Abstract
The control of translation and mRNA degradation is important in the regulation of eukaryotic gene expression. In general, translation and steps in the major pathway of mRNA decay are in competition with each other. mRNAs that are not engaged in translation can aggregate into cytoplasmic mRNP granules referred to as processing bodies (P-bodies) and stress granules, which are related to mRNP particles that control translation in early development and neurons. Analyses of P-bodies and stress granules suggest a dynamic process, referred to as the mRNA Cycle, wherein mRNPs can move between polysomes, P-bodies and stress granules although the functional roles of mRNP assembly into higher order structures remain poorly understood. In this article, we review what is known about the coupling of translation and mRNA degradation, the properties of P-bodies and stress granules, and how assembly of mRNPs into larger structures might influence cellular function.
In eukaryotic cells, translation and mRNA decay are coordinated. Nontranslating mRNAs can accumulate in P-bodies (with mRNA decay machinery) and stress granules (with translation initiation components).
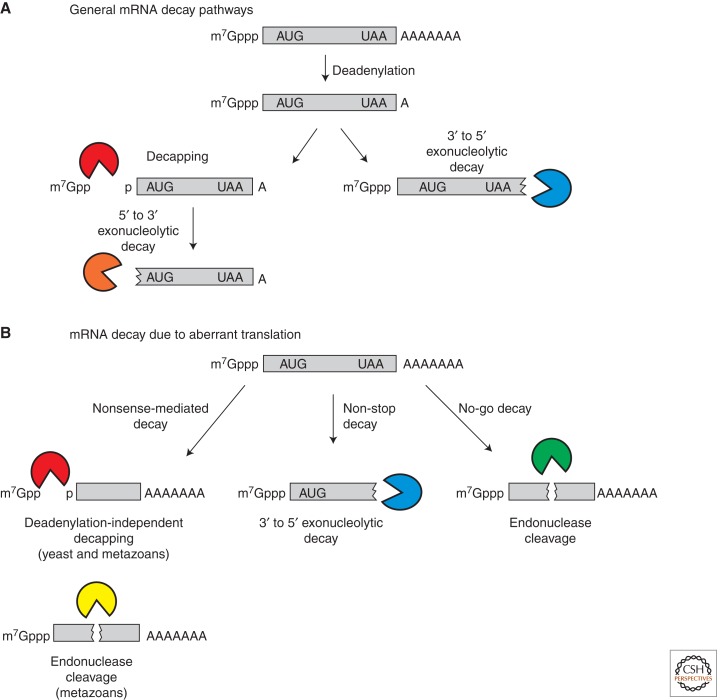
The translation and decay of mRNAs play key roles in the control of eukaryotic gene expression. The determination of eukaryotic mRNA decay pathways has allowed insight into how translation and mRNA degradation are coupled. Degradation of eukaryotic mRNAs is generally initiated by shortening of the 3′ poly (A) tail (Fig. 1A) (reviewed in Parker and Song 2004; Garneau et al. 2007) by the major mRNA deadenylase, the Ccr4/Pop2/Not complex (Daugeron et al. 2001; Tucker et al. 2001; Thore et al. 2003). Following deadenylation, mRNAs can be degraded 3′ to 5′ by the exosome (Anderson and Parker 1998; Wang and Kiledjian 2001). However, more commonly, mRNAs are decapped by the Dcp1/Dcp2 decapping enzyme and then degraded 5′ to 3′ by the exonuclease, Xrn1 (Decker and Parker 1993; Hsu and Stevens 1993; Muhlrad et al. 1994, 1995; Dunckley and Parker 1999; van Dijk et al. 2002; Steiger et al. 2003). In metazoans, a second decapping enzyme, Nudt16, also contributes to mRNA turnover (Song et al. 2010).
Figure 1.
Eukaryotic mRNA decay pathways. (A) General mRNA decay pathways. (B) Specialized decay pathways that degrade translationally aberrant mRNAs.
The processes of mRNA decay and translation are interconnected in eukaryotic cells in many ways. For example, quality control mechanisms exist to detect aberrancies in translation, which then lead to mRNAs being degraded by specialized mRNA decay pathways (Fig. 1B). Nonsense-mediated decay (NMD) is one such mRNA quality control system that degrades mRNAs that terminate translation aberrantly. In yeast, aberrant translation termination leads to deadenylation-independent decapping (Muhlrad and Parker 1994), whereas in metazoan cells NMD substrates can be both decapped and endonucleolytically cleaved and degraded (reviewed in Isken and Maquat 2007). A second quality control system for mRNA translation is referred to as no-go decay (NGD) and leads to endonucleolytic cleavage of mRNAs with strong stalls in translation elongation (Doma and Parker 2006; reviewed in Harigaya and Parker 2010). Another mechanism of mRNA quality control is the rapid 3′ to 5′ degradation of mRNAs that do not contain translation termination codons, which is referred to as non-stop decay (NSD) (Frischmeyer et al. 2002; van Hoof et al. 2002). The available evidence suggests these specialized mechanisms function primarily on aberrant mRNAs that are produced by defects in splicing, 3′ end formation, or damage to RNAs.
The main pathway of mRNA degradation is also in competition with translation initiation. Competition between the two processes was first suggested by the observation that removal of the poly (A) tail and the cap structure, both of which stimulate translation initiation, were the key steps in mRNA degradation. In addition, inhibition of translation initiation by strong secondary structures in the 5′UTR, translation initiation inhibitors, a poor AUG context, or mutations in initiation factors increases the rates of deadenylation and decapping (Muhlrad et al. 1995; Muckenthaler et al. 1997; Lagrandeur and Parker 1999; Schwartz and Parker 1999). Moreover, the cap binding protein eIF4E, known to stimulate translation initiation, inhibits the decapping enzyme, Dcp1/Dcp2, both in vivo and in vitro (Schwartz and Parker 1999; Schwartz and Parker 2000). Finally, many mRNA specific regulatory factors, (e.g., miRNAs or PUF proteins), both repress translation and accelerate deadenylation and decapping (reviewed in Wickens et al. 2002; Behm-Ansmant et al. 2006; Franks and Lykke-Anderson 2008; Shyu et al. 2008).
In the simplest model, the competition between translation and mRNA degradation can be understood through changes in the proteins bound to the cap and poly (A) tail that then influence the accessibility of these structures to deadenylases and decapping enzymes. For example, given that the Ccr4/Pop2/Not deadenylase complex is inhibited by poly (A)-binding protein (Pab1) (Tucker et al. 2002), the effects of translation on deadenylation are most likely through dynamic changes in the association of Pab1 binding with the poly (A) tail. One possibility is that defects in translation initiation either directly or indirectly decrease Pab1 association with the poly (A) tail. Deadenylation is also affected by aspects of translation termination. For instance, premature translation termination in yeast accelerates poly (A) shortening as part of the process of NMD (Cao and Parker 2003; Mitchell and Tollervey 2003). The coupling of translation termination to deadenylation has been suggested to occur through direct interactions of the translation termination factor eRF3 with Pab1 (Cosson et al. 2002), which may lead to Pab1 transiently dissociating from the poly (A) tail. Interestingly, in yeast, once the poly (A) tail reaches an oligo (A) length of 10–12 residues, a length that reduces the affinity of Pab1, the mRNA can become a substrate for decapping and for binding of the Pat1/Lsm1-7 complex (Tharun and Parker 2001; Chowdhury et al. 2007), which enhances the rate of decapping. This exchange of the Pab1 protein for the Pat1/Lsm1-7 complex is part of the mechanism that allows decapping to be promoted following deadenylation.
