Summary
Autophagy is a fundamental biological process of the eukaryotic cell contributing to diverse cellular and physiological functions including cell-autonomous defense against intracellular pathogens. Here we screened the Rab family of membrane trafficking regulators for effects on autophagic elimination of Mycobacterium tuberculosis var. bovis BCG and found that Rab8b and its downstream interacting partner, innate immunity regulator TBK-1, are required for autophagic elimination of mycobacteria in macrophages. TBK-1 was necessary for autophagic maturation. TBK-1 coordinated assembly and function of the autophagic machinery and phosphorylated the autophagic adaptor p62 (sequestosome 1) on Ser-403, a residue essential for its role in autophagic clearance. A key pro-inflammatory cytokine, IL-1β, induced autophagy leading to autophagic killing of mycobacteria in macrophages and this IL-1β activity was dependent on TBK-1. Thus, TBK-1 is a key regulator of immunological autophagy and is responsible for the maturation of autophagosomes into lytic bactericidal organelles.
Introduction
Autophagy is a homeostatic process highly conserved in eukaryotic cells where it acts as a cytoplasmic biomass quantity and quality control system (Mizushima et al., 2011). Its functions encompass programmed cell survival and cell death, normally skewed toward cell survival through provision of energy and nutrients and ridding the cytoplasm of toxic macromolecular aggregates, faulty organelles (Mizushima et al., 2011) and invading microorganisms (Deretic, 2012b; Levine et al., 2011).
The cell-autonomous antimicrobial defense functions of autophagy, demonstrated initially in the case of streptococci (Nakagawa et al., 2004) and Mycobacterium tuberculosis (Gutierrez et al., 2004; Ponpuak et al., 2010), have been extended to a wide variety of microbes with a caveat that most highly adapted pathogens have evolved specific protective mechanisms against autophagic elimination of microbes (Deretic and Levine, 2009). Other studies have uncovered orderly intersections between autophagy and innate and adaptive immunity, T cell development, differentiation and homeostasis, and inflammatory responses (Deretic, 2012b; Levine et al., 2011). Autophagy suppresses endogenous, cell-autonomous promoters of inflammation. (Deretic, 2012b; Levine et al., 2011). Specific autophagic factors, such as Atg5-Atg12, have been shown to inhibit RIG-I receptor signaling (Jounai et al., 2007) whereas Atg9 has been reported to negatively regulate trafficking, assembly and activation of TBK-1 (TANK-binding kinase 1), which, among its key functions, controls type I interferon response elicited by intracellular double stranded DNA (Saitoh et al., 2009). In the context of anti-inflammatory function, autophagy plays an inhibitory role in inflammasome and IL-1β activation (Dupont et al., 2011; Nakahira et al., 2011; Zhou et al., 2011). Finally, a number of genetic links have been found in human populations between autophagy and idiopathic inflammatory or infectious diseases such as tuberculosis with significant inflammatory components and tissue damage (Deretic, 2012b; Levine et al., 2011).
Given the interconnectedness of autophagy and immunity, it is likely that the immune manifestations of autophagy are affected not only by the induction of autophagy but also by the completion of the autophagic pathway. The formation of the autophagic organelles of the sensu stricto autophagy pathway (also referred to as macroautophagy) depends on multiple sources of membrane and regulatory factors (Mizushima et al., 2011). The key stages of autophagy however are not restricted to the formation of autophagosomal membranes and include the sequestration of the earmarked cargo by the autophagic adaptors (Bjorkoy et al., 2005; Thurston et al., 2009; Wild et al., 2011), and the less understood process of the maturation of autophagic organelles into autolysosomes where the captured material is degraded (Korolchuk et al., 2011; Liang et al., 2008; Matsunaga et al., 2009; Zhong et al., 2009).
Here, we approached the far less studied processes governing autophagic flux by a systematic screening of Rabs, the central regulators of membrane trafficking and organellar identity in eukaryotic cells (Stenmark, 2009). We showed that Rab8b and its downstream effector TBK-1 played a key role in orchestrating autophagic maturation and cell-autonomous defense against mycobacteria. Furthermore TBK-1 phosphorylated a key autophagy adapter p62 (sequestosome 1) (Bjorkoy et al., 2005), the founding member of a new subfamily of pattern recognition receptors (PRRs) termed Sequestosome-like receptors (SLRs) (Deretic, 2012a) at the Ser-403 residue essential for its autophagic clearance function. Finally, we showed that the major proinflammatory cytokine IL-1β induced autophagy that killed intracellular Mycobacterium tuberculosis in a TBK-1 dependent manner. Thus, whereas the autophagy initiation is conrolled, as previously shown (Criollo et al., 2011), by the canonical members of the Iκ B family of kinases (IKK), autophagic maturation is controlled by the IKK-related kinase TBK-1.
Results
Rab screen for autophagic killing of M. tuberculosis var. bovis BCG reveals a role for Rab8b
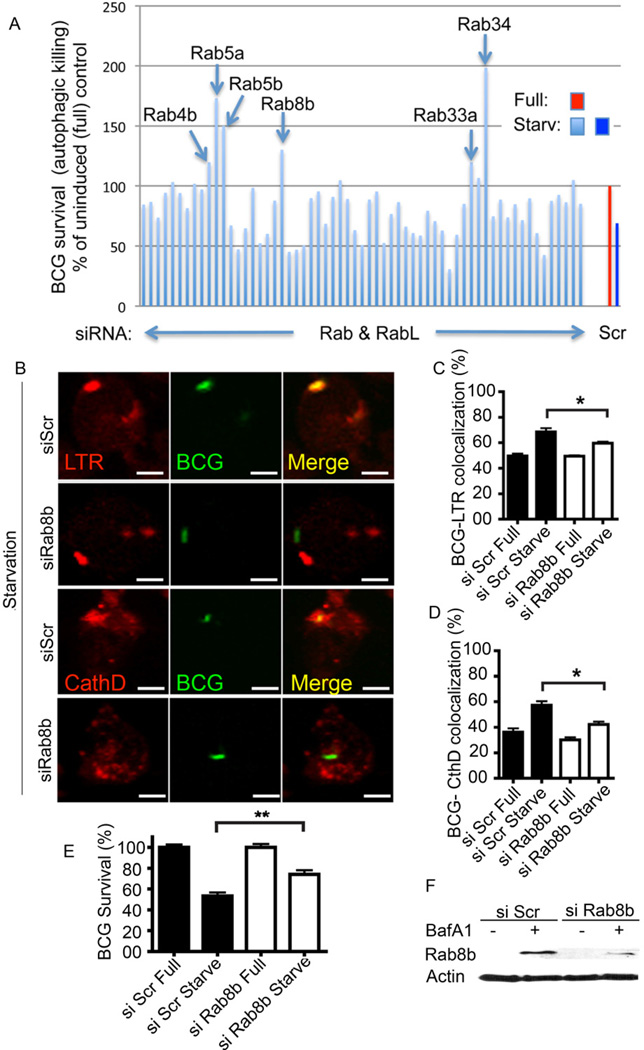
We screened a library of siRNAs to all murine Rabs and Rab-like factors (Fig. 1A and Suppl. Table S1) for effects on the previously characterized autophagic killing of Mycobacterium tuberculosis and M. tuberculosis var. bovis BCG (BCG) (Alonso et al., 2007; Gutierrez et al., 2004; Lam et al., 2012). The previously implicated endocytic Rabs (e.g. Rab5) showed a role in accordance with observations in other autophagy-dependent systems (Ravikumar et al., 2008). Additional Rabs displayed a range of effects on autophagic killing of BCG, including those Rabs previously implicated in autophagy, e.g., Rab7, Rab32, Rab33b, (Hirota and Tanaka, 2009; Itoh et al., 2011; Jager et al., 2004) with the notable exception of Rab24 (Olkkonen et al., 1993). Our preliminary observations indicated that Rab8b and Rab34 (data not shown) affected autophagic maturation. Here, we focused on Rab8b. Rab8b knockdown caused a decrease in conversion of BCG phagosomes to degradative compartments following induction of autophagy (Gutierrez et al., 2004; Ponpuak et al., 2010) (Fig. 1B–D) and autophagic killing of mycobacteria (Fig. 1E). In side-by-side comparisons, we observed only a trend with Rab8a (Suppl. Fig. S1A), whereas Rab8b siRNA reproducibly diminished autophagic killing of BCG in a statistically significant fashion (Fig. 1E). Thus, both Rab8a and Rab8b likely play a role in autophagy but Rab8b had a dominant effect at least in our model system measuring autophagic killing of mycobacteria. Rab8b furthermore influenced general markers of autophagy in resting cells: a knockdown of Rab8b reduced the number of LC3 puncta in a high content imaging analysis of cells stably expressing fluorescent protein fusion with LC3 (Suppl. Fig. S1B) and impeded basal clearance of the autophagic adaptor p62 (Suppl. Fig. S1C).
Fig. 1. Analysis of murine Rab factors in cell-autonomous autophagic elimination of mycobacteria and the role of Rab8b.
