modifications in studies of nucleotide excision repair (reviewed in ref. 1). Another deviation from the regular duplex we observed in the DNA from the structure of the Tm-UvrA–DNA complex is a widened minor groove at the midpoint, with a width of ~10 Å compared with 7.4 Å for regular B-form DNA. All described DNA deformations are coupled with unwinding of the double helix (Fig. 4 and Supplementary Movies 1 and 2). On the basis of the sum of the twist angle values32, the middle portion of the DNA (unstacked bases and five steps on both sides) is unwound by ~20°. Many types of DNA damage (for example, psoralen and cisplatin adducts and photoproducts) cause unwinding of DNA in its free unbound form (refs. 29 (and references there in),33,34). The unwinding angle for different lesions varies, and for the majority of them falls between 10° and 30° (refs. 26,29). The angle we observed in our structure is within this range. Because of the different values of unwinding angle observed for various modifications recognized by UvrA, the unwinding has to be adjusted during binding by the protein so that the DNA conformation can be complementary to the surface of UvrA.
Although many modifications induce DNA bending, unwinding seems to be the universal feature of DNA lesions. Therefore, unwinding of DNA has been suggested to be used by NER for damage recognition29. NMR studies have also shown that damaged DNA has more conformational heterogeneity than regular DNA and therefore higher deformability35, which also seems to be the case for the midpoint of the DNA in the Tm-UvrA structure. UvrA may detect the deformations induced by the DNA damage itself (for example, in the case of psoralen or PAH-DNAs; Fig. 4b), but it may also sense the less rigid structure of the region around the lesion by inducing unwinding and bending that are complementary to the protein surface. Biochemical experiments have shown that UvrA can unwind DNA: a change in the plasmid’s linking number has been observed after its incubation with UvrA and topoisomerase36. In conclusion, our structure supports the hypothesis that UvrA uses an indirect readout mechanism to recognize a wide range of DNA modifications.
Comparison with ADP-bound structures of UvrA
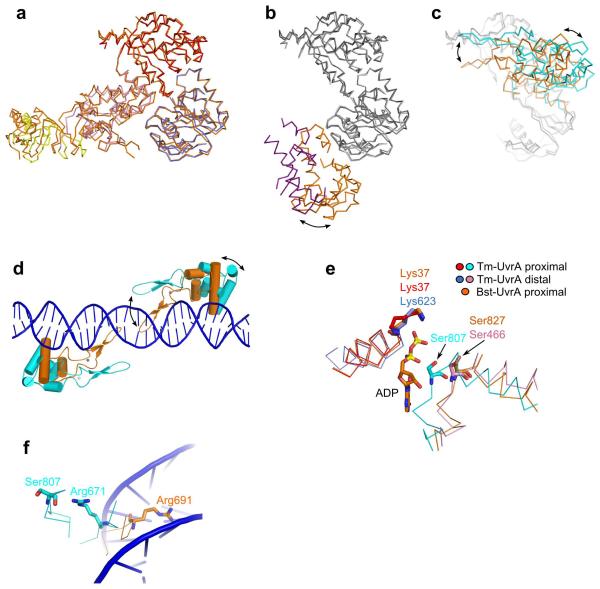
When the Bst-UvrA–ADP structure and our Tm-UvrA–DNA structure are superimposed using the positions of Cα atoms from ATP-binding domains I and II (308 of 340 observed pairs of atoms were used in the superimposition), the r.m.s. deviation is only 0.74 Å, indicating that the ATP-binding domains form a rigid core of the structure (Fig. 5a). The dimer interface also does not differ substantially between the two structures. The positions of signature domain I and its insertion, which is responsible for UvrB binding, are only slightly different (Fig. 5a). However, the position of the DNA-binding insertion domain differs substantially (Fig. 5b). As we indicated above, this difference can be caused by DNA binding, but crystal contacts can also contribute to it. In the previously reported UvrA and UvrA2 structures, the position of the DNA-binding domain was the most varied, indicating its general mobility19,20. Signature domain II also has a different position in DNA-free and DNA-bound UvrA structures (Fig. 5c). In the Tm-UvrA–DNA structure, the signature domains II from the two subunits of the dimer are farther away from each other, and the conformation of the dimer is more open (Fig. 5d). Many important DNA interactions are mediated by signature domain II, and only the open dimer conformation is complementary to the deformed unwound DNA observed in our structure. Biochemical and atomic force microscopy studies show that nucleotides bound at the active sites influence damage discrimination. UvrA in nucleotide-free form (such as in our structure of the complex) or with a mixture of ATP and ADP has the highest binding preference for damaged DNA over the regular duplex37-39. In the presence of ADP, the specificity is lower, and the binding is even less specific in the presence of non-hydrolyzable analogs of ATP. These effects could be explained by ‘locking’ of one of the conformations of signature domain II upon binding of a particular nucleotide. The functional significance of ATP binding and hydrolysis and the role of the resulting conformational changes in DNA damage discrimination by UvrA are not entirely clear. More structures in different ligand-bound states need to be determined to elucidate these mechanisms.