A similar mRNP dynamic is also likely to occur on the cap structure. Specifically, the competition between translation initiation and decapping suggests that prior to decapping, translation initiation factors are exchanged for decapping factors, thereby assembling a distinct “decapping” mRNP that is no longer capable of translation initiation (Tharun and Parker 2001). This idea is supported by the observation that some decapping activators also function as translational repressors (Coller and Parker 2005; Pilkington and Parker 2008; Nissan et al. 2010). Thus, mRNA decapping appears to occur in two steps, first inhibition of translation initiation and exchange of translation factors for the general repression/degradation machinery, and a second step whereby the mRNA is actually degraded. Thus, by understanding the changes in mRNP states between actively translating mRNAs and mRNAs that are translationally repressed and possibly stored or ultimately degraded we will better understand how the fate of mRNAs is controlled in the cytoplasm.
KEY PROTEINS PROMOTING DECAPPING AND REPRESSING TRANSLATION INITIATION
A conserved set of proteins function to modulate the transition between mRNAs engaged in translation initiation and an mRNP competent for mRNA decapping. Based on experiments in yeast, several of these conserved protein factors, referred to as either decapping enhancers or activators, function to stimulate the rate of decapping in vivo (Table 1). The core set of proteins affecting decapping includes Dhh1/Rck, a DEAD-box helicase, Pat1, Edc3, Scd6/Rap55, and the Lsm1-7 complex. Some of these decapping activators promote decapping by inhibiting translation initiation. For example, Dhh1, a member of the DEAD box family of ATPases, represses translation in vitro and its overexpression in cells inhibits translation and leads to the accumulation of cytoplasmic mRNP granules (Coller and Parker 2005; Swisher et al. 2010; Carroll et al. 2011). Similarly, Pat1, Scd6, and Stm1 (which affects the decapping of some mRNAs [Balagopal and Parker 2009]) repress translation both in vivo and in vitro (Pilkington and Parker 2008; Nissan et al. 2010; Balagopal and Parker 2011).
Table 1.
Components of P-bodies and stress granules
| Name | Function | References |
|---|---|---|
| Components found predominantly in P-bodies | ||
| Ccr4/Pop2/Not complex | Deadenylase | Sheth and Parker 2003; Cougot et al. 2004 |
| Dcp1 | Decapping enzyme subunit | Sheth and Parker 2003; Cougot et al. 2004 |
| Dcp2 | Decapping enzyme | Sheth and Parker 2003; Cougot et al. 2004 |
| Edc1 and 2 | Decapping activators | Neef and Thiele 2009 |
| Edc3 | Decapping activator | Kshirsagar and Parker 2004; Fenger-Gron et al. 2005 |
| eIF4E-T | Translation repressor | Andrei et al. 2005; Ferraiuolo et al. 2005 |
| eRF1 and eRF3 | Translation termination | Buchan et al. 2008 |
| GW182 | miRNA function | Eystathioy et al. 2003 |
| Hedls/Ge-1 | Decapping activator | Fenger-Gron et al. 2005; Yu et al. 2005 |
| Lsm1-7 complex | Decapping activator | Ingelfinger et al. 2002; Sheth and Parker 2003 |
| Pat1/PatL1 | Translation repressor/decapping activator | Sheth and Parker 2003; Scheller et al. 2007 |
| Upf1-3 | Nonsense-mediated decay | Sheth and Parker 2006; Durand et al. 2007 |
| Components found predominantly in stress granules | ||
| 40S ribosomal subunit | Translation | Kedersha et al. 2002; Grousl et al. 2009 |
| Ataxin-2/Pbp1 | Translation/mRNA processing | Nonhoff et al. 2007; Buchan et al. 2008 |
| DDX3/Ded1 | RNA helicase | Chalupnikova et al. 2008; Lai et al. 2008; Hilliker et al. 2011 |
| eIF2α | Translation initiation | Kedersha et al. 2002; Kimball et al. 2003 |
| eIF3 | Translation initiation | Kedersha et al. 2002; Grousl et al. 2009 |
| eIF4A | Translation initiation | Low et al. 2005; Buchan et al. 2011 |
| eIF4B | Translation initiation | Low et al. 2005; Buchan et al. 2011 |
| eIF4G | Translation initiation | Kedersha et al. 2005; Hoyle et al. 2007 |
| FMRP | Translation, repression/miRNA function | Mazroui et al. 2002; Kim et al. 2006 |
| G3BP | Scaffolding protein, endoribonuclease | Tourriere et al. 2001 |
| Pabp | polyA-binding protein | Kedersha et al. 1999; Hoyle et al. 2007 |
| RACK1 | Signaling scaffold protein | Arimoto et al. 2008 |
| TIA-1/TIAR/Pub1/Ngr1 | Translation repression/mRNA stability | Kedersha et al. 1999; Buchan et al. 2008 |
| Components found in both P-bodies and stress granules | ||
| Agonaute proteins | miRNA function | Sen and Blau 2005; Leung et al. 2006 |
| Dhh1/Rck/p54/ | Translation repressor/decapping activator | Sheth and Parker 2003; Wilczynska et al. 2005 |
| eIF4E | Translation initiation | Andrei et al. 2005; Ferraiuolo et al. 2005; Hoyle et al. 2007 |
| FAST | Fas activated serine/ threonine phosphoprotein | Kedersha et al. 2005 |
| Rap55/Scd6 | Translation repressor | Yang et al. 2006; Teixeira and Parker 2007 |
| Xrn1 | 5′ to 3′ exonuclease | Sheth and Parker 2003; Kedersha et al. 2005 |
Decapping activators can inhibit translation at different steps. For example, the Pat1, Dhh1, and Scd6 proteins all appear to block translation before the formation of a 48S preinitiation complex (Coller and Parker 2005; Nissan et al. 2010). For Scd6, this translation repression appears to occur by direct binding to eIF4G and inhibition of the joining of the 43S complex (Rajyaguru et al. 2012). In contrast, the Stm1 protein, which promotes decapping of a subset of yeast mRNAs (Balagopal and Parker 2009), inhibits translation after formation of an 80S complex, likely through direct interactions with the ribosome (Balagopal and Parker 2011). An unresolved issue is how inhibition of translation initiation by these factors leads to decapping. One possibility is that by stalling initiation, it simply gives more time for dissociation of the translation initiation factors to allow for decapping complexes to associate with the mRNA. Alternatively, such a transition may involve an ordered exchange of factors on the mRNA, which is suggested by decapping activators, such as Pat1 and Scd6 that can directly interact with translation factors and the decapping enzyme (Nissan et al. 2010; Fromm et al. 2011). An important area for future research is determining how mRNPs are remodeled from a translationally active state to allow decapping complexes to form and degrade the mRNA. Moreover, a quantitative kinetic analysis of the binding interactions between mRNAs and translation initiation components and decapping factors would shed light on the apparent competition between translation initiation and decapping.