A. Sixty-two Rab or Rablike (RabL) factors encoded by the Mus musculus genome were knocked down by siRNA in RAW264.7 macrophages (details and identity of each bar in Suppl. Table S1), macrophages infected with M. tuberculosis var. bovis BCG, autophagy induced by starvation (Starv), and autophagic killing of BCG quantified by colony forming unit (CFU) counting. Scr, scrambled (control) siRNA. Bars, BCG survival (relative to Scr siRNA) following induction (4 h) of autophagy by starvation for 4 h. B–D. Effect of Rab8b knockdown on maturation of BCG phagosomes into autophagolysosomes. LTR, Lysotracker Red (acidotropic dye); CathD, cathepsin D. Images: LTR (red fluorescence), BCG (green fluorescence) and merged fields (yellow, colocalization of LTR and BCG). Bar graphs, quantitative colocalization analysis of images. (% of total green profiles showing red fluoresecence) E,F. Validation (E, % survival measured and represented as in A); F, Rab8b knockodown analysis by immunoblotting).of the role for Rab8b in autophagic killing of BCG. BCG survival, % of BCG CFU recovered from RAW 264.7 macrophages pretreated with siRNAs; si Scr, control scrambled siRNA; si Rab8b, Rab8b siRNA. Full, full medium; Starve, autophagy induced by starvation. [Data, means ± se (n≥3; †, p≥0.05*, p<0.05; **, p<0.01; ANOVA).
TBK-1 contributes to autophagic elimination of mycobacteria
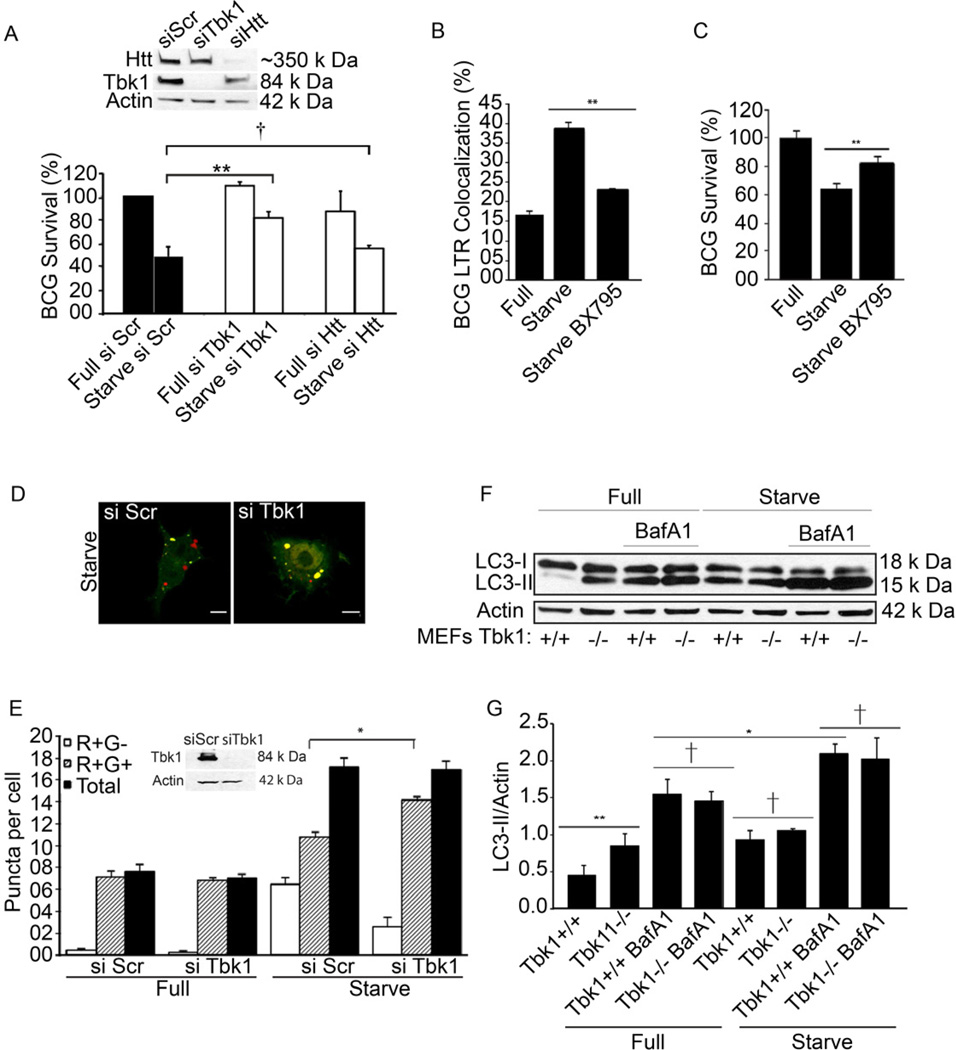
Among the few known interacting partners of Rab8b is FIP-2 (also known as optineurin). FIP-2 in turn associates with the wild-type (non-pathogenic) huntingtin (Htt) (Hattula and Peranen, 2000), and TBK-1 (Morton et al., 2008), a protein broadly conserved within Coelomata. TBK-1 is a pivotal regulator of innate immunity strategically positioned at the interface with cellular pro-survival pathways (Ou et al., 2011). Htt and TBK-1 have been thus far implicated as an autophagic degradation substrate (Sarkar et al., 2007) and, in the case of TBK-1, as being indirectly inhibited by intracellular trafficking imposed by the autophagy factor Atg9 (Saitoh et al., 2009) or playing a role in modifying an autophagic adaptor for Salmonella (Wild et al., 2011). We hence considered a model whereby these factors could play an active role in the regulation of the autophagic pathway. We first tested whether the Rab8b phenotype was exerted via Htt as a putative effector (known to functionally affect peripheral myeloid cells in Huntington’s disease patients (Bjorkqvist et al., 2008). However, an Htt knockdown in macrophages did not result in a statistically significant loss of autophagic killing of BCG (Fig. 2A). Thus, Htt was not critical for autophagic elimination of mycobacteria. In contrast to Htt, a knockdown of TBK-1 caused a deficit in autophagic killing of BCG (Fig. 2A), indicating that Rab8b effects on autophagy and elimination of mycobacteria may be exerted via TBK-1. This was confirmed using the TBK-1 inhibitor BX795 (Clark et al., 2009). When macrophages were treated with BX795, this abrogated starvation-induced BCG phagosome maturation (Fig. 2B) and autophagic killing of BCG (Fig. 2C). Thus, of the candidate effectors tested, TBK-1 was the one required for BCG killing.
Fig. 2. Role of TBK-1 in autophagic maturation.
A. RAW 264.7 macrophages were pretreated with siRNAs, infected with BCG, and autophagy induced by starvation (Starve). BCG survival (% of BCG) was analyzed and represented as in Fig. 1. Full, full medium (control conditions); si Scr, scrambled siRNA (control siRNA) si TBK-1, siRNA to TBK-1. Si Htt, siRNA to huntingtin (Htt). Immunoblot, analysis of TBK-1 and Htt knockdowns. B. Effect of TBK-1 inhibitor BX795 on acidification of BCG-containing organelles following induction of autophagy. RAW 264.7 macrophages were pretreated with 10 nM BX795, infected, and induced for autophagy by starvation. C. Autophagic killing of BCG in RAW 264.7 macrophages pretreated with BX795. BCG survival, % CFU recovered from RAW 264.7 cells. D,E. RAW 264.7 expressing RFP-GFP-LC-3 and pretreated with scrambled or TBK-1 siRNAs were untreated (Full) or induced (Starv) for autophagy. Puncta; R+G+ (RFP+ GFP+), early autophagic organelles; R+G- (RFP+ GFP-), late autophagic organelles. Images, merged red and green channels. Inset in E, immunoblot analysis of TBK-1 knockdown. F,G. Effects of TBK-1 on LC3-II levels and degradation during autophagic maturation. Tbk-1−/− and Tbk-1+/+ MEFs were uninduced and induced for autophagy, treated or not treated with bafilomycin A1 (BafA1) to inhibit autophagic degradation of LC3-II. Data, means ± se (n≥3; †, p≥0.05*, p<0.05; **, p<0.01; ANOVA).
TBK-1 is necessary for autophagosome maturation
We next tested how TBK-1 affected autophagy and found that TBK-1 knockdown did not affect formation of autophagosomes but suppressed their maturation (autophagic flux) into autolysosomes, as revealed by the tandem RFP-GFP-LC3 reporter (Fig. 2D,E), which identifies early autophagic organelles as RFP+GFP+ and matured autolysosomal organelles as RFP+GFP− since GFP (but not RFP) fluorescence is sensitive to lumenal acidification (Kimura et al., 2007; Pankiv et al., 2007). A loss of TBK-1 reduced the number of RFP+GFP− acidified autophagic organelles and increased RFP+GFP+ puncta, consistent with inhibition of autophagic maturation (Fig. 2D,E). A positive role of TBK-1 in autophagosomal maturation was confirmed by LC3 immunoblots in Tbk1−/− MEFs (Fig. 2F,G). MEFs lacking TBK-1 showed accumulation of LC3-II (a marker of early autophagic organelles) under both basal (full medium) and induced (starvation) conditions. The increase in LC3-II in the absence of TBK-1 cannot be explained by de-repression of autophagy initiation since the amounts of LC3-II protected from degradation by bafilomycin A1 were slightly lower in or equal to Tbk1−/− cells (Fig. 2F,G and Suppl. Fig. S2A). To confirm that the phenotype was due to TBK-1 deficiency, we inhibited with BX795 TBK-1 in wild type (Tbk1+/+) MEFs and tested effects of BX795 on starvation-induced autophagy. As with genetically deficient Tbk1−/− MEFs, pharmacological inhibition of TBK-1 led to diminished autophagic maturation as determined by LC3-II blots (Suppl. Fig. S2B). BX795 did not affect autophagy in Tbk1−/− MEFs (Suppl. Fig. S2C). Although attempts to complement the absence of TBK-1 in Tbk1−/− MEFs by transfection with Tbk-1 expression constructs were hampered by low transfection efficiency, overexpresion of Tbk-1 transgene in RAW264.7 cells caused alterations in LC3-II amounts, which diminished faster with time in Tbk-1 transgene-expressing cells relative to untransfected cells (Suppl. Fig. S2D). TBK-1 was also important for delivery of the lysosomal hydrolase Cathepsin D to autophagolysosomal compartments. These organelles were purified from macrophages induced for autophagy by starvation using magnetic beads as previously described (Ponpuak et al., 2010) and probed for Cathepsin D levels. A knockdown of TBK-1 diminished cathepsin D delivery to one third of normal (Suppl. Fig. S2D). This was specific for TBK-1 as a knockdown of another Rab8b-optineurin interacting partner, Htt, did not reduce cathepsin D delivery (Suppl. Fig. S2E). TBK-1 was also needed for delivery of Cathepsin D to conventional phagolysosomes (Suppl. Fig. S2F) mirroring the Rab8a-dependent delivery of cathepsins from the TGN to the lysosomal compartments (del Toro et al., 2009). Thus, TBK-1 is required for autophagosomal maturation.