Figure 5.
Comparison of Bst-UvrA–ADP and Tm-UvrA–DNA structures. (a) Structure of Tm-UvrA–DNA complex (Tm-UvrA–DNA colored by domain as in Fig. 2, Bst-UvrA–ADP in orange) was superimposed on the Bst-UvrA structure using the positions of Cα atoms of the two ATP-binding domains. Only the most invariant core of the structures, ATP-binding domains I and II, signature domains I and UvrB-binding domains, are shown. Only one subunit of the dimer is shown for clarity. (b) Comparison of positions of DNA-binding domains (purple, Tm-UvrA–DNA; orange, Bst-UvrA–ADP). ATP-binding domains are light gray for Tm-UvrA and dark gray for Bst-UvrA. (c) Comparison of position of signature II domains (cyan, Tm-UvrA–DNA; orange, Bst-UvrA–ADP). In a–c the structures are in the same orientation. (d) Position of signature II domains from Bst-UvrA–ADP structure (orange) and Tm-UvrA–DNA structure (cyan) relative to DNA from the latter model. The zinc finger, its loop and the helical core of the domains’ structure are in ribbon representation for both subunits of the dimers. Zinc ions are gray spheres with darker shade for Bst-UvrA. The DNA is blue. (e) Close-up of ATP-binding sites from Tm-UvrA (domains colored as in Fig. 2) and Bst-UvrA (orange). Backbone trace of helices containing Walker A motif (left) and signature motif (right) is in ribbon representation. Catalytic lysine from Walker A motif, conserved serine from signature motif and ADP from Bst-UvrA structure are sticks. (f) Position of conserved arginine located near the of DNA interface and proximal active site. Protein structure is colored as in e.
The third zinc finger in the UvrA sequence is inserted into signature domain II. Different positioning of this domain in ADP-bound and DNA-complex structures leads to a large difference in the position of this zinc finger and its loop: the position of the tip of the loop differs by 21 Å (Fig. 5d). In the DNA complex, the zinc finger is located away from the nucleic acid and does not form any contacts with it. In contrast, when DNA from our Tm-UvrA structure is modeled into the APD-bound structure, the zinc finger and its loop clash with the DNA backbone. Deletion of the zinc finger loop increases the affinity for fluorescein-adducted DNA but at the same time leads to a loss of specificity for the modified DNA40. To explain these results, we are tempted to speculate that undeformed DNA, which is not stretched in the middle and which is shorter, could bind to the closed dimer conformation observed in the Bst-UvrA–ADP structure. However, the closed conformation would lead to clashes of the third zinc finger and its loop with the undeformed and/or undamaged DNA, reducing its binding. Deletion of the finger’s loop would remove some of the clashes, allowing the binding of undeformed and/or undamaged DNA. Therefore, the zinc finger could serve as an additional element ensuring specific binding of the damaged and deformed DNA.