A second role of decapping activators is to promote the assembly of a larger decapping complex, which might also indirectly prevent translation initiation by limiting the interaction of translation initiation factors with the mRNA. The core set of decapping components shows an extensive network of direct interactions as determined by protein binding experiments with recombinant proteins and supported by coimmunoprecipitation and two hybrid analyses (Decker et al. 2007; Nissan et al. 2010). Based on coimmunoprecipitation results and the dependence of interactions on RNA, there appear to be two complexes that assemble on mRNAs targeted for decapping. One complex consists of the Pat1 protein, the Lsm1-7 complex, and Xrn1 (Bouveret et al. 2000; Tharun et al. 2000; Tharun and Parker 2001). This complex is thought to assemble on the 3′ end of deadenylated mRNAs based on its binding specificity in vitro (Chowdury et al. 2007) and the exonuclease trimming of deadenylated mRNAs in the absence of Pat1 or Lsm1 (Boeck et al. 1998; Tharun et al. 2000; He and Parker 2001). A second set of interacting proteins consists of Dcp1, Dcp2, Edc3 or Scd6 and Dhh1, although whether all these factors can associate at the same time remains to be determined. Within and between these complexes, Pat1 and Edc3 appear to play important scaffolding roles and interact with many components of the decapping machinery (Decker et al. 2007; Nissan et al. 2010).
A third role of decapping activators is to directly stimulate decapping by Dcp2. For example, the Edc3 and Pat1 proteins directly bind Dcp2 and enhance its activity in purified systems (Harigaya et al. 2010; Nissan et al. 2010). Similarly, the paralogs Edc1 and Edc2 in yeast, which are high copy suppressors of temperature- sensitive alleles in Dcp1 or Dcp2 (Dunckley et al. 2001), bind RNA and stimulate Dcp2 either in extracts or in reconstituted systems (Schwartz et al. 2003; Steiger et al. 2003). Edc1, and presumably Edc2 as well, directly bind Dcp1 to stimulate the decapping enzyme by enhancing both Km and kcat of Dcp2 (Borja et al. 2011).
Taken together, these proteins appear to modulate the mRNP composition on mRNAs to create two different classes of mRNPs, those associated with translation initiation factors and capable of recruiting ribosomes, and those associated with translational repressors and the mRNA degradation machinery. This dynamic between two different functional states also appears to be modulated by sequence specific RNA binding factors. For example, the Puf5 protein acts to repress translation and promote degradation at least in part by directly recruiting the Ccr4/Pop2/Not mRNA deadenylase complex through interactions with Pop2 (Goldstrum and Wickens 2006). Similarly, recent results show that miRNA-mediated translation repression and mRNA degradation are promoted by direct interactions between GW182 and the Not1 protein, which is a component of the Ccr4/Pop2/Not complex (Braun et al. 2011; Chekulaeva et al. 2011; Fabian et al. 2011).
NONTRANSLATING mRNAS CAN ASSEMBLE INTO RNA–PROTEIN GRANULES
In eukaryotic cells, nontranslating mRNAs can accumulate in two types of cytoplasmic mRNP granules: P-bodies, which contain the mRNA decay machinery (reviewed in Anderson and Kedersha 2006; Parker and Sheth 2007; Franks and Lykke-Andersen 2008), and stress granules, which contain many translation initiation components (Table 1) (reviewed in Buchan and Parker 2009). Moreover, stress granules and P-bodies are related to neuronal RNA granules and germ granules, which play important roles in the localization and control of mRNAs in neurons and embryos (reviewed in Kiebler and Bassell 2006; Seydoux and Braun 2006).
P-bodies are present in unstressed cells but are further induced in response to stresses or other conditions that lead to the inhibition of translation initiation (Kedersha et al. 2005; Teixeira et al. 2005). P-bodies are dynamic complexes whose assembly is dependent on, and proportional to, the pool of nontranslating mRNA (Liu et al. 2005a; Pillai et al. 2005; Teixeira et al. 2005). In addition to nontranslating mRNA, P-bodies contain the conserved core of proteins involved in mRNA decay and translation repression. These factors include the decapping enzyme complex Dcp1/Dcp2; the decapping activators Edc3 and the Lsm1-7 complex; factors that function to repress translation as well as to activate decapping including Dhh1/RCK/p54, Pat1 and Scd6/RAP55; the 5′ to 3′ exonuclease, Xrn1, and the Ccr4/Pop2/Not deadenylase complex (reviewed in Anderson and Kedersha 2006; Eulalio et al. 2007a; Parker and Sheth 2007). P-bodies can also contain mRNAs and proteins involved in NMD (Sheth and Parker 2006; reviewed in Franks and Lykke-Anderson 2008; Shyu et al. 2008). Metazoan P-bodies contain additional factors including proteins and miRNAs involved in the miRNA repression pathway (Eulalio et al. 2008; Lian et al. 2009). Translation initiation factors and ribosomal proteins are generally excluded from P-bodies with the exception of eIF4E in mammalian P-bodies. However, eIF4E is likely associated with repressed nontranslating mRNA in P-bodies given that P-bodies also contain eIF4E-T, which inhibits eIF4E function (Andrei et al. 2005; Ferraiuolo et al. 2005).
Stress granules are a second type of cytoplasmic mRNP granule that can be juxtaposed or overlap with P-bodies in both yeast and mammalian cells (Kedersha et al. 2005; Brengues et al. 2007; Hoyle et al. 2007; Buchan et al. 2008). Like P-bodies, stress granules are dynamic complexes whose assembly is dependent on the pool of nontranslating mRNAs. Although they share some components in common with P-bodies, stress granules typically contain translation initiation factors eIF4E, eIF4G, eIF4A, eIF4B, poly-A binding protein (Pabp), eIF3, eIF2, and the 40S ribosomal subunit (reviewed in Buchan and Parker 2009). Their composition suggests that stress granules are aggregates of mRNPs stalled in the process of translation initiation. Indeed, stress granules were first observed under stress conditions, in which translation initiation is often inhibited (Kedersha et al. 1999). However, it is now clear that stress granule formation is not limited to stress conditions, but can occur in response to a variety of blocks in translation initiation. For example, inhibition of translation initiation using drugs, knock down of translation initiation factors, or overexpression of translation repressors have all been shown to induce stress granules (reviewed in Buchan and Parker 2009). Interestingly, not all blocks to translation initiation induce stress granule assembly. For example, stress granules fail to assemble in response to depletion of eukaryotic initiation factor 3 (eIF3) subunits or to reduction in 60S subunit joining (Ohn et al. 2008; Mokas et al. 2009) suggesting there is a defined window within which translation needs to be stalled for an mRNP to be targeted to stress granules. Depending on conditions, stress granules can contain many other protein components including RNA helicases, regulators of translation and mRNA stability, and factors involved in cell signaling (reviewed in Buchan and Parker 2009; Kedersha and Anderson 2009).
HOW DO CYTOPLASMIC mRNP GRANULES ASSEMBLE?