TBK-1 associates with Rab8b and colocalizes with Rab8b on autophagic organelles
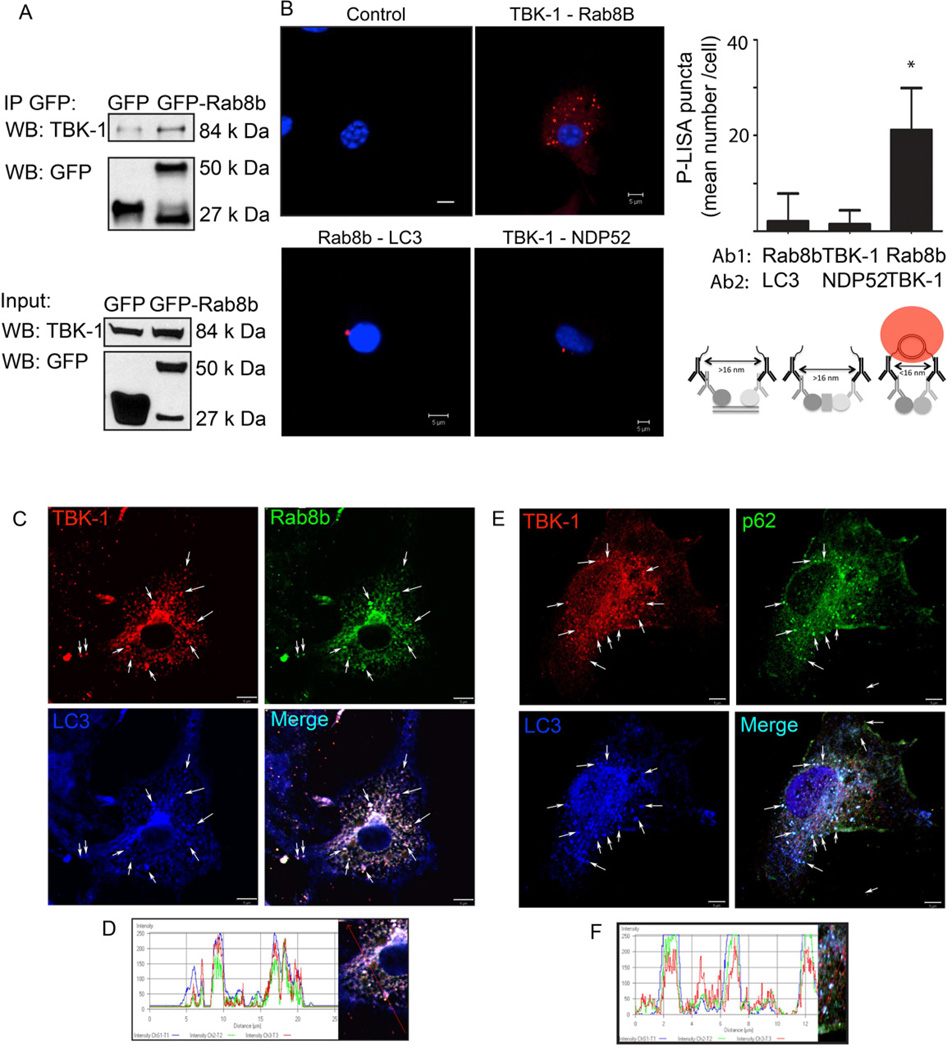
We next turned to the mechanisms of how TBK-1 affects autophagy. If Rab8b and TBK-1 cooperate in the control of the autophagic pathway, we reasoned that they might associate. This was examined first in co-immunoprecipitation experiments with GFP-Rab8b (Fig. 3A). The interaction between Rab8b and TBK-1 was tested further using an independent method, termed proximity ligation in situ assay (P-LISA), designed to detect endogenous protein-protein interactions in whole cells (Soderberg et al., 2006). When murine bone marrow macrophages (BMM) were tested by P-LISA for Rab8b-TBK-1 interactions, a positive result was obtained (Fig. 3B: antibody 1 to Rab8b + antibody 2 to TBK-1). When TBK-1 and NDP52 proteins were tested, they were negative for direct interaction. This can be explained at the very least by the existence of Nap and Sintbad proteins as intervening adaptors linking TBK-1 with NDP52 (Thurston et al., 2009) (Fig. 3B: antibody 1 to TBK-1 + antibody 2 to NDP52).
Fig. 3. Rab8b and TBK-1 interact and colocalize with autophagic organelles.
A. HEK 293T cell extracts, transiently transfected with control (EGFP), GFPRab8b (Wt, wild type) and GFP-Rab8b Q67L (Q67L, constitutively active) expression constructs were immunoprecipitated with anti-GFP antibody; immunoblots of immune complexes and inputs were probed with anti-TBK-1 and anti-GFP antibodies. B. Proximity ligation in situ assay (P-LISA) for proteinprotein interactions in murine primary bone marrow macrophages (BMM). BMMs were fixed, incubated with primary antibodies against two proteins tested (Ab1 and Ab2), and P-LISA oligonucleotides added. After nucleic acid ligation, rolling circle amplification, and hybridization with fluorescent oligonucleotides (and counterstaining of nuclei with DAPI), red fluorescent dots (see scheme for distance requirements between probes for positive signal) were imaged by confocal microscopy. Control, no primary antibodies. TBK-1-Rab8b, antibody 1 (Ab1): anti-TBK-1, antibody 2 (Ab2): anti-Rab8b. Rab8b-LC3, Ab1: anti-Rab8b, Ab2: anti-LC3. TBK-1-NDP52, Ab1: anti-TBK-1, Ab2: anti-NDP52. Graph, quantitative analysis of P-LISA red-fluorescent dots (indicating close proximity of proteins recognized by Ab1 and Ab2). Cells (≤ 25 per sample) with positive red fluorescing dots were morphometrically analyzed and the mean number of red fluorescing dots per cell quantified. Up to 10% of the cells in any given sample were positive for red dots. Values, means ± sd; * p<0.05 (t-test). C. Colocalization analysis of Rab8b and its downstream effector, TBK-1 with the basal autophagic machinery factor LC3. Fluorescence; endogenous TBK-1 (red, Alexa568), Rab8B (green, Alexa488), LC3 (blue, Alexa633). Cells (BMM) were induced for autophagy by starvation in the presence of bafilomycin A1 to inhibit autophagic maturation and degradation. Arrows, colocalization of Rab8b, TBK-1, and LC3. D. Representative line tracing of three fluorescence channels in images in A. E. Analysis of TBK-1 localization relative to the autophagic adaptor sequestosome 1/p62 and LC3. Cells (BMM), treatments, labels and graphs as in C and D. Arrows, colocalization of TBK-1, p62 and LC3. Cells (BMM) were induced for autophagy by starvation in the presence of bafilomycin A1 to inhibit autophagic maturation. F. Representative line tracing of fluorescence channels in images in C. Data, representative of ≥3 independent experiments.
When we examined intracellular localization of Rab8b and TBK-1 relative to autophagic organelles, TBK-1 colocalized with endogenous LC3 and Rab8b (Fig. 3C,D). Although Rab8b and LC3 colocalized on intracellular profiles, they were negative for direct interactions by the P-LISA assay (Fig. 3B: antibody 1 to Rab8b + antibody 2 to LC3B). The autophagic adaptor p62, demonstrated to be critical for autophagic killing of M. tuberculosis (Ponpuak et al., 2010) also colocalized with TBK-1 (Fig. 3E,F). Supplementary Fig. S3 shows colocalization analyses of TBK-1 and LC3 with Rab8b (Suppl. Fig. S3A) and with p62 (Suppl. Fig. S3B), under different conditions: basal, induced autophagy, presence or absence of bafilomycin A1. The colocalization and a striking similarity in the overall intracellular organellar distribution were enhanced in the presence of LC3- and p62-sparing activity of bafilomycin A1 (Suppl. Fig. S3A,B). We conclude that Rab8b and TBK-1 localize to autophagosomal organelles in keeping with their role in regulating autophagic flux.