Another difference between the DNA-free and DNA-bound structures is the conformation of the signature motif loop forming the ATPase active site (Fig. 5e). In the proximal active site of the structure of the DNA complex, the loop is closer to the nucleotide-binding site; this could lead to its tighter binding and might explain the lower KM values for ATPase activity we observed in the presence of the DNA (Supplementary Table 1). An additional variation between the two structures is the positioning of Arg671 (Fig. 5f and Supplementary Fig. 3). An equivalent of this residue was substituted in B. caldotenax UvrA, leading to a reduction in its DNA affinity by a factor of three28. Arg671 is located in a loop in the vicinity of the proximal active site, and in the Tm-UvrA–DNA complex this loop is inserted into the minor groove of the DNA, forming van der Waals contacts with the nucleic acid. The side chain of Arg671 points away from the DNA (Fig. 5f), so it does not participate in the binding of nucleic acid directly, but rather stabilizes the conformation of the DNA-interacting loop. The side chain of Arg671 interacts with the signature motif loop and seems to stabilize its conformation. We therefore suspected that it might be an additional element of the mechanism coupling the DNA binding to the modulation of the ATPase activity. However, the kinetic parameters of the R671A variant of Tm-UvrA in the presence and absence of DNA were indistinguishable from the wild-type protein (data not shown). Therefore, Arg671 is not essential for the coupling of DNA binding and ATP hydrolysis.
DISCUSSION
Our structure provides the first glimpse into the mechanism of DNA damage recognition by the bacterial NER protein UvrA and indicates a unique mechanism of damage sensing. DNA repair enzymes for which structures of DNA complex are available form specific interactions with the lesion site and very often flip out modified bases. One example is yeast protein Rad4 and its human ortholog XPC, which are the functional equivalents of UvrA in eukaryotes. Together with Rad23 they form a complex that detects DNA damage. According to crystal structures, Rad4 inserts a β-hairpin between the two DNA strands, leading to a flipping out of two bases opposite the lesion41. The flipped-out bases are stabilized by extensive contacts with the protein, but the lesion itself does not interact with Rad4 and is disordered in solution, allowing the protein to accommodate various bulky adducts. The flipping out leads to a distortion of the geometry of the double helix: bending by ~40°, stretching and unwinding. UV-DDB is another eukaryotic NER complex comprising the DDB1 and DDB2 proteins, which are important for the repair of photoproducts. Similarly to Rad4, UV-DDB uses a hairpin in DDB2, which inserts itself into the double helix of the DNA and flips out two modified bases42. In contrast to Rad4, UV-DDB binds the flipped-out modified bases in a lesion-binding pocket, allowing for higher specificity for particular types of lesions. The DNA is unwound by ~20° and bent by 40°. For both Rad4 and UV-DDB the key element of lesion detection is the sensing of weakened base-pairing and stacking, which facilitate the insertion of β-hairpins into the double helix41,42.
The mechanism of action of UvrA is different. The modified region of the DNA does not form specific contacts with the protein. Instead, the conformation of the double helix is probed by protein interactions on both sides of the damage site. The dimeric structure of UvrA, which is unique among DNA repair enzymes, is ideally suited for this purpose. In contrast to Rad4 and UV-DDB, UvrA does not use base-flipping, and the base-pairing in the DNA from the Tm-UvrA–DNA complex is maintained. UvrA does not seem to probe the stability of the base interactions but rather senses unwinding and bending of the DNA and the deformability of the global conformation of the double helix. This indirect readout mechanism allows UvrA to detect various DNA lesions and achieve broad specificity. A complication in this mechanism is that it does not distinguish which of the two DNA strands is damaged and should be incised. This role is probably played by UvrB, which verifies the presence of the damage and loads the UvrC nuclease, directing its cleavage to the modified strand.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/nsmb/.