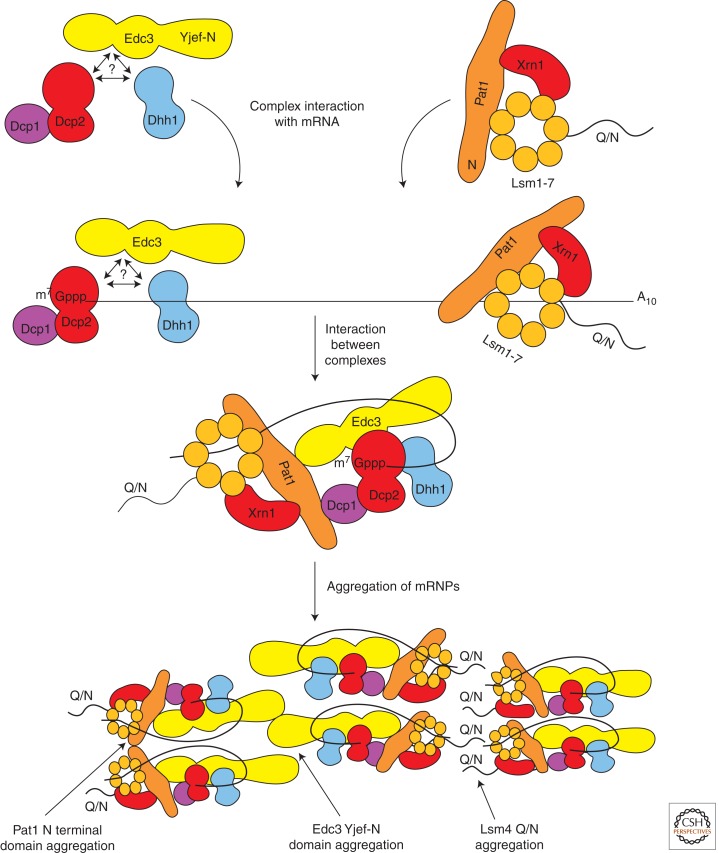
The dependence on nontranslating mRNA on the formation of both P-bodies and stress granules suggests that they assemble first through the formation of translationally repressed mRNPs, which then aggregate into larger structures by specific protein–protein interaction domains. In yeast, P-body assembly may involve the recruitment of preexisting protein complexes to the mRNA given, as described above, that two sub-complexes of core P-body components copurify under a variety of conditions and appear to interact independent of RNA (Fig. 2). (Bouveret et al. 2000; Tharun et al. 2000; Tharun and Parker 2001; Fenger-Grøn et al. 2005; Gavin et al. 2006; Teixeira and Parker 2007). Together these observations suggest that the Dcp1/Dcp2/Dhh1/Edc3 complex, or an alternative Dcp1/Dcp2/Dhh1/Scd6 complex (Fromm et al. 2011), and the Pat1, Xrn1, and the Lsm1-7p complex are recruited onto mRNA as two groups, though the exact order of recruitment is unknown. Moreover, because Edc3, Dcp2, Scd6, and Dhh1 can interact with Pat1, these two complexes are proposed to interact to form a larger RNA–protein complex (Pilkington and Parker 2008; Nissan et al. 2010). Interestingly, because the Pat1-Lsm1-7p complex has been proposed to bind the 3′ end (Chowdhury and Tharun 2009), and the decapping enzyme binds to the cap, this suggests a possible “closed-loop” model for mRNPs that assemble to form P-bodies. A similar rich interaction network between P-body components is likely to be the basis for formation of “P-body mRNPs” in metazoans although some of the specific interactions have been replaced or swapped between partners and additional factors also contribute.
Figure 2.
Model for P-body assembly in yeast. First, P-body factors are recruited to the mRNA as complexes. Second, interactions between P-body proteins in the complexes lead to the formation of a “closed-loop” structure. Finally, mRNPs aggregate via the Q/N domain of Lsm4, the Yjef-N domain of Edc3, or the amino-terminal domain of Pat1 to form microscopically visible cytoplasmic granules.
P-bodies are then formed from these individual mRNPs aggregating into larger structures. In yeast, aggregation of mRNPs into P-bodies has been shown to be primarily dependent on a self-interaction domain (referred to as a Yjef-N domain) in the Edc3 protein and a glutamine/asparagine (Q/N) rich prion-like domain in the Lsm4 carboxyl terminus (Decker et al. 2007; Mazzoni et al. 2007; Reijns et al. 2008). Because the YjeF domain of Edc3 is conserved (Ling et al. 2008), it is likely that Edc3 will contribute to assembly of metazoan P-bodies. However, because depletion of Edc3 does not block P-body assembly in Drosophila S2 cells (Eulalio et al. 2007b), one anticipates that Q/N domains, and possibly other mechanisms, contribute to metazoan P-body assembly. Interestingly, multiple proteins in metazoan P-bodies contain Q/N rich domains including GW182, which functions in miRNA-mediated repression, and Ge-1/Hedls, a component of the metazoan decapping enzyme. Moreover, depletion of either of these proteins leads to decreased P-bodies in human and Drosophila cells (Liu et al. 2005b; Yu et al. 2005; Eulalio et al. 2007b). Finally, the Pat1 protein contributes to P-body aggregation (Buchan et al. 2008), possibly because of its role as a scaffold interacting with multiple P-body components including the Lsm1-7 complex, which is dependent on Pat1 for its localization to P-bodies (Texeira and Parker 2007). An unresolved and intriguing issue is why these protein components do not aggregate all the time. One possibility is that interaction with mRNA might in some manner promote the aggregation interactions that lead to P-bodies.
Multiple mechanisms involving protein–protein interactions between RNA-binding proteins have been implicated in the assembly of initiation-stalled mRNPs into stress granules. One mechanism of stress granule assembly is through the self-aggregation of QN-rich prion-like domains in the RNA binding proteins TIA-1 and TIA-R, and their orthologs (Kedersha et al. 1999, 2000; Gilks et al. 2004). Because aggregation of QN-rich prion domains is reversed by specific heat shock protein function (Rikhvanov et al. 2007), one possibility is that stress granule and P-body assembly may be promoted during stress because of accumulation of unfolded proteins, which may titrate heat shock proteins, thus driving the equilibrium of QN-rich domains toward an aggregated state. A second mechanism that contributes to stress granule assembly is the dimerization of G3BP protein (Tourriere et al. 2003). Assembly factors important under one stress condition can be unimportant during other stresses. For example, TIA-1, and its yeast homolog Pub1, facilitate stress granule assembly in response to arsenite and glucose deprivation, respectively (Gilks et al. 2004; Buchan et al. 2008), but not in response to other stresses such as heat shock (Lopez de Silanes 2005; Grousl et al. 2009). Therefore, the nature of the stress, which shapes the nontranslating mRNP pool, likely defines the assembly rules for stress granule formation.
Posttranslational modifications likely play a role in controlling the assembly of both P-bodies and stress granules (reviewed in Hilliker and Parker 2008; Buchan and Parker 2009). For example, phosphorylation of G3PB inhibits stress granule assembly (Tourriere et al. 2003). Recently, Pat1 was identified as a target for PKA phosphorylation and its phosphorylation interferes with the assembly of P-body aggregates (Ramachandran et al. 2011). Modification of proteins with O-Glc-NAc also enhances stress granule formation (Ohn et al. 2008). Finally, methylation, or the ability to bind methyl groups via Tudor domains, is necessary for localization of specific stress granule components (de Leeuw et al. 2007; Goulet et al. 2008), or their ability to drive stress granule formation when overexpressed (Hua and Zhou 2004). Methylation and Tudor domains have also been implicated in the assembly of other RNA granules (Thomson and Lasko 2004; Arkov et al. 2006; Chuma et al. 2006).