Assembly of membranous compartments containing Rab8b, TBK-1 and autophagy factors
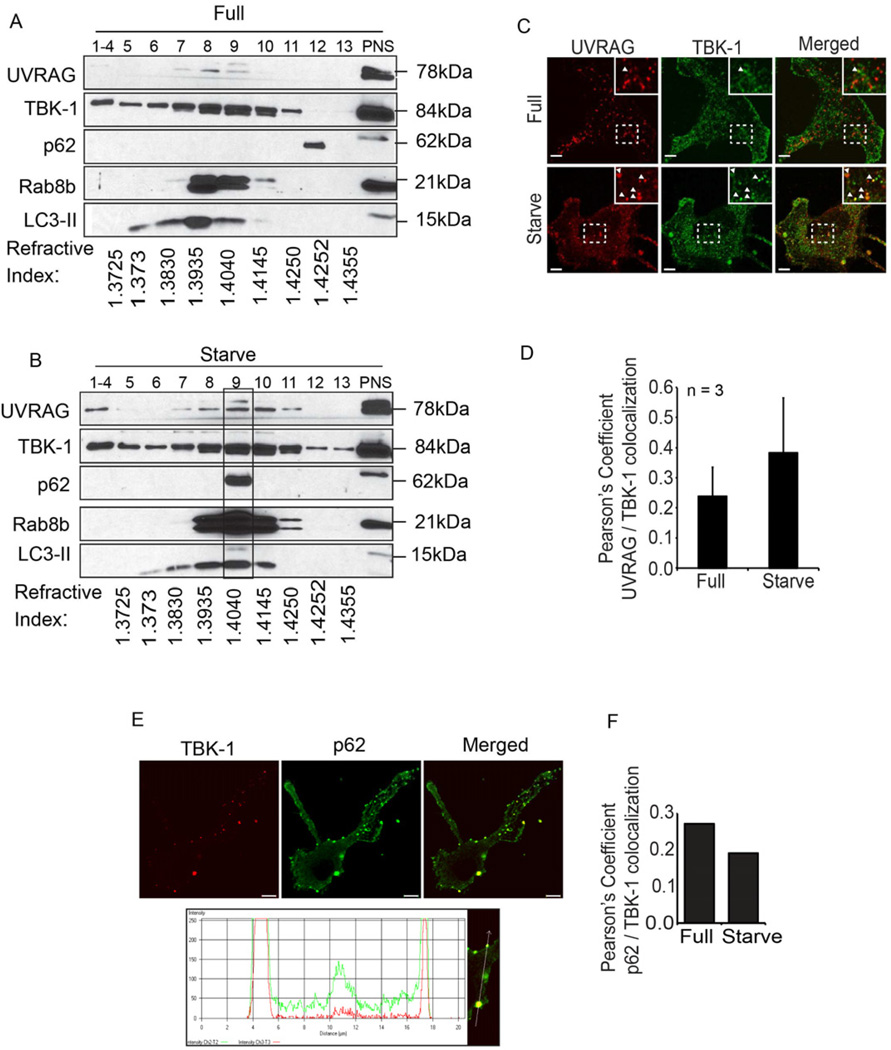
When subcellular membranous compartments were separated by isopycnic centrifugation in sucrose gradients, induction of autophagy resulted in redistribution of multiple components engaged in autophagy causing them to cofractionate with TBK-1 and Rab8b (Fig. 4A,B). These were LC3-II and the autophagic adaptor p62 (Bjorkoy et al., 2005; Johansen and Lamark, 2011) and NDP52 (Thurston et al., 2009), as well as UVRAG, a component of the Beclin 1- hVPS34 complex II specific for autophagosomal maturation into lytic compartments (Liang et al., 2008). In keeping with its sucrose gradient cofractionation with TBK-1 (Fig. 4A,B), UVRAG colocalized with TBK-1 as evident from Pearson’s colocalization coefficient (Fig. 4C,D). The autophagic adaptor p62 (Bjorkoy et al., 2005) colocalized with TBK-1 (Fig. 4E,F), congruent with the biochemical analyses of intracellular organelles (Fig. 4A,B). Without bafilomycin A1, upon induction of autophagy by starvation, p62-TBK-1 colocalization was reduced (Fig. 4E,F) in keeping with p62 being consumed during autophagy. These findings indicate convergence of Rab8b and TBK-1 containing compartments with autophagic machinery components.
Fig. 4. TBK-1 co-fractionates and colocalizes with intracellular membranous organelles containing autophagic adaptors and machinery.
A,B. Analysis by subcellular fractionation of Rab8b, TBK-1 and components of autophagic machinery (UVRAG, p62, LC3-II) in resting cells (Full, full medium) or upon induction of autophagy by starvation (Starve). Membranous organelles from RAW 264.7 macrophages uninduced (A) or induced (B) for autophagy were subjected to subcellular fractionation by isopycnic sucrose density gradient centrifugation, fractions collected, and proteins analyzed by immunolotting. PNS, postnuclear supernatant. 1–4, pooled fractions. Rectangle over fraction 9, convergence in autophagic organelles (LC3-II) of: Rab8b, TBK-1, UVRAG (Beclin 1 interacting protein specific for autophagosomal maturation), and autophagic adaptor p62. Refractive indexes below the lanes reflect sucrose density of each fraction. C. Images; confocal microscopy analysis of endogenous UVRAG (Alexa568) and endogenous TBK-1 (Alexa488). Cells, BMM, uninduced (Full) and induced (Starvation) for autophagy. Arrows, coloclaization; insets, enlarged areas. D. Pearson’s coefficient for TBK-1 and UVRAG colocalization (Starv, starvation). E. Analysis of TBK-1 localization relative to autophagic adaptor p62 in BMM. Images: endogenous TBK-1 (Alexa568; red), p62 (Alexa488; green) and merged. Line tracing, analysis of colocalization of TBK-1 (red tracing) and p62 (green tracing). F. Pearson’s colocalization coefficients for TBK-1-p62. (Starv, starvation). Data, means ± se (n=3, three independent experiments with at least 5 images analyzed per experiment; †, p≥0.05; **, p<0.01; ANOVA).
TBK-1 affects p62 clearance
If TBK-1 is needed for autophagic maturation, we reasoned that it might affect clearance of the autophagic adaptor p62, as it has been reported to be a good marker of autophagic maturation, matching or exceeding in performance LC3- based assays (Larsen et al., 2010). Using antibody to endogenous p62 we employed high content quantitative imaging analysis and detected significant increase in p62 puncta in BMMs treated with TBK-1 inhibitor BX795 (Fig. 5A–C). Protein blot analysis confirmed that p62 amounts increased when TBK-1 was inhibited (Fig. 5D), in keeping with the high content image analysis of p62. TBK-1 was also necessary to authorize p62 for autophagic degradation, since p62 accumulated in Tbk1−/− cells; the surplus p62 seen in Tbk1−/− MEFs showed a laddering pattern (Suppl. Fig. S4) neither previously reported nor observed here in the presence of TBK-1. Furthermore, ubiquitinated proteins accumulated in Tbk1−/− cells revealed by pull-down assays in cellular extracts using TUBE-2 (tandem ubiquitin binding entities 2), which recognizes polyubiquitinated proteins and protects them from deubiquitinating enzymes and proteasome during isolation (Hjerpe et al., 2009) (Fig. 5E) Thus, p62 is not being cleared by autophagy when TBK-1 is unavailable and accumulates in the cell, TBK-1 is needed to enable the entry of p62 into autophagic degradative pathway, and several ubiquitinated cargo do not enter degradative pathways in the absence of TBK-1.
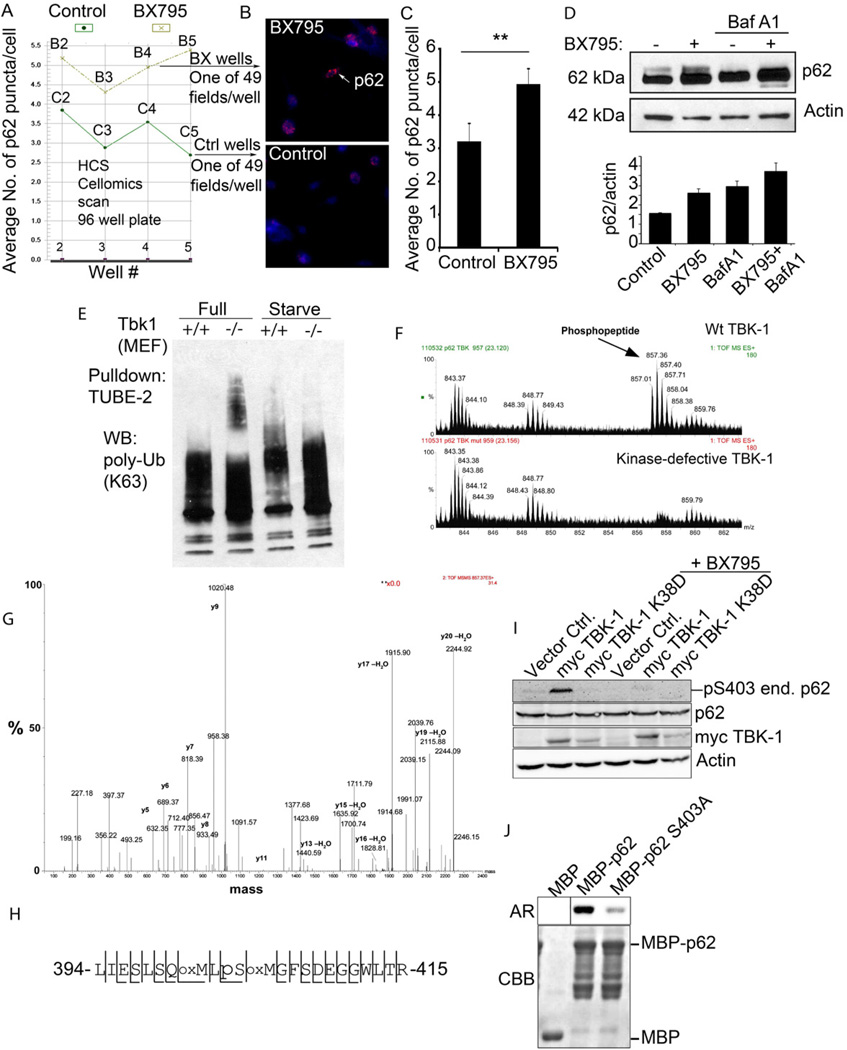
Fig. 5. TBK-1 controls p62 phosphorylation, and affects autophagic clearance of p62 and its cargo capture, delivery and degradation.