ONLINE METHODS
Crystallization and structure determination
Protein preparation is described in detail in Supplementary Methods. Tm-UvrA was produced either in His-tagged or untagged version. HPLC-purified oligonucleotides were purchased from Metabion. Before crystallization the untagged protein was mixed with the oligonucleotide at a molar ratio of 1.2:1 substrate/protein dimer. The final protein concentration was 5.5 mg ml−1. The complexes were mixed with the reservoir solution at equal volume and crystallized by sitting-drop vapor diffusion at 18 °C. The length of the oligonucleotide for crystallization was selected based on published DNase I footprinting data38. The first crystals were grown with 30-mer palindromic dsDNA with fluorescein-dT in various positions but they did not diffract X-rays, so we carried out systematic optimization of the length of the oligonucleotides and the position of fluorescein modification. In the end we obtained two crystal forms of Tm-UvrA with a 32-mer DNA containing a fluorescein-modified thymine in position 14 (5′-AGTGATCAGTGGTXC CGGAACCACTGATCACT-3′, where X denotes the position of the modified thymine). They grew in 49–52% (v/v) 2-methyl-2,4-pentanediol (MPD), 0.1–0.2 M ammonium phosphate and 0.1 M Tris, pH 8.5. The first form belonged to space group C2 and diffracted X-rays poorly, to a resolution of ~5 Å. The second crystal form grew in the same condition but belonged to space group P42. The first diffraction data from these crystals were collected at a microfocus beamline 23-2 at ESRF and the 2.9 Å data set at beamline 14-1 at BESSY. The diffraction data were processed using HKL2000 (ref. 43). The structure of the complex was solved using the molecular replacement method using the Bst-UvrA structure (PDB 2R6F)19 and BALBES software44. Initially, only the two ATP-binding domains and one signature domain were found. The rest of the protein and the model of nucleic acid were built manually in Coot45. The resulting model was refined using phenix.refine46 interspersed with manual building in Coot. Several regions of the protein, located mostly in the mobile DNA- and UvrB-binding insertions, could not be traced because of the lack of interpretable electron density (residues 61–68, 155–202, 294–303, 330–336, 349–350 and 366–368). In the final model 92% of the residues are in the most favored region of the Ramachandran plot and 0.7% are in the disallowed region as defined by MolProbity47. Structure analyses were carried out in PyMol (Schrödinger). Surface potentials were calculated with APBS48. Figures were prepared using PyMol. Morphing movies were created in Chimera49 and PyMol. The DNA conformation was analyzed in Curves+50.
ATPase activity assay
ATPase activity assays were carried out using His-tagged Tm-UvrA and malachite green colorimetric method51. The reactions were carried out in buffer containing 20 mM HEPES, pH 7.0, 75 mM NaCl, 5% (v/v) glycerol, 1 mM DTT and 10 mM MgCl2. ATP concentration was varied from 0 to 1 mM, the UvrA active site concentration was 1 μM and the DNA concentration was 2 μM. Reaction mixtures were incubated at 50 °C and 25 μl aliquots were with-drawn at selected time points. Perchloric acid (25 μl of 0.6 M) was added to stop the hydrolysis reaction. Then, an equal volume of malachite green solution was added. A655 was measured in 96-well plates using a plate reader. Phosphate concentration was calculated using standard curve of potassium phosphate (monobasic), measured for each experiment. Km and kcat values were calculated by curve fitting of the data in GraphPad Prism 5.0 (GraphPad Software) using the equation:
where vmax is the maximum velocity and c is substrate concentration. kcat was calculated by dividing the νmax value by the concentration of the enzyme’s catalytic sites. Reported values are an average of five independent measurements.
Nitrocellulose filter-binding assay
Equilibrium binding constants were determined in a filter binding assay52. Substrate oligonucleotides were radiolabeled with [γ-33P]ATP (Hartmann Analytic) and T4 polynucleotide kinase (Fermentas). His-tagged Tm-UvrA–DNA mixtures (50 μl total volume in 150 mM NaCl, 20 mM HEPES, pH 7.0, 5% (v/v) glycerol, 1 mM DTT, 0.5 mM EDTA) containing a constant concentration of oligonucleotide (100 pM) and varying concentration of protein (0.5–50 nM) were incubated for 30 min at 25 °C. No nucleotide was added to the mixture. Reaction mixtures were filtered through a 0.22-μm nitrocellulose filter (Whatman) in a Dot-Blot apparatus (Bio-Rad). Each well was washed three times with 200 μl of binding buffer. Dried filters were exposed to a phosphorimager screen overnight. Images were scanned on a Storm Phosphorimager (GE Healthcare) and retained radioactivity was quantified using ImageQuant software (GE Healthcare). The data were fitted with GraphPad Prism 5.0 to a one-site binding equation:
where c is the protein concentration, Bmax is the maximum binding and Kd is the dissociation constant.
Incision assay
Oligonucleotides (for sequences, see Fig. 1e) were radioactively labeled on the 5′ end as described above. For 3′ end labeling, 20 U of terminal deoxynucleotidyl transferase (Fermentas) and [α-32P]GTP (Hartmann Analytic) were added to the oligonucleotides and the reaction was incubated at 37 °C for 30 min. The oligonucleotides were next treated with 1,000 U of RNase T1 (Fermentas). Both 5′ and 3′ end labeled oligonucleotides were purified on desalting spin columns (Roche). The 3′ end labeled oligonucleotides were additionally purified by TBE-urea PAGE.