Posttranslational modification of mRNP components is an ideal mechanism to modulate mRNA function during a stress, in which rapid and reversible protein modifications allow adaptation to stress without new protein synthesis. Elucidating the key physiological targets of various modifications, and the mechanisms underlying their effects, will therefore be an important future goal.
DYNAMICS OF mRNPs IN THE CYTOPLASM
Several observations argue that cytoplasmic mRNAs can cycle between polysomes, P-bodies, and stress granules. First, inhibition of translation initiation by drugs, stresses, or mutations leads to loss of mRNAs from polysomes and a corresponding increase of mRNAs in P-bodies and stress granules (Kedersha et al. 2005; Teixeira et al. 2005; Anderson and Kedersha 2006). Second, trapping mRNAs in polysomes by blocking translation elongation decreases P-bodies and stress granules even during continued stress, which suggests that mRNAs in these compartments are in dynamic equilibrium with polysomes (Kedersha et al. 2000; Cougot et al. 2004; Teixeira et al. 2005; Mollet et al. 2008). This is consistent with the dynamic nature of P-bodies and stress granules based on fluorescence recovery after photobleaching (FRAP) studies (Andrei et al. 2005; Kedersha et al. 2005). Third, P-bodies and stress granules physically interact, often docking together in mammalian cells during stress (Kedersha et al. 2005; Wilczynska et al. 2005) or partially overlapping in yeast (Brengues and Parker 2007; Hoyle et al. 2007; Buchan et al. 2008). Finally, mRNAs within P-bodies and stress granules in yeast and mammalian cells can return to translation (Brengues et al. 2005; Anderson and Kedersha 2006; Bhattacharyya et al. 2006).
The mechanisms and directionality of mRNA movement between P-bodies and stress granules remain unresolved. During glucose deprivation in yeast, stress granules form after P-bodies, they primarily assemble on preexisting P-bodies, and are dependent on existing P-bodies for their efficient assembly (Buchan et al. 2008). This suggests that yeast mRNAs exiting translation first form a P-body mRNP, and then mRNAs, which are targeted for reentry into translation, undergo mRNP remodeling to load translation initiation factors, thereby forming the type of mRNP that accumulates in stress granules. Some evidence however suggests that stress granules also form independently of P-bodies. First, in mammalian cells and with sodium azide stress in yeast, stress granules often form independently of visible P-bodies (Kedersha et al. 2005; Mollet et al. 2008, Buchan et al. 2011). Second, depletion of some factors in mammalian cells prevents P-body formation without affecting stress granule formation, suggesting the two processes can be uncoupled (Ohn et al. 2008). One possibility is that mRNAs may exchange in a bidirectional manner between stress granules and P-bodies and the specific mRNA, cell type, or condition may affect the predominant flow of bulk mRNA.
The movement of mRNAs between polysomes, stress granules, and P-bodies implies transitions between different mRNP states through specific rearrangements and exchanges of proteins on individual mRNAs. This may be facilitated by RNA helicases as exemplified by the Ded1 protein. Ded1 acts to assemble an mRNP intermediate that is stalled in translation initiation and accumulates in stress granules, which it then resolves in an ATP-dependent manner to allow the mRNA to reenter translation (Hilliker et al. 2011). Although P-bodies and stress granules represent microscopically visible aggregates of different mRNPs, the simplest model is that these mRNP transitions can occur independently of the larger aggregates. An important area of future work will be to determine how mRNPs in either stress granules or P-bodies are remodeled to affect their fate and how that impinges on the control of gene expression and response to stress.
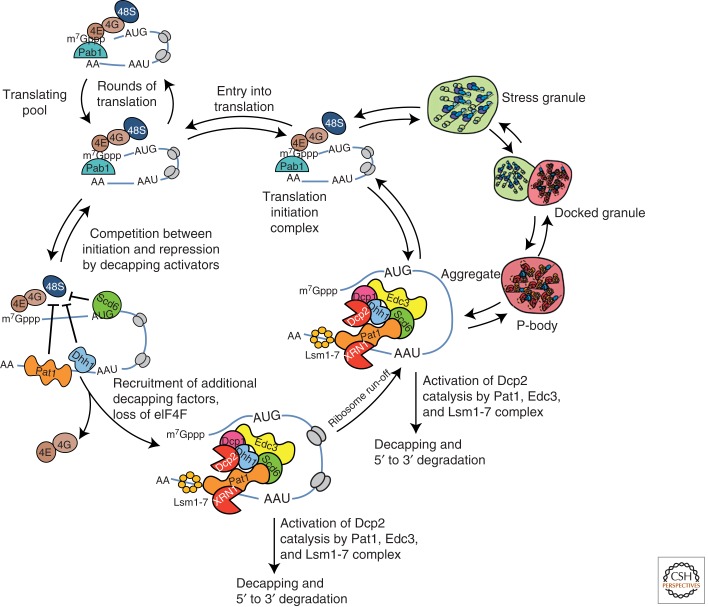
A WORKING MODEL: THE mRNA CYCLE
The analyses of P-bodies and stress granules suggest a working model for the metabolism of cytoplasmic mRNA termed the mRNA Cycle (Fig. 3). In this model, mRNAs present in polysomes undergo repeated rounds of translation initiation, elongation, and termination to produce polypeptides. In response to defects in translation initiation and/or termination, or through specific recruitment, mRNAs found in polysomes interact with proteins that repress translation initiation such as Dhh1/Rck and Pat1. At this stage, we envision run off of the elongating ribosomes, recruitment of the remainder of the decapping machinery including the decapping enzyme, and decapping followed by transcript degradation, although the relative timing of these events is unclear. After assembly of the decapping machinery individual mRNPs may aggregate into a P-body. mRNAs that are stalled in translation initiation but fail to recruit P-body components might accumulate in stress granules after elongating ribosomes run off the mRNA.
Figure 3.
Model of the “mRNA cycle.” Showing the dynamic movement of mRNA between polysomes, P-bodies, and stress granules, and the possible mRNP transitions between the different states of the mRNA.
In this model, we suggest that mRNAs complexed with the decapping machinery can be degraded, aggregate into a P-body, or undergo an mRNP rearrangement wherein the degradation machinery is exchanged for translation initiation factors. Such mRNAs could then go on to initiate translation and enter polysomes. When translation initiation is inhibited, however, these mRNPs could accumulate in the stress granule state before eventually entering polysomes. Further, the state at which translation initiation is limiting might define the composition of the stress granule and therefore stress granule composition might vary in different organisms or in response to different stresses. One anticipates that specific mRNAs may preferentially accumulate in stress granules, P-bodies, or polysomes depending on their relative rates of transitions between these different biochemical states.
WHY DO mRNPs AGGREGATE?