A–C. High content imaging analysis (using Cellomics high-content microscopy system) of p62 puncta (endogenous, revealed by imunofluorescence) in BMM with or without treatment with TBK-1 inhibitor BX795. Panel A shows output from Cellomics high-content microscopy and analysis software comparing the number of p62 puncta between TBK-1 inhibitor-treated (BX795) and control (DMSO) BMM. Vertical axis denotes the mean number of p62 puncta per cell and horizontal axis denotes the position of the well (B, BX795 series; C, control series) on the plate. Between 754 and 2395 cells were analyzed per well. Panel C shows t test (data, means ± se; **, p<0.01) from cumulative data treating only whole wells as independent samples (n=4). D. Analysis of the effects of TBK-1 pharmacological inhibitor, BX795 on p62 levels. Tbk-1+/+ MEFs were treated with BX795; bafilomycin A1 (BafA1) to inhibit autophagic degradation. Densitometric analyses of p62 levels normalized against actin levels were plotted (n=2; error bars, range). E. Analysis of TBK-1 role in autophagic clearance of polyubiquitinated proteins. Cell lysates from Tbk1+/+ and Tbk1−/− MEFs uninduced and induced for autophagy by starvation were incubated with TUBE2 agarose beads and bound material pulled down. Protein blots were probed for K63 polyubiquitin chains. F–H. Identification of TBK-1-dependent S303 phosphorylation of the UBA domain of p62. In vivo phosphorylation of p62 UBA domain following cotransfection of GFP-p62D69A (D69A mutation prevents oligomerization with endogenous p62) and expression constructs of TBK-1 wild type or kinase defective form. Immunoprecipitated (GFP-p62) material was subjected to tandem mass spectrometry. A triply charged ion with the mass 857.01 was selected for fragmentation. This ion was identified as the phosphorylated LIESLSQMLpSMGFSDEGGWLTR peptide (shown in panel H) from p62. Panel G shows MS spectra from LC-MS, showing the phosphopeptide of 857.01 m/z observed in p62 phosphorylated by TBK-1. The peptide was not observed when GFP-p62 was co-transfected with the kinase-defective K38D mutant of TBK-1. Spectra are taken from the same retention time in both runs, confirmed by the unspecific peaks observed in both spectra. I. HEK293 cells transfected with vector control, myc-TBK-1 or myc-TBK-1 K38D were left untreated or were treated for 2 h with 1 µM BX795. Cell extracts were immunoblotted with antibodies against phospho-p62 (S303), p62, myc and actin. Abbreviations: end. p62, endogenous p62. J. MBP or MBP-tagged p62 proteins were expressed and affinity-purified from E. coli. TBK-1 mediated phosphorylation was assessed by incubating recombinant MBP, MBP-p62 or MBP-p62 S303A with recombinant active TBK-1 in the presence of [γ-32P] ATP for 10 min at 30°C. The reaction products were analyzed by autoradiography (AR). CBB, Coomassie Brilliant Blue staining.
TBK-1 phosphorylates p62 on Ser-403
To determine whether TBK-1 modified (e.g. phosphorylated) p62 in vivo and to examine the potential sites involved we co-expressed GFP-p62D69A (mutant preventing oligomerization of p62) and myc-TBK1 in HEK293 cells, immunoprecipitated GFP-p62 from cellular extracts and carried out mass spectrometry analyses on the immunoprecipitated material. As a control, we cotransfected GFP-p62D69A with myc-TBK-1K38D, a kinase defective mutant. By manual inspection of the mass spectrometry data, a peptide of 857.01 m/z (2568.03 Da) was detected in GFP-p62D69A expressing cells cotransfected with myc-TBK-1, but not when GFP-p62D69A was co-expressed with the kinase defective mutant myc-TBK-1K38D (Fig. 5F). The 857.01 m/z (2568.03 Da) peptide was selected for tandem mass spectrometry and identified as the LIESLSQMLpSMGFSDEGGWLTR phosphopeptide from p62, where serine 403 in the UBA domain of p62 is phosphorylated (Fig. 5G,H). The unphosphorylated peptide with the mass 2488.1 was observed in both samples. The Ser-403 site corresponds to the TBK-1 consensus sequence SxxxpS. Thus, TBK-1 affects p62 phosphorylation of the ubiquitin associated (UBA) domain, providing a specific link between TBK-1 and autophagic adaptor post-translational modifications. Phosphorylation of Ser-403 strongly increases affinity of the UBA domain of p62 for K48 and K63-linked ubiquitin chains, and promotes autophagic clearance of p62 and polyubiquitinated protein aggregates (Matsumoto et al., 2011). Using a phosphospecific antibody developed by Matsumoto et al., (2011), we monitored phosphorylation of Ser-403 in vivo and in vitro (Fig. 5I,J). TBK-1 phosphorylated Ser-403 of endogenous p62 in HEK293 cells transfected with a myc-TBK-1 expression vector (Fig. 5I). This was not observed when HEK293 cells were transfected with kinase defective TBK-1 or when mycTBK-1 expressing cells where treated with the TBK-1 inhibitor BX795 (Fig. 5I). When purified proteins were examined in a phosphorylation in vitro assay, TBK-1 directly phosphorylated p62 at the Ser-403 site (Fig. 5J). Thus, TBK-1 phosphorylates p62 at Ser-403, a residue required for autophagic function of p62.
IL-1β induces autophagy
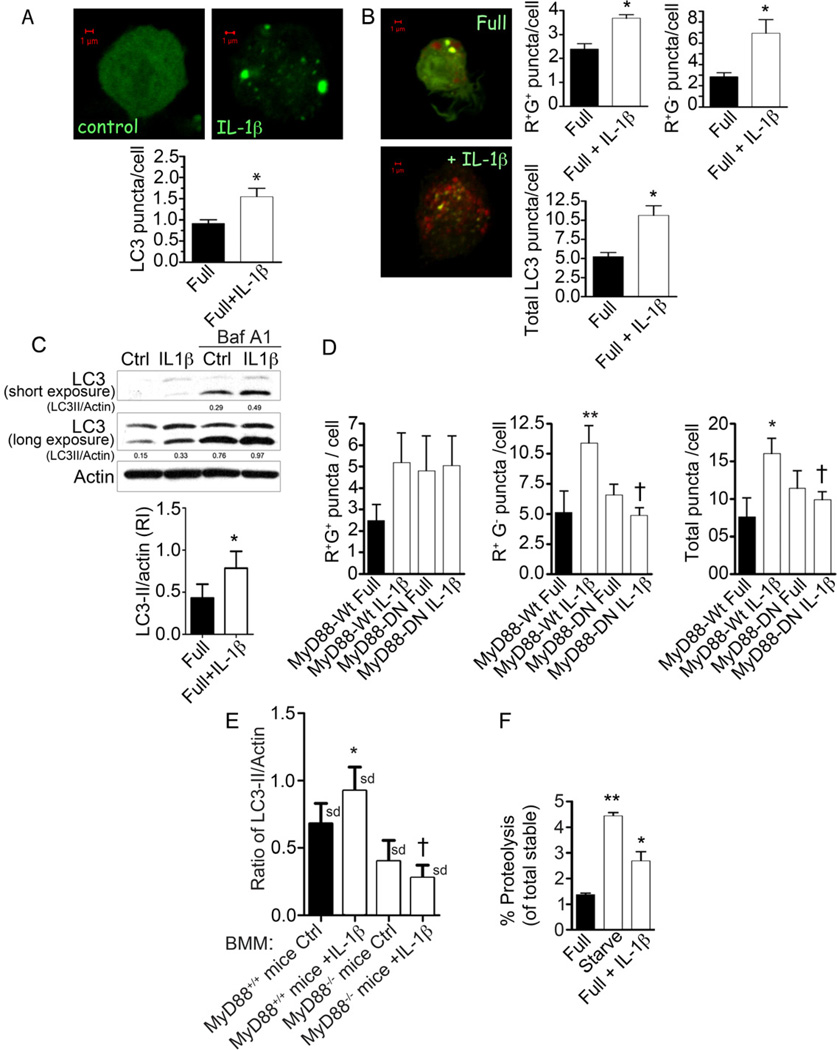
In order to provide an immunologically relevant context in which to test the role of TBK-1 in autophagy, we first examined whether IL-1β could induce autophagy in macrophages and contribute to control of M. tuberculosis; this has not been previously tested albeit IL-1β signaling is important in control of intracellular mycobcteria (Fremond et al., 2007; Mayer-Barber et al., 2011). When RAW264.7 macrophages transiently expressing EGFP-LC3 were treated with mouse IL-1β this induced LC3 puncta formation (Fig. 6A). When RAW264.7 cells transiently expressing the tandem RFP-GFP-LC3 probe were stimulated with IL-1β (Fig. 6B), an increase in both early autophagosomal (RFP+GFP+) and mature, acidified puncta (RFP+GFP−) was detected indicating that IL-1β induces both autophagic initiation and maturation resulting in progression through the autophagic pathway. Induction of autophagy by IL-1β was confirmed by immunoblot analysis of lipidated LC3-II levels in the presence of autophagic maturation inhibitor bafilomycin A1, since LC3-II levels were increased in IL-1β-stimulated cells (Fig. 6C). IL-1β induced autophagy in primary cells, including murine bone marrowderived macrophages (BMM) (Suppl. Fig. S5A) and human peripheral blood monocyte-derived macrophages (Suppl. Fig. S5B). Induction of autophagy by IL- 1β was dependent on MyD88 as shown by expression of dominant negative MyD88 (Fig. 6D) and by analyzing BMM from MyD88-deficient mice (Fig. 6E). A progression through the autophagic pathway was confirmed by detecting proteolysis of long-lived proteins in macrophages stimulated with IL-1β (Fig. 6F). Thus, IL-1β induces autophagic pathway in macrophages.