The incision assay was done essentially as described53 and a fluorescein-modified 50-mer oligonucleotide (F2650) from that study was used as a positive control. Before the initiation of incision assay, oligonucleotides (2 nM) were incubated with untagged Tm-UvrA (20 nM) and T. maritima UvrB (100 nM) at 65 °C for 30 min in a buffer containing 50 mM Tris-HCl, pH 7.5, 50 mM KCl, 10 mM MgCl2, 1 mM ATP and 5 mM DTT. T. maritima UvrC was next added to a final concentration of 50 nM and the reactions were continued for 2 h at 55 °C. The reactions were stopped by addition of an equal volume of the sample buffer (formamide, 10 mM EDTA, xylene cyanol F, bromophenol blue) and heating at 95 °C for 5 min. The reaction products were resolved on 20% (w/v) TBE-urea denaturing polyacrylamide gels and visualized by phoshorimaging using the FLA-7000 system (Fuji).
Supplementary Material
ACKNOWLEDGMENTS
We thank T. Sixma and J. Bujnicki for critical reading of the manuscript, M. Rychlik, M. Cybulska and J. Dyttus for excellent technical assistance, the staff of beamline 23-2 at European Synchrotron Radiation Facility (ESRF) and beamline 14-1 at Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung (BESSY) for assistance with data collection, K. Górecka and M. Figiel for their help with data collection and P. Afonine and S. Ramón Maiques for help with structure refinement. This work was supported by a European Molecular Biology Organization Installation Grant to M.N. The access to ESRF was financed by the Polish Ministry of Science and Higher Education (project no. ESRF/73/2006). The research leading to these results has also received funding from the European Community’s Seventh Framework Program under grant agreement no. 226716.
Footnotes
Accession codes. Protein Data Bank: Atomic coordinates have been deposited with accession code 3PIH.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
AUTHOR CONTRIBUTIONS
M.J. purified Tm-UvrA, obtained its complex crystals and carried out the biochemical experiments. K.S. and E.N. carried out the biochemical experiments. A.T. carried out the initial expression and purification studies. M.N. collected the data. M.J., E.N. and M.N. refined and analyzed the structure. M.N. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Truglio JJ, Croteau DL, Van Houten B, Kisker C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem. Rev. 2006;106:233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 2.Boyce RP, Howard-Flanders P. Release of ultraviolet light-induced thymine dimers from DNA in E. coli K-12. Proc. Natl. Acad. Sci. USA. 1964;51:293–300. doi: 10.1073/pnas.51.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howard-Flanders P, Boyce RP, Theriot L. Three loci in Escherichia coli K-12 that control the excision of pyrimidine dimers and certain other mutagen products from DNA. Genetics. 1966;53:1119–1136. doi: 10.1093/genetics/53.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sancar A, Reardon JT. Nucleotide excision repair in E. coli and man. Adv. Protein Chem. 2004;69:43–71. doi: 10.1016/S0065-3233(04)69002-4. [DOI] [PubMed] [Google Scholar]
- 5.Lehmann AR. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie. 2003;85:1101–1111. doi: 10.1016/j.biochi.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 6.Cleaver JE, Lam ET, Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat. Rev. Genet. 2009;10:756–768. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- 7.Sancar A, Rupp WD. A novel repair enzyme: UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell. 1983;33:249–260. doi: 10.1016/0092-8674(83)90354-9. [DOI] [PubMed] [Google Scholar]
- 8.Machius M, Henry L, Palnitkar M, Deisenhofer J. Crystal structure of the DNA nucleotide excision repair enzyme UvrB from Thermus thermophilus. Proc. Natl. Acad. Sci. USA. 1999;96:11717–11722. doi: 10.1073/pnas.96.21.11717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakagawa N, et al. Crystal structure of Thermus thermophilus HB8 UvrB protein, a key enzyme of nucleotide excision repair. J. Biochem. 1999;126:986–990. doi: 10.1093/oxfordjournals.jbchem.a022566. [DOI] [PubMed] [Google Scholar]