An unresolved issue is the significance of mRNP aggregation into P-bodies and stress granules. The presence of the mRNA decay machinery and mRNA decay intermediates in P-bodies is consistent with the possibility that mRNA degradation can occur in these structures (Sheth and Parker 2003). This possibility is supported by the observation that P-bodies increase in number and size when mRNA decay is inhibited at the stage of decapping by deletion of Dcp1 or after decapping by the deletion of Xrn1 (Sheth and Parker 2003; Cougot et al. 2004). However, to date, aggregation of mRNPs into microscopically visible P-bodies has been shown not to be required for mRNA decapping in yeast (Decker et al. 2007), for translation repression during stress in both yeast and mammals (Decker et al. 2007; Kwon et al. 2007; Buchan et al. 2008; Ohn et al. 2008), or for mRNA stability during stress, at least in yeast (Buchan 2008). In addition, in metazoans, depletion of microscopically visible P-bodies does not seem to affect miRNA-mediated repression, decay of messages containing ARE elements, or decay of transcripts subject to NMD (Chu and Rana 2006; Stoecklin et al. 2006; Eulalio et al. 2007b). In contrast, deletion of the Lsm4 carboxy-terminal domain, which promotes P-body assembly (Decker et al. 2007; Mazzoni et al. 2007; Reijns et al. 2008), can affect mRNA degradation in at least some strains and/or conditions (Reijns et al. 2008), which suggests that aggregation of individual mRNPs into larger structures may have some role in mRNA degradation. In addition, there is some evidence that P-body aggregation may play a role in the long-term survival of yeast cells during stationary phase (Ramachandran et al. 2011). Moreover, because aggregation into RNP granules is a conserved feature of eukaryotic cells it is anticipated to have some role. One possibility is that mRNP aggregation has consequences for the control of translation and/or degradation but these functions are either limited to a subset of mRNAs or conditions, or are performed by granules below the detection limit of the light microscope.
More generally, the formation of RNP granules such as stress granules and P-bodies is expected to have specific consequences both by increasing the local concentration of factors within granules, and by depleting them from the bulk cytosol. For comparison, Cajal bodies improve the assembly of spliceosomal small nuclear ribonucleoprotein particles (snRNPs) by increasing the local concentrations of U4/U6 (Klingauf et al. 2006). By analogy, the concentration of Dcp2 in P-bodies might facilitate its interaction with mRNAs when Dcp2 is limiting, or the concentration of translation initiation factors in stress granules might drive the formation of productive translation complexes. In addition, an important role of RNP granules may be to remove factors from the cytosol. For example, formation of stress granules may sequester RACK1 away from MAP kinases, thereby limiting signal transduction and apoptosis (Arimoto et al. 2008). Moreover, the aggregation of mRNPs into stress granules and P-bodies may provide a buffering system for maintaining a proper ratio of translation capacity to the pool of mRNAs that are translating (discussed in Coller and Parker 2004). An excessive amount of mRNAs within the translating pool may compete for limiting translation factors and thereby prevent effective translation of many mRNAs.
CONCLUDING REMARKS
Although considerable advances have been made in the understanding of the mechanisms of mRNA decapping and the subcellular distribution of different mRNPs, there are several outstanding questions that need to be addressed. One key issue will be to understand the molecular functions of decapping activators and how they affect translation mechanisms as well as the recruitment and stimulation of the decapping enzyme. A second important challenge is to understand the significance of the aggregation of mRNPs into P-bodies and stress granules, which is likely to contribute to our growing understanding of the importance of sub-cellular organization. Finally, it will be critical to understand the mechanisms and rates of the transitions of mRNPs between polysomes, P-bodies, and stress granules. Here it will be critical to understand the frequency and directionality of these exchanges, the molecular mechanisms that move mRNAs from one state to another, and how these states differ on individual mRNAs, thereby impacting the control of gene expression.
ACKNOWLEDGMENTS
We thank R. Buchan, S. Jain, and V. Balagopal for helpful discussions and A. Webb for technical support in the preparation of the manuscript. This work is supported by funds from the National Institutes of Health (Grant R37 GM45443) and the Howard Hughes Medical Institute.
Footnotes
Editors: John W.B. Hershey, Nahum Sonenberg, and Michael B. Mathews
Additional Perspectives on Protein Synthesis and Translational Control available at www.cshperspectives.org
REFERENCES
- Anderson P, Kedersha N 2006. RNA Granules. J Cell Biol 172: 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JS, Parker RP 1998. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J 17: 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Luhrmann R 2005. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 11: 717–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M 2008. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat Cell Biol 10: 1324–1332 [DOI] [PubMed] [Google Scholar]
- Arkov A, Wang JY, Ramos A, Lehmann R 2006. The role of Tudor domains in germline development and polar granule architecture. Development 133: 4053–4062 [DOI] [PubMed] [Google Scholar]
- Balagopal V, Parker R 2009. Stm1 modulates mRNA decay and Dhh1 function in Saccharomyces cerevisae. Genetics 181: 93–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopal V, Parker R 2011. Stm1 modulates translation after 80S formation in Saccharomyces cerevisiae. RNA 17: 835–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E 2006. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 20: 1885–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W 2006. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125: 1111–1124 [DOI] [PubMed] [Google Scholar]
- Boeck R, Lapeyre B, Brown CE, Sachs AB 1998. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol Cell Biol 18: 5062–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borja MS, Piotukh K, Freund C, Gross JD 2011. Dcp1 links coactivators of mRNA decapping to Dcp2 by proline recognition. RNA 17: 278–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret E, Rigaut G, Shevchenko A, Wilm M 2000. A Sm-like protein complex that participates in mRNA degradation. EMBO J 19: 1661–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun JE, Huntzinger E, Fauser M, Izzaurralde E 2011. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell 44: 120–133 [DOI] [PubMed] [Google Scholar]
- Brengues M, Parker R 2007. Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol Biol Cell 18: 2592–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M, Teixeira D, Parker R 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310: 486–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R 2009. Eukaryotic stress granules: The ins and outs of translation. Mol Cell 36: 932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Muhlrad D, Parker R 2008. P-bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol 183: 441–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Yoon J-H, Parker R 2011. Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J Cell Sci 124: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Parker R 2003. Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell 113: 533–545 [DOI] [PubMed] [Google Scholar]
- Carroll JS, Munchel SE, Weis K 2011. The DExD/H box ATPase Dhh1 functions in translational repression, mRNA decay, and processing body dynamics. J Cell Biol 194: 527–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalupnikova K, Lattmann S, Selak N, Iwamoto F, Fujiki Y, Nagamine Y 2008. Recruitment of the RNA helicase RHAU to stress granules via a unique RNA-binding domain. J Biol Chem 283: 35186–35198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M, Mathys H, Zipprich JT, Attig J, Colic M, Parker R, Filipowicz W 2011. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat Struct Mol Biol 18: 1218–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A, Tharun S 2009. Activation of decapping involves binding of the mRNA and facilitation of the post-binding steps by the Lsm1-7-Pat1 complex. RNA 15: 1837–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A, Mukhopadhyay J, Tharun S 2007. The decapping activator Lsm1p 7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA 13: 998–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CY, Rana TM 2006. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol 4: e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuma S, Hosokawa M, Kitamura K, Kasai S, Fujioka M, Hiyoshi M, Takamune K, Noce T, Nakatsuji N 2006. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc Natl Acad Sci 103: 15894–15899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R 2004. Eukaryotic mRNA decapping. Annu Rev Biochem 73: 861–890 [DOI] [PubMed] [Google Scholar]