Fig. 6. IL-1β induces autophagy in macrophages.
A. RAW264.7 macrophages were transiently transfected with EGFP-LC3 and treated with 10 ng/ml murine IL- 1β for 2 h, and assayed for LC3 puncta formation by confocal microscopy (only puncta ≥ 1µm were scored as positive). B. RAW264.7 macrophages transfected with mRFP-GFP-LC3 tandem probe, treated with 10 ng/ml IL-1β for 2 h were scored for number (per transfected cell) of RFP+GFP+ puncta (R+G+; early autophagosomes), RFP+GFP− (R+Gβ− ; autolysosomes), and total LC3 puncta. C. Immunoblot analysis of endogenous LC3 conversion to lipidated form (LC3-II) in RAW 264.7 cells upon treatment with 10 ng/ml IL-1β for 2 h, in the absence or presence of bafilomycin A1. Graph, ratio of LC3-II to actin intensity in immunoblots from bafilomycin A1-treated samples. D. RAW264.7 murine macrophages were co-transfected with tandem mRFP-GFP-LC3 probe and expression constructs containing either wild-type MyD88 (MyD88-WT) or a dominant-negative mutant of MyD88 (MyD88-DN). Following stimulation with 10 ng/ml IL-1β for 2 h, LC3 puncta were quantified as in B. E. Induction of autophagy in response to IL-1β is abrogated in bone marrow-derived macrophages (BMM) from MyD88-deficient (Myd88−/−) mice, measured by ratios of LC3-II band relative to actin following treatments of BMMs and immunoblotting of cellular extracts. F. Proteolysis of stable proteins (radiolabeled by a pulsechase protocol) upon stimulation of RAW264.7 cells with 10 ng/ml IL-1β for 2 h (Full + IL-1β) relative to control (Full) or starvation-induced autophagy (Starve).. Data, means ± se, except in E where data are means ± sd (n≥3; †, p≥0.05; *, p<0.05; **, p<0.01; ANOVA).
TBK-1 is required for IL-1β-induced autophagic elimination of M. tuberculosis
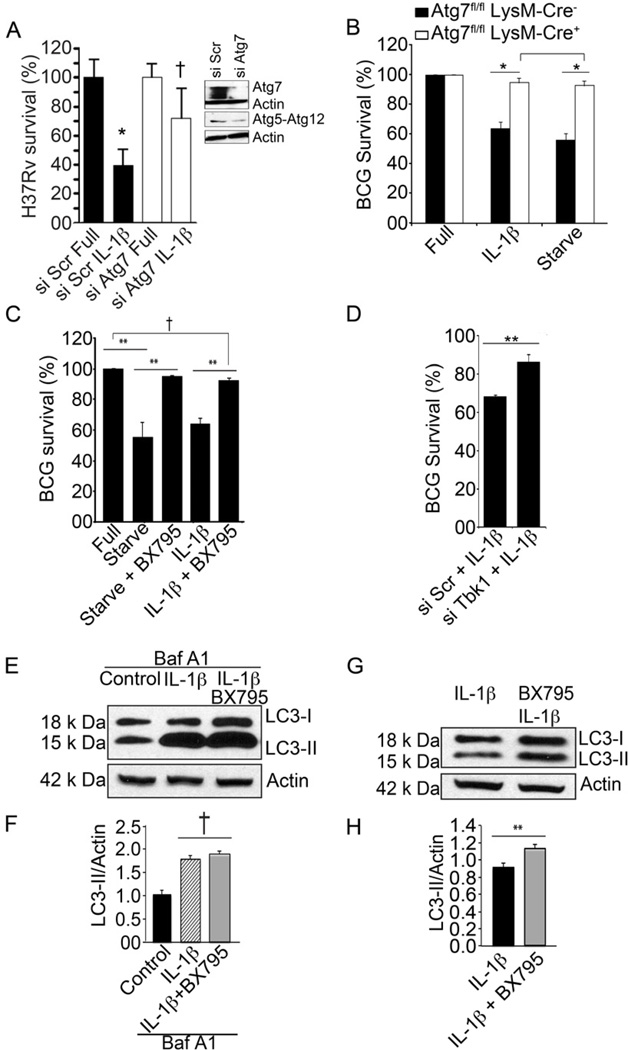
We next used mycobacterial killing as a measure of autophagic cell-autonomus defense output (Ponpuak et al., 2010). IL-1β induced killing of M. tuberculosis, whereas elimination of the tubercle bacillus was reduced showing a trend in cells that were subjected to Atg7 knock down (Fig. 7A), suggesting that IL-1β-induced autophagy can eliminate intracellular M. tuberculosis. We further tested the role of autophagy in IL-1β-mediated killing of mycobacteria by using BMMs from mice in which Atg7 was deleted in the myeloid lineage (Atg7fl/fl LysM-Cre+) and compared them to their Atg7+ littermates (Atg7fl/fl LysM-Cre−). When Atg7+ BMM were infected with BCG and induced for autophagy by either IL-1β or starvation, BCG was eliminated with similar efficiency under both conditions (Fig. 7B). In BMM lacking Atg7 (Atg7fl/fl LysM-Cre+) the absence of this core autophagy factor abrogated autophagic killing of BCG by either IL-1β or starvation (Fig. 7B). Thus, IL-1β-induced killing of intracellular mycobacteria was mediated through autophagy.
Fig. 7. Requirement for TBK-1 in IL-1β mediated autophagic killing of mycobacteria.
A. RAW264.7 macrophages were knocked down for Atg7 (by Atg7 siRNA transfection 48 h prior to infection), infected with M. tuberculosis H37Rv for 1h, washed and then left untreated or treated with 10 ng/ml recombinant murine IL-1β for 2 h after which they were lysed and plated for CFU determination, and survival expressed relative to sample transfected with control scrambled siRNA and not treated with IL-1β. Immunoblots, Atg7 knockdown and levels of Atg5-Atg12 complexes. B. BCG survival, % CFU of M. tuberculosis var. bovis BCG recovered from bone marrow macrophages derived from Atg7fl/fl LysM-Cre− and Atg7fl/fl LysM-Cre+ mice treated with 50 ng/ml IL-1β (16 h preinfection + 4 h post-infection). Data, means ± se (n=3, *, p<0.05; t-test. C. BCG survival, % CFU recovered from RAW264.7 macrophages treated with IL-1β with and without 10 nM BX795. D. BCG survival in infected RAW264.7 (and knocked down or not for TBK-1) macrophages stimulated with IL-1β. E,F. RAW264.7 macrophages were incubated in full medium (Control) or induced by adding IL-1β to full medium. Cells were pretreated with 10 nM BX795 where indicated. Macrophages were treated with or without bafilomycin A1 (BafA1) to inhibit autophagic degradation of LC3-II, and cellular extracts analyzed by immunoblotting. Graphs, densitometric analyses of LC3-II levels normalized to actin levels (LC3-II/Actin). G,H. Cells, treatments and analysis as in E and F but in the absence of bafilomycin A1. Data, means ± se (n=3; †, p≥0.05; **, p<0.01; ANOVA).
Having established that IL-1β induces autophagy with its physiological and cell-autonomous immunity outputs, we asked whether TBK-1 was important for these processes. RAW264.7 macrophages infected with mycobacteria were stimulated with IL-1β or by starvation in the presence or absence of TBK-1 inhibitor BX795. Either starvation or IL-1β caused mycobacterial killing, whereas BX795 (Fig. 7C) or TBK-1 siRNA (Fig. 7D) reduced their autophagic elimination. As with starvation, TBK-1 inhibition did not affect initiation of autophagy (Fig. 7E,F) but suppressed autophagic maturation (Fig. 7G,H). In conclusion, TBK-1 is important for conversion of autophagic organelles into mature and bactericidal organelles, for both physiologically (starvation) and immunologically (IL-1β ) induced autophagy.
Discussion
In this study, we have uncovered the role for TBK-1 in the maturation of autophagic organelles and shown its immunological significance in the context of cell-autonomous antimycobacterial functions of autophagy. We have additionally established that IL-1β induces autophagy in a MyD88-dependent fashion, that autophagy in macrophages is an anti-M. tuberculosis effector mechanism of IL- 1β, and that TBK-1 is necessary for the completion of these defense processes against mycobacteria downstream of IL-1β.
TBK-1 is a member of the IKK family of kinases that are well recognized as regulators of innate immunity (Perkins, 2007). IKKs generally fall into two categories: canonical (IKK-α, IKK-β ) and IKK-related kinases (IKKε and TBK-1). The canonical IKKs, IKKα and IKKβ, have been recently implicated in the induction of autophagy (Criollo et al., 2011). The findings that TBK-1 controls autophagy maturation extend the observations with IKKα and IKKβ, whereas together they align autophagy with these classical regulators of innate immunity responses. The role of TBK-1 in autophagy maturation suggests a sequential action in autophagy of the canonical IKKs at the initiating stages and TBK-1 at the completion end of the process.