- 10.Theis K, Chen PJ, Skorvaga M, Van Houten B, Kisker C. Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair. EMBO J. 1999;18:6899–6907. doi: 10.1093/emboj/18.24.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Truglio JJ, et al. Structural basis for DNA recognition and processing by UvrB. Nat. Struct. Mol. Biol. 2006;13:360–364. doi: 10.1038/nsmb1072. [DOI] [PubMed] [Google Scholar]
- 12.Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Truglio JJ, et al. Structural insights into the first incision reaction during nucleotide excision repair. EMBO J. 2005;24:885–894. doi: 10.1038/sj.emboj.7600568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karakas E, et al. Structure of the C-terminal half of UvrC reveals an RNase H endonuclease domain with an Argonaute-like catalytic triad. EMBO J. 2007;26:613–622. doi: 10.1038/sj.emboj.7601497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hopfner KP, Tainer JA. Rad50/SMC proteins and ABC transporters: unifying concepts from high-resolution structures. Curr. Opin. Struct. Biol. 2003;13:249–255. doi: 10.1016/s0959-440x(03)00037-x. [DOI] [PubMed] [Google Scholar]
- 16.Linton KJ. Structure and function of ABC transporters. Physiology (Bethesda) 2007;22:122–130. doi: 10.1152/physiol.00046.2006. [DOI] [PubMed] [Google Scholar]
- 17.Lamers MH, et al. The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 18.Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 19.Pakotiprapha D, et al. Crystal structure of Bacillus stearothermophilus UvrA provides insight into ATP-modulated dimerization, UvrB interaction, and DNA binding. Mol. Cell. 2008;29:122–133. doi: 10.1016/j.molcel.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Timmins J, et al. Structural and mutational analyses of Deinococcus radiodurans UvrA2 provide insight into DNA binding and damage recognition by UvrAs. Structure. 2009;17:547–558. doi: 10.1016/j.str.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 21.Pakotiprapha D, Liu Y, Verdine GL, Jeruzalmi D. A structural model for the damage-sensing complex in bacterial nucleotide excision repair. J. Biol. Chem. 2009;284:12837–12844. doi: 10.1074/jbc.M900571200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh EY, Claassen L, Thiagalingam S, Mazur S, Grossman L. ATPase activity of the UvrA and UvrAB protein complexes of the Escherichia coli UvrABC endonuclease. Nucleic Acids Res. 1989;17:4145–4159. doi: 10.1093/nar/17.11.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thiagalingam S, Grossman L. The multiple roles for ATP in the Escherichia coli UvrABC endonuclease-catalyzed incision reaction. J. Biol. Chem. 1993;268:18382–18389. [PubMed] [Google Scholar]
- 24.DellaVecchia MJ, et al. Analyzing the handoff of DNA from UvrA to UvrB utilizing DNA-protein photoaffinity labeling. J. Biol. Chem. 2004;279:45245–45256. doi: 10.1074/jbc.M408659200. [DOI] [PubMed] [Google Scholar]
- 25.Waters TR, Eryilmaz J, Geddes S, Barrett TE. Damage detection by the UvrABC pathway: crystal structure of UvrB bound to fluorescein-adducted DNA. FEBS Lett. 2006;580:6423–6427. doi: 10.1016/j.febslet.2006.10.051. [DOI] [PubMed] [Google Scholar]
- 26.Van Houten B, Snowden A. Mechanism of action of the Escherichia coli UvrABC nuclease: clues to the damage recognition problem. Bioessays. 1993;15:51–59. doi: 10.1002/bies.950150108. [DOI] [PubMed] [Google Scholar]
- 27.Moolenaar GF, et al. The effect of the DNA flanking the lesion on formation of the UvrB-DNA preincision complex. Mechanism for the UvrA-mediated loading of UvrB onto a DNA damaged site. J. Biol. Chem. 2000;275:8038–8043. doi: 10.1074/jbc.275.11.8038. [DOI] [PubMed] [Google Scholar]
- 28.Croteau DL, DellaVecchia MJ, Perera L, Van Houten B. Cooperative damage recognition by UvrA and UvrB: identification of UvrA residues that mediate DNA binding. DNA Repair (Amst.) 2008;7:392–404. doi: 10.1016/j.dnarep.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellon SF, Coleman JH, Lippard SJ. DNA unwinding produced by site-specific intrastrand cross-links of the antitumor drug cis-diamminedichloroplatinum(II) Biochemistry. 1991;30:8026–8035. doi: 10.1021/bi00246a021. [DOI] [PubMed] [Google Scholar]