- Coller J, Parker R 2005. General translational repression by activators of mRNA decapping. Cell 122: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson B, Couturier A, Chabelskaya S, Kiktev D, Inge-Vechtomov S, Philippe M, Zhouravleva G 2002. Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI(+)] propagation. Mol Cell Biol 22: 3301–3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N, Babajko S, Seraphin B 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol 165: 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugeron MC, Mauxion F, Seraphin B 2001. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res 29: 2448–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Parker R 1993. A turnover pathway for both stable and unstable mRNAs in yeast: Evidence for a requirement for deadenylation. Genes Dev 7: 1632–1643 [DOI] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, Parker R 2007. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol 179: 437–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leeuw F, Zhang T, Wauquier C, Huez G, Kruys V, Gueydan C 2007. The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. Exp Cell Res 313: 4130–4144 [DOI] [PubMed] [Google Scholar]
- Doma MK, Parker R 2006. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440: 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley T, Parker R 1999. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J 18: 5411–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley T, Parker R 2001. Two related proteins, Edc1p and Edc2p, stimulate mRNA decapping in Saccharomyces cerevisiae. Genetics 157: 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand S, Cougot N, Mahuteau-Betzer F, Nguyen CH, Grierson DS, Bertrand E, Tazi J, Lejuene F 2007. Inhibition of nonsense-mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P-bodies. J Cell Biol 178: 1145–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E 2007a. P-bodies: At the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E 2007b. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol 27: 3970–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E 2008. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol 15: 346–353 [DOI] [PubMed] [Google Scholar]
- Eystathioy T, Jakymiw A, Chan EK, Séraphin B, Cougot N, Fritzler MJ 2003. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA 9: 1171–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Cieplak MK, Frank F, Morita M, Green J, Srikumar T, Nagar B, Yamamoto T, Raught B, Duchaine TF, et al. 2011. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat Struct Mol Biol 18: 1211–1217 [DOI] [PubMed] [Google Scholar]
- Fenger-Grøn M, Fillman C, Norrild B, Lykke-Andersen J 2005. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Molecular Cell 20: 905–915 [DOI] [PubMed] [Google Scholar]
- Ferraiuolo MA, Basak S, Dostie J, Murray EL, Schoenberg DR, Sonenberg N 2005. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J Cell Biol 170: 913–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J 2008. The control of mRNA decapping and P body formation. Mol Cell 32: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer PA, van Hoof A, O'Donnell K, Guerrerio AL, Parker R, Dietz HC 2002. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295: 2258–2261 [DOI] [PubMed] [Google Scholar]
- Fromm SA, Truffault V, Kamenz J, Braun JE, Hoffmann NA, Izaurralde E, Sprangers R 2011. The structural basis of Edc3- and Scd6-mediated activation of the Dcp1:Dcp2 mRNA decapping complex. EMBO J 31: 279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ 2007. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 8: 113–126 [DOI] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dümpelfeld B, et al. 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature 440: 631–636 [DOI] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 15: 5383–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm AC, Hook BA, Seay DJ, Wickens M 2006. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol 13: 533–539 [DOI] [PubMed] [Google Scholar]
- Goulet I, Boisvenue S, Mokas S, Mazroui R, Côté J 2008. TDRD3, a novel Tudor domain-containing protein, localizes to cytoplasmic stress granules. Hum Mol Genet 17: 3055–3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grousl T, Ivanov P, Frydlova I, Vasicova P, Janda F, Vojtova J, Malinska K, Malcova I, Novakova L, Janoskova D, Hasek J, et al. 2009. Robust heat shock induces eIF2α-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J Cell Sci 122: 2078–2088 [DOI] [PubMed] [Google Scholar]
- Harigaya Y, Parker R 2010. No-go decay: A quality control mechanism for RNA in translation. RNA 1: 132–141 [DOI] [PubMed] [Google Scholar]
- Harigaya Y, Jones BN, Muhlrad D, Gross JD, Parker R 2010. Identification and analysis of the interaction between Edc3 and Dcp2 in Saccharomyces cerevisiae. Mol Cell Biol 30: 1446–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Parker R 2001. The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3′ termini from partial degradation. Genetics 158: 1445–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker A, Parker R 2008. Stressed out? Make some modifications! Nat Cell Biol 10: 1129–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker A, Gao Z, Jankowsky E, Parker R 2011. The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol Cell 43: 962–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyle NP, Castelli LM, Campbell SG, Holmes LEA, Ashe MP 2007. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J Cell Biol 179: 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CL, Stevens A 1993. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol 13: 4826–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Zhou J 2004. Survival motor neuron protein facilitates assembly of stress granules. FEBS Lett 572: 69–74 [DOI] [PubMed] [Google Scholar]
- Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T 2002. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA 8: 1489–1501 [PMC free article] [PubMed] [Google Scholar]
- Isken O, Maquat LE 2007. Quality control of eukaryotic mRNA: Safeguarding cells from abnormal mRNA function. Genes Dev 21: 1833–1856 [DOI] [PubMed] [Google Scholar]
- Kedersha N, Anderson P 2009. Regulation of translation by stress granules and processing bodies. Prog Mol Biol Transl Sci 90: 155–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 α to the assembly of mammalian Stress Granules. J Cell Biol 147: 1431–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol 151: 1257–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Chen S, Gilks N, Li W, Miller IJ, Stahl J, Anderson P 2002. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol Biol Cell 13: 195–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol 169: 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler MA, Bassell GJ 2006. Neuronal RNA granules: Movers and makers. Neuron 51: 685–90 [DOI] [PubMed] [Google Scholar]
- Kim SH, Dong WK, Weiler IJ, Greenough WT 2006. Fragile X mental retardation protein shifts between polyribosomes and stress granules after neuronal injury by arsenite stress or in vivo hippocampal electrode insertion. J Neurosci 26: 2413–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball SR, Horetsky RL, Ron D, Jefferson LS, Harding HP 2003. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am J Physiol Cell Physiol 284: C273–84 [DOI] [PubMed] [Google Scholar]
- Klingauf M, Stanek D, Neugebauer KM 2006. Enhancement of U4/U6 small nuclear ribonucleoprotein particle association in Cajal bodies predicted by mathematical modeling. Mol Biol Cell 17: 4972–4981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kshirsagar M, Parker R 2004. Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics 166: 729–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S, Zhang Y, Matthias P 2007. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev 21: 3381–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrandeur T, Parker R 1999. The cis acting sequences responsible for the differential decay of the unstable MFA2 and stable PGK1 transcripts in yeast include the context of the translational start codon. RNA 5: 420–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MC, Lee YH, Tarn WY 2008. The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol Biol Cell 19: 3847–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Calabrese JM, Sharp PA 2006. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci 103: 18125–18130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian S, Li S, Abadal G, Pauley B, Fritzler M, Chan EK 2009. The C-terminal half of human Ago2 binds to multiple GW-rich regions of GW182 and requires GW182 to mediate silencing. RNA 15: 804–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling SH, Decker CJ, Walsh MA, She M, Parker R, Song H 2008. Crystal structure of human Edc3 and its functional implications. Mol Cell Biol 28: 5965–5976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R 2005a. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol 7: 719–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Rivas FV, Wohlschlegel J, Yates JR 3rd, Parker R, Hannon GJ 2005b. A role for the P-body component GW182 in microRNA function. Nat Cell Biol 7: 1261–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López de Silanes I, Galbán S, Martindale JL, Yang X, Mazan-Mamczarz K, Indig FE, Falco G, Zhan M, Gorospe M 2005. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol Cell Biol 25: 9520–9531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low WK, Dang Y, Schneider-Poetsch T, Shi Z, Choi NS, Merrick WC, Romo D, Liu JO 2005. Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol Cell 20: 709–722 [DOI] [PubMed] [Google Scholar]