Whereas much effort has been devoted to understanding the initiation and elongation stages of autophagy (Mizushima et al., 2011) the control of the flux and conversion of autophagic organelles into degradative compartments is just as important since in most cases autophagy exerts its physiological and immunological functions by degrading or processing the captured intracellular material. TBK-1 enables the execution of autophagosomal maturation and likely couples this stage of autophagy with cargo capture. This is evidenced by the in vivo dependence on TBK-1 of the phosphorylation of the key Ser-403 residue within the UBA domain of one of the principal autophagic adaptors, p62/seqestosome 1. While our study was in revision, phosphorylation of Ser-403 by CK2 was shown to increase affinity of p62 UBA for polyubiquitin chains on its cargo (Matsumoto et al., 2011). Our findings are congruent with these observations and suggest that TBK-1 also targets and phosphorylates directly the Ser-403 site, thus linking immunological and physiological inputs with p62 UBA activation in vivo.
A recent study implicating TBK-1 via optineurin-sponsored events in the autophagic control of Salmonella upon escape into the cytosol (Wild et al., 2011) and our present work are in keeping with the report (Radtke et al., 2007) uncovering the role of TBK-1 in control of intracellular bacteria, in addition to its well appreciated role in antiviral defenses. O’Riordan and colleagues (Radtke et al., 2007) pointed to the role of TBK-1 as keeping the cytosolic bacteria in check by a mechanism that at the time was characterized as prevention of bacterial escape into or multiplication in the cytosol but was deemed not to involve autophagy due to an apparent increase in LC3 puncta in Tbk-1−/− cells. Given that TBK-1, as shown here, is primarily involved in autophagosomal maturation, accumulation of LC3 in the absence of TBK-1 observed by O’Riordan (Radtke et al., 2007) and colleagues remarkably fits our data showing that TBK-1 controls flux and progression through the autophagic pathway (causing disappearance of the initially formed LC3 puncta via degradation in autolysosomes), rather than its initiation (characterized by appearance of LC3 puncta). Thus, all presently existing data are compatible with the role of TBK-1 in autophagic maturation and its role as an antimicrobial mechanism that can eliminate cytoplasmic bacteria.
The Rab screen performed here, which has led to the investigations of TBK-1 downstream of Rab8b, relied on the cell-autonomous antimicrobial defense functionality that depended, as previously defined (Ponpuak et al., 2010), on the execution of the entire autophagic pathway. Of the two Rab8 GTPases only Rab8b showed statistically significant effect on autophagic killing of BCG. TBK-1 and Rab8b were found in shared protein complexes, and following induction of autophagy, TBK-1 and Rab8b colocalized and cofractionated with autophagic organelles. These relationships likely underlie the role of Rab8b and TBK-1 in autophagic control. Optineurin interacts with both Rab8b and TBK-1 but in our studies we could not establish that optineurin physically or functionally links Rab8b and TBK-1. Since optineruin binds Rab8a as well as Rab8b, it is furthermore unlikely that optineurin underlies the stronger Rab8b engagement with autophagy maturation as detected here. Of note, Rab8a, does associate with a subset of LC3 compartments, however they specialize in the alternative secretory pathway of inflammasome substrates such as IL-1β (Dupont et al., 2011).
TBK-1 turned out to be important in cell-autonomous defense against mycobacteria upon macrophage activation by the inflammatory cytokine IL-1β. We have shown in this study that IL-1β induces autophagy in a MyD88- dependent fashion and that one of the outputs of this proinflammatory cytokine is to promote autophagic killing of M. tuberculosis. This is in keeping with the reports that IL-1β, IL-1R, and MyD88 are important in elimination of M. tuberculosis (Fremond et al., 2007; Mayer-Barber et al., 2011). However, the relationships between autophagy and IL-1 signaling are more complex and reciprocal. Autophagy is necessary to keep down the endogenous sources of inflammasome and IL-1β activation (Dupont et al., 2011; Nakahira et al., 2011; Zhou et al., 2011). Autophagy does that by eliminating unkempt mitochondria that otherwise leak ROS or mitochondrial DNA implicated as agonists increasing tonic inflammasome activation levels (Nakahira et al., 2011; Zhou et al., 2011). Autophagy has also been implicated in acute extracellular delivery of IL-1β and HMGB1 (Dupont et al., 2011) indicating a possible role for autophagy-based unconventional secretion in amplifying autocrine and paracrine signals of the alarmins IL-1β and HMGB-1. Thus, autophagy and IL-1β have intimate relationship, resulting in context-dependent positive and negative feed-back loops. Whether TBK-1 primarily plays a role in promoting autophagic flux during IL-1β- and starvation-induced autophagy that has been uncovered in the present study, or also throttles autophagy-based positive and negative interactions with inflammasome and its clients, including IL-1β, remains to be determined.
Experimental Procedures
Cells and siRNA knockdowns
For autophagy effects, cells (RAW264.7, BMM, HeLa, MEF) were uninduced (in full medium) or induced for autophagy by starvation with EBSS for 90 min for autophagic assays wheras 4 h of induction was used in bacterial killing experiments (parallel controls with Beclin 1 or Atg7 knockdowns were used to ascertain autophagy authenticity). Rabs and Rab-like factors knockdowns details are given in Suppl. Table S1. Sources of RAW264.7, HeLa, MEFs and Atg7fl/fl LysM-Cre+ BMM are described in supplementary Experimental Procedures.
Subcellular fractionation and stable protein turn-over
Subcellular compartments were separated by isopycnic density equilibrium centrifugation in sucrose gradients. Cells were homogenized in 250 mM sucrose, 20 mM HEPES-NaOH and 0.5 mM EGTA, pH 7.5 (SHE). The post nuclear supernatant was layered on top of a pre-formed sucrose gradient consisting of 60%, 50%, 40%, 35%, 30%, 25%, 20%, 15% sucrose from top to bottom. The sample was centrifuged at 100,000g in a Beckman SW 40 rotor at 4 °C for 18 hours. Equivalent density fractions (verified for refractive index match) were analyzed with antibodies to UVRAG (MBL), TBK-1 (AbCam), LC3 (Sigma), p62 (Promega) and Rab8b (custom made); staining was revealed with Super Signal West Dura chemiluminescent substrate (Pierce). Long-lived protein turnover was measured as described previously (Roberts and Deretic, 2008). Anti-phospho-S303 p62 antibody (Matsumoto et al., 2011) was from Nobuyuki Nukina.
Phosphorylation analysis of p62
HEK293 cells were transfected using Metafectene Pro (Biontex) with GFP-p62 D69A (a mutant in the PB1 domain, to prevent oligomerization) and myc-TBK-1 or as a control TBK-1 kinase defective mutant myc-TBK-1 K38D. Immunoprecipitation of GFP-p62 D69A from cell extracts prepared from two 10-cm plates) using a custom made GFP antibody was performed as described previously (Lamark et al., 2003). Following SDS PAGE, gel bands containing GFP-p62(D69A) were excised and subjected to ingel reduction, alkylation, and tryptic digestion using 2–10 ng/µl trypsin (V511A; Promega) (Shevchenko et al., 1996). Peptide mixtures containing 0.1% formic acid were loaded onto a nanoACQUITY UltraPerformance LC (Waters), containing a 5-µm Symmetry C18 Trap column (180 µm × 20 mm; Waters) in front of a 1.7-µm BEH130 C18 analytical column (100 µm × 100 mm; Waters). Peptides were separated with a gradient of 5–95% acetonitrile, 0.1% formic acid, with a flow of 0.4 µl/min eluted to a Q-TOF Ultima mass spectrometer (Micromass/Waters). Each sample was run in ms and data dependent tandem ms mode. Peak lists were generated from MS/MS by the ProteinLynx Global server software (version 2.2; Waters). The resulting pkl files were searched against the Swiss-Prot 57.15 protein sequence databases using an in-house Mascot server (Matrix Sciences). Peptide mass tolerances used in the search were 100 ppm, and fragment mass tolerance was 0.1 Da. Mascot analysis confirmed that the sample contained p62. The Mascot analysis also confirmed that p62 was phosphorylated on Serine 403 after cotransfection with myc-TBK-1. For in vitro kinase assays recombinant maltose binding protein (MBP) and MBPp62 proteins were purified from E. coli. Recombinant TBK-1 (50 ng; Millipore) was incubated with recombinant MBP, MBP-p62 or MBP-p62 S303A in the presence of 10 mM ATP and 5mCi [γ-32P]ATP using kinase buffer (40 mM Tris– HCl pH 7.5, 10 mM MgCl2, 1 mM DTT supplemented with Calbiochem phosphatase inhibitor cocktail set II) at 30 °C. Th e reaction was terminated by boiling in SDS sample buffer after 10 min. Samples were separated by SDS– PAGE, the gels were stained with coomassie brilliant blue, dried and analysed by autoradiography.
TUBE2 pulldown
Tandem ubiquitin binding entities (TUBE) in the form of TUBE2-agarose beads (Lifesensors) were used as described (Hjerpe et al., 2009) to pull down polyubiquitinated proteins from cell lysates precleared with agarose beads for 1 h at 4oC and incubated with 20 µl of pre-equilibrated TUBE2-agarose beads in 20 mM Tris, pH 8.0, 0.15M NaCl, 0.1% Tween-20 (TBS-T) with nutation overnight at 4oC. Agarose beads were washed three times with TBS-T, eluted with 25 µl 2X Laemmeli buffer and subjected to SDS PAGE and immunoblot analysis.