- 30.Spielmann HP. Dynamics in psoralen-damaged DNA by 1H-detected natural abundance 13C NMR spectroscopy. Biochemistry. 1998;37:5426–5438. doi: 10.1021/bi972536b. [DOI] [PubMed] [Google Scholar]
- 31.Suri AK, Mao B, Amin S, Geacintov NE, Patel DJ. Solution conformation of the (+)-trans-anti-benzo[g]chrysene-dA adduct opposite dT in a DNA duplex. J. Mol. Biol. 1999;292:289–307. doi: 10.1006/jmbi.1999.2974. [DOI] [PubMed] [Google Scholar]
- 32.Kabsch W, Sander C, Trifonov EN. The ten helical twist angles of B-DNA. Nucleic Acids Res. 1982;10:1097–1104. doi: 10.1093/nar/10.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park H, et al. Crystal structure of a DNA decamer containing a cis-syn thymine dimer. Proc. Natl. Acad. Sci. USA. 2002;99:15965–15970. doi: 10.1073/pnas.242422699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiesehahn G, Hearst JE. DNA unwinding induced by photoaddition of psoralen derivatives and determination of dark-binding equilibrium constants by gel electrophoresis. Proc. Natl. Acad. Sci. USA. 1978;75:2703–2707. doi: 10.1073/pnas.75.6.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Isaacs RJ, Spielmann HP. A model for initial DNA lesion recognition by NER and MMR based on local conformational flexibility. DNA Repair (Amst.) 2004;3:455–464. doi: 10.1016/j.dnarep.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 36.Oh EY, Grossman L. The effect of Escherichia coli Uvr protein binding on the topology of supercoiled DNA. Nucleic Acids Res. 1986;14:8557–8571. doi: 10.1093/nar/14.21.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazur SJ, Grossman L. Dimerization of Escherichia coli UvrA and its binding to undamaged and ultraviolet light damaged DNA. Biochemistry. 1991;30:4432–4443. doi: 10.1021/bi00232a009. [DOI] [PubMed] [Google Scholar]
- 38.Van Houten B, Gamper H, Hearst JE, Sancar A. Analysis of sequential steps of nucleotide excision repair in Escherichia coli using synthetic substrates containing single psoralen adducts. J. Biol. Chem. 1988;263:16553–16560. [PubMed] [Google Scholar]
- 39.Wagner K, Moolenaar G, van Noort J, Goosen N. Single-molecule analysis reveals two separate DNA-binding domains in the Escherichia coli UvrA dimer. Nucleic Acids Res. 2009;37:1962–1972. doi: 10.1093/nar/gkp071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Croteau DL, et al. The C-terminal zinc finger of UvrA does not bind DNA directly but regulates damage-specific DNA binding. J. Biol. Chem. 2006;281:26370–26381. doi: 10.1074/jbc.M603093200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 42.Scrima A, et al. Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell. 2008;135:1213–1223. doi: 10.1016/j.cell.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 44.Long F, Vagin AA, Young P, Murshudov GN. BALBES: a molecular-replacement pipeline. Acta Crystallogr. D Biol. Crystallogr. 2008;64:125. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Afonine PV, Grosse-Kunstleve RW, Adams PD. The Phenix refinement framework. CCP4 Newsl. 2005;42:8. [Google Scholar]
- 47.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 50.Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henkel RD, VandeBerg JL, Walsh RA. A microassay for ATPase. Anal. Biochem. 1988;169:312–318. doi: 10.1016/0003-2697(88)90290-4. [DOI] [PubMed] [Google Scholar]
- 52.Conlan LH, Dupureur CM. Dissecting the metal ion dependence of DNA binding by PvuII endonuclease. Biochemistry. 2002;41:1335–1342. doi: 10.1021/bi015843x. [DOI] [PubMed] [Google Scholar]
- 53.Jiang GH, Skorvaga M, Croteau DL, Van Houten B, States JC. Robust incision of Benoz[a]pyrene-7,8-dihyrodiol-9,10-epoxide-DNA adducts by a recombinant thermoresistant interspecies combination UvrABC endonuclease system. Biochemistry. 2006;45:7834–7843. doi: 10.1021/bi052515e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.