- Mazroui R, Huot ME, Tremblay S, Filion C, Labelle Y, Khandjian EW 2002. Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum Mol Genet 11: 3007–3017 [DOI] [PubMed] [Google Scholar]
- Mazzoni C, D'Addario I, Falcone C 2007. The C-terminus of the yeast Lsm4p is required for the association to P-bodies. FEBS Lett 581: 4836–4840 [DOI] [PubMed] [Google Scholar]
- Mitchell P, Tollervey D 2003. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′→5′ degradation. Mol Cell 11: 1405–1413 [DOI] [PubMed] [Google Scholar]
- Mokas S, Mills JR, Garreau C, Fournier MJ, Robert F, Arya P, Kaufman RJ, Pelletier J, Mazroui R 2009. Uncoupling stress granule assembly and translation initiation inhibition. Mol Biol Cell 20: 2673–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet S, Cougot N, Wilczynska A, Dautry F, Kress M, Bertrand E, Weil D 2008. Translationally repressed mRNA transiently cycles through stress granules during stress. Mol Biol Cell 19: 4469–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckenthaler M, Gunkel N, Stripecke R, Hentze MW 1997. Regulated poly(A) tail shortening in somatic cells mediated by cap-proximal translation repressor proteins and ribosome association. RNA 3: 983–995 [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Parker R 1992. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev 6: 2100–2111 [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Parker R 1994. Premature translational termination triggers mRNA decapping. Nature 370: 578–581 [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Decker CJ, Parker R 1994. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev 8: 855–866 [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Decker CJ, Parker R 1995. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol 15: 2145–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef DW, Thiele DJ 2009. Enhancer of decapping proteins 1 and 2 are important for translation during heat stress in Saccharomyces cerevisiae. Mol Microbiol 73: 1032–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan T, Rajyaguru P, She M, Song H, Parker R 2010. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol Cell 39: 773–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonhoff U, Ralser M, Welzel F, Piccini I, Balzereit D, Yaspo ML, Lehrach H, Krobitsch S 2007. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol Biol Cell 18: 1385–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P 2008. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol 10: 1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Sheth U 2007. P-bodies and the control of mRNA translation and degradation. Mol Cell 25: 635–646 [DOI] [PubMed] [Google Scholar]
- Parker R, Song H 2004. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol 11: 121–127 [DOI] [PubMed] [Google Scholar]
- Pilkington GR, Parker R 2008. Pat1 contains distinct functional domains that promote P-body assembly and activation of decapping. Mol Cell Biol 28: 1298–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W 2005. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science 309: 1573–1576 [DOI] [PubMed] [Google Scholar]
- Rajyaguru P, She M, Parker R 2012. Scd6 targets eIF4G to repress translation: RGG motif proteins as a class of eIF4G-binding proteins. Mol Cell 45: 244–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V, Shah KH, Herman PK 2011. The cAMP-dependent protein kinase signaling pathway is a key regulator of P body foci formation. Mol Cell 43: 973–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns MA, Alexander RD, Spiller MP, Beggs JD 2008. A role for Q/N-rich aggregation-prone regions in P-body localization. J Cell Sci 121: 2463–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikhvanov EG, Romanova NV, Chernoff YO 2007. Chaperone effects on prion and nonprion aggregates. Prion 1: 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller N, Resa-Infante P, de la Luna S, Galao RP, Albrecht M, Kaestner L, Lipp P, Lengauer T, Meyerhans A, Díez J 2007. Identification of PatL1, a human homolog to yeast P-body component Pat1. Biochim Biophys Acta 1773: 1786–92 [DOI] [PubMed] [Google Scholar]
- Schwartz DC, Parker R 1999. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol 19: 5247–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz DC, Parker R 2000. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol Cell Biol 20: 7933–7942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D, Decker CJ, Parker R 2003. The enhancer of decapping proteins, Edc1p and Edc2p, bind RNA and stimulate the activity of the decapping enzyme. RNA 9: 239–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Blau HM 2005. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol 7: 633–636 [DOI] [PubMed] [Google Scholar]
- Seydoux G, Braun RE 2006. Pathway to totipotency: Lessons from germ cells. Cell 127: 891–904 [DOI] [PubMed] [Google Scholar]
- Sheth U, Parker R 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R 2006. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 125: 1095–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu AB, Wilkinson MF, van Hoof A 2008. Messenger RNA regulation: To translate or to degrade. EMBO J 27: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MG, Li Y, Kiledjian M 2010. Multiple mRNA decapping enzymes in mammalian cells. Mol Cell 40: 423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger M, Carr-Schmid A, Schwartz DC, Kiledjian M, Parker R 2003. Analysis of recombinant yeast decapping enzyme. RNA 9: 231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G, Mayo T, Anderson P 2006. ARE-mRNA degradation requires the 5′–3′ decay pathway. EMBO Rep 7: 72–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swisher KD, Parker R 2010. Localization to, and effects of Pbp1, Pbp4, Lsm12, Dhh1, and Pab1 on stress granules in Saccharomyces cerevisiae. PLoS ONE 5: e10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Parker R 2007. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol Biol Cell 18: 2274–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11: 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S, Parker R 2001. Targeting an mRNA for decapping: Displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol Cell 8: 1075–1083 [DOI] [PubMed] [Google Scholar]
- Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R 2000. Yeast Sm-like proteins function in mRNA decapping and decay. Nature 404: 515–518 [DOI] [PubMed] [Google Scholar]
- Thomson T, Lasko P 2004. Drosophila tudor is essential for polar granule assembly and pole cell specification, but not for posterior patterning. Genesis 40: 164–170 [DOI] [PubMed] [Google Scholar]
- Thore S, Mauxion F, Seraphin B, Suck D 2003. X-ray structure and activity of the yeast Pop2 protein: A nuclease subunit of the mRNA deadenylase complex. EMBO Rep 4: 1150–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H, Gallouzi IE, Chebli K, Capony JP, Mouaikel J, van der Geer P, Tazi J 2001. RasGAP-associated endoribonuclease G3BP: Selective RNA degradation and phosphorylation-dependent localization. Mol Cell Biol 21: 7747–7760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H, Chebli K, Zekri L, Courslaud B, Blanchard JM, Bertrand E, Tazi J 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol 160: 823–831 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104: 377–386 [DOI] [PubMed] [Google Scholar]
- Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R 2002. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J 21: 1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Séraphin B 2002. Human Dcp2: A catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J 21: 6915–6924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoof A, Frischmeyer PA, Dietz HC, Parker R 2002. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295: 2262–2264 [DOI] [PubMed] [Google Scholar]
- Wang Z, Kiledjian M 2001. Functional link between the mammalian exosome and mRNA decapping. Cell 107: 751–762 [DOI] [PubMed] [Google Scholar]
- Wickens M, Bernstein DS, Kimble J, Parker R 2002. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet 18: 150–157 [DOI] [PubMed] [Google Scholar]
- Wilczynska A, Aigueperse C, Kress M, Dautry F, Weil D 2005. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J Cell Sci 118: 981–992 [DOI] [PubMed] [Google Scholar]
- Yang WH, Yu JH, Gulick T, Bloch KD, Bloch DB 2006. RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules. RNA 12: 547–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JH, Yang WH, Gulick T, Bloch KD, Bloch DB 2005. Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA 11: 1795–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]