High content quantitative microscopy
For quantitative autophagic puncta analyses, HeLa cells stably expressing fluorescent protein, mRFP-GFP-LC3 were employed. Cellomics Array Scan (Thermo Scientific) was used to acquire images by computer-driven collection of 49 valid fields per well with cells in 96 well plates detecting GFP fluorescent puncta (for LC3) or endogenous p62 puncta revealed by fluorescent antibody staining, and data morphometrically and statistically analyzed using puncta-counting application within the iDev software (Thermo Scientific).
Mycobacterial survival, phagosome and phagolysosome purification purification
Microbiological analyses of bacterial viability were carried out as previously described (Ponpuak et al., 2010). RAW 264.7 cells were used to isolate phagosomes or magnetic bead autophagolysosomes (Ponpuak et al., 2010) as previousy described.
Confocal microscopy and proximity ligation in situ assay (P-LISA)
Images were collected using a Zeiss LSM 510 Meta confocal microscope (laser wavelength, 488 nm, 543 nm and 633 nm). Antibodies against endogenous proteins TBK-1 (AbCam), Rab8b (AbCam & custom made), p62 (AbCam), NDP52 (Millipore), LC3 (Sigma) and UVRAG (MBL) were used for indirect immunofluorescence analysis. Procedures used for P-LISA (Soderberg et al., 2006) followed strictly manufacturer’s instructions (Olink Bioscience, Uppsala, Sweden). Details on P-LISA principles and procedures and other methods are given in Supplementary Materials.
Supplementary Material
Acknowledgments
We thank I. Vergne, R. Elmaoued, J. Naylor, and M. Mudd for assistance. We are grateful to N. Nukina for anti-phospho-S303 antibody, to H. Virgin for Atg7fl/fl LysM-Cre+ mice, and D. Rubinsztein for mRFP-GFP-LC3 HeLa cells. This work was supported by grants R01 AI069345, RC1AI086845, and R01 AI042999 from National Institutes of Health and a Bill and Melinda Gates Grand Challenge Explorations grant to V. D. and grants from the Norwegian Research Council and the Norwegian Cancer Society to T. J.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alonso S, Pethe K, Russell DG, Purdy GE. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc Natl Acad Sci U S A. 2007;104:6031–6036. doi: 10.1073/pnas.0700036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkqvist M, Wild EJ, Thiele J, Silvestroni A, Andre R, Lahiri N, Raibon E, Lee RV, Benn CL, Soulet D, et al. A novel pathogenic pathway of immune activation detectable before clinical onset in Huntington's disease. J Exp Med. 2008;205:1869–1877. doi: 10.1084/jem.20080178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K, Plater L, Peggie M, Cohen P. Use of the pharmacological inhibitor BX795 to study the regulation and physiological roles of TBK1 and IkappaB kinase epsilon: a distinct upstream kinase mediates Ser-172 phosphorylation and activation. J Biol Chem. 2009;284:14136–14146. doi: 10.1074/jbc.M109.000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criollo A, Niso-Santano M, Malik SA, Michaud M, Morselli E, Marino G, Lachkar S, Arkhipenko AV, Harper F, Pierron G, et al. Inhibition of autophagy by TAB2 and TAB3. The EMBO journal. 2011;30:4908–4920. doi: 10.1038/emboj.2011.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Toro D, Alberch J, Lazaro-Dieguez F, Martin-Ibanez R, Xifro X, Egea G, Canals JM. Mutant huntingtin impairs post-Golgi trafficking to lysosomes by delocalizing optineurin/Rab8 complex from the Golgi apparatus. Mol Biol Cell. 2009;20:1478–1492. doi: 10.1091/mbc.E08-07-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V. Autophagy as an innate immunity paradigm: expanding the scope and repertoire of pattern recognition receptors. Current opinion in immunology. 2012a;24:21–31. doi: 10.1016/j.coi.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V. Autophagy: an emerging immunological paradigm. J Immunol. 2012b;189:15–20. doi: 10.4049/jimmunol.1102108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1beta. The EMBO journal. 2011;30:4701–4711. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fremond CM, Togbe D, Doz E, Rose S, Vasseur V, Maillet I, Jacobs M, Ryffel B, Quesniaux VF. IL-1 receptor-mediated signal is an essential component of MyD88-dependent innate response to Mycobacterium tuberculosis infection. J Immunol. 2007;179:1178–1189. doi: 10.4049/jimmunol.179.2.1178. [DOI] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Hattula K, Peranen J. FIP-2, a coiled-coil protein, links Huntingtin to Rab8 and modulates cellular morphogenesis. Curr Biol. 2000;10:1603–1606. doi: 10.1016/s0960-9822(00)00864-2. [DOI] [PubMed] [Google Scholar]
- Hirota Y, Tanaka Y. A small GTPase, human Rab32, is required for the formation of autophagic vacuoles under basal conditions. Cell Mol Life Sci. 2009 doi: 10.1007/s00018-009-0080-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V, England P, Rodriguez MS. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO reports. 2009;10:1250–1258. doi: 10.1038/embor.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Kanno E, Uemura T, Waguri S, Fukuda M. OATL1, a novel autophagosome-resident Rab33B-GAP, regulates autophagosomal maturation. The Journal of cell biology. 2011;192:839–853. doi: 10.1083/jcb.201008107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen EL. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, Ishii KJ, Kawai T, Akira S, Suzuki K, et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U S A. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Korolchuk VI, Saiki S, Lichtenberg M, Siddiqi FH, Roberts EA, Imarisio S, Jahreiss L, Sarkar S, Futter M, Menzies FM, et al. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol. 2011 doi: 10.1038/ncb2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam KK, Zheng X, Forestieri R, Balgi AD, Nodwell M, Vollett S, Anderson HJ, Andersen RJ, Av-Gay Y, Roberge M. Nitazoxanide Stimulates Autophagy and Inhibits mTORC1 Signaling and Intracellular Proliferation of Mycobacterium tuberculosis. PLoS Pathog. 2012;8:e1002691. doi: 10.1371/journal.ppat.1002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamark T, Perander M, Outzen H, Kristiansen K, Overvatn A, Michaelsen E, Bjorkoy G, Johansen T. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J Biol Chem. 2003;278:34568–34581. doi: 10.1074/jbc.M303221200. [DOI] [PubMed] [Google Scholar]
- Larsen KB, Lamark T, Overvatn A, Harneshaug I, Johansen T, Bjorkoy G. A reporter cell system to monitor autophagy based on p62/SQSTM1. Autophagy. 2010;6:784–793. doi: 10.4161/auto.6.6.12510. [DOI] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 Phosphorylation of p62/SQSTM1 Regulates Selective Autophagic Clearance of Ubiquitinated Proteins. Molecular Cell. 2011;44:279–289. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. Two Beclin 1- binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- Mayer-Barber KD, Andrade BB, Barber DL, Hieny S, Feng CG, Caspar P, Oland S, Gordon S, Sher A. Innate and Adaptive Interferons Suppress IL-1alpha and IL-1beta Production by Distinct Pulmonary Myeloid Subsets during Mycobacterium tuberculosis Infection. Immunity. 2011;35:1023–1034. doi: 10.1016/j.immuni.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. The role of atg proteins in autophagosome formation. Annual review of cell and developmental biology. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Morton S, Hesson L, Peggie M, Cohen P. Enhanced binding of TBK1 by an optineurin mutant that causes a familial form of primary open angle glaucoma. FEBS Lett. 2008;582:997–1002. doi: 10.1016/j.febslet.2008.02.047. [DOI] [PubMed] [Google Scholar]
- Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nature immunology. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olkkonen VM, Dupree P, Killisch I, Lutcke A, Zerial M, Simons K. Molecular cloning and subcellular localization of three GTP-binding proteins of the rab subfamily. J Cell Sci. 1993;106(Pt 4):1249–1261. doi: 10.1242/jcs.106.4.1249. [DOI] [PubMed] [Google Scholar]
- Ou YH, Torres M, Ram R, Formstecher E, Roland C, Cheng T, Brekken R, Wurz R, Tasker A, Polverino T, et al. TBK1 Directly Engages Akt/PKB Survival Signaling to Support Oncogenic Transformation. Mol Cell. 2011;41:458–470. doi: 10.1016/j.molcel.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Perkins ND. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nature reviews Molecular cell biology. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- Ponpuak M, Davis AS, Roberts EA, Delgado MA, Dinkins C, Zhao Z, Virgin HWt, Kyei GB, Johansen T, Vergne I, et al. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity. 2010;32:329–341. doi: 10.1016/j.immuni.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke AL, Delbridge LM, Balachandran S, Barber GN, O'Riordan MX. TBK1 protects vacuolar integrity during intracellular bacterial infection. PLoS Pathog. 2007;3:e29. doi: 10.1371/journal.ppat.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Imarisio S, Sarkar S, O'Kane CJ, Rubinsztein DC. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J Cell Sci. 2008;121:1649–1660. doi: 10.1242/jcs.025726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts EA, Deretic V. Autophagic proteolysis of long-lived proteins in nonliver cells. Methods Mol Biol. 2008;445:111–117. doi: 10.1007/978-1-59745-157-4_6. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, et al. Atg9a controls dsDNAdriven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Perlstein EO, Imarisio S, Pineau S, Cordenier A, Maglathlin RL, Webster JA, Lewis TA, O'Kane CJ, Schreiber SL, et al. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nat Chem Biol. 2007;3:331–338. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Analytical chemistry. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nature methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009 doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.