Abstract
Xylan is a major component of the plant cell wall and the most abundant noncellulosic component in the secondary cell walls that constitute the largest part of plant biomass. Dicot glucuronoxylan consists of a linear backbone of β(1,4)-linked xylose residues substituted with α(1,2)-linked glucuronic acid (GlcA). Although several genes have been implicated in xylan synthesis through mutant analyses, the biochemical mechanisms responsible for synthesizing xylan are largely unknown. Here, we show evidence for biochemical activity of GUX1 (for GlcA substitution of xylan 1), a member of Glycosyltransferase Family 8 in Arabidopsis (Arabidopsis thaliana) that is responsible for adding the glucuronosyl substitutions onto the xylan backbone. GUX1 has characteristics typical of Golgi-localized glycosyltransferases and a Km for UDP-GlcA of 165 μm. GUX1 strongly favors xylohexaose as an acceptor over shorter xylooligosaccharides, and with xylohexaose as an acceptor, GlcA is almost exclusively added to the fifth xylose residue from the nonreducing end. We also show that several related proteins, GUX2 to GUX5 and Plant Glycogenin-like Starch Initiation Protein6, are Golgi localized and that only two of these proteins, GUX2 and GUX4, have activity as xylan α-glucuronosyltransferases.
Plant cell walls consist of crystalline cellulose microfibrils embedded in a matrix of pectins and hemicelluloses. Some cell types also have lignin in their walls. Xylans are the major hemicelluloses in dicot secondary cell walls and in both primary and secondary cell walls in grasses (Carpita, 1996; Scheller and Ulvskov, 2010). In Arabidopsis (Arabidopsis thaliana), xylan consists of a linear backbone of β(1,4)-linked d-Xyl residues, some of which are acetylated at the C2 or C3 position. About one in eight Xyl residues are substituted with α(1,2)-linked d-GlcA or 4-O-methyl-d-GlcA (Brown et al., 2007). Ara substitutions on the xylan backbone are also common in plants, especially in grasses, but have not been described in Arabidopsis. Xylans from grasses are unique in having some of the Ara residues on xylan esterified with ferulic acid; this feature is not found in other plants (Carpita, 1996; Ebringerová et al., 2005). Although the exact functions of these substitutions are not known, it is thought that they influence the solubility of xylan and its interaction with other cell wall components such as cellulose and lignin (Fry, 1986; Carpita, 1996). In addition, GlcA and 4-O-methyl-d-GlcA substitutions inhibit the enzymatic degradation of xylan into monosaccharides (Mortimer et al., 2010).
Xylans are the second most abundant polymer after cellulose in grasses and in dicot woody tissue, two sources of biomass that may potentially be used for conversion into biofuels. In addition to making up a large percentage of the sugars available for fermentation, xylans affect biomass recalcitrance because they are cross-linked to lignin through ester linkages to ferulate and 4-O-methyl-d-GlcA (Watanabe and Koshijima, 1988; Ebringerová and Heinze, 2000). Xylan structure, therefore, is an important consideration when engineering plants for improved saccharification and fermentation properties. However, the genes responsible for xylan synthesis have only begun to be discovered in the last few years through forward and reverse genetics in Arabidopsis. Three genes, Irregular Xylem9 (IRX9), IRX10, and IRX14, are thought to encode glycosyltransferases responsible for synthesizing the xylan backbone, as mutations in these genes cause a dwarf phenotype, collapsed xylem vessels, and a reduction in xylan content (Liepman et al., 2010). The related genes IRX9-like (IRX9L), IRX10L, and IRX14L apparently function redundantly to synthesize xylan, as double irx9/irx9l, irx10/irx10l, and irx14/irx14l mutants exhibit more severe reductions in xylan than the single mutants and because they can fully complement the double mutants (Keppler and Showalter, 2010; Wu et al., 2010).
Dicot xylans contain the tetrasaccharide (→4)-β-d-Xylp-(1→4)-β-d-Xylp-(1→3)-α-l-Rhap-(1→2)-α-d-GalpA-(1→4)-d-Xylp at their reducing ends. This tetrasaccharide was first identified in birch (Betula spp.) and spruce (Picea spp.; Johansson and Samuelson, 1977; Andersson et al., 1983) and has more recently been found in Arabidopsis (Peña et al., 2007). Although the function of this tetrasaccharide is unclear, it has been proposed to function in the initiation or termination of xylan synthesis (Peña et al., 2007; York and O’Neill, 2008). The genes IRX8, Fragile Fiber8 (FRA8/IRX7), FRA8 Homolog (F8H/IRX7L), and PARVUS/GATL1 are thought to encode glycosyltransferases involved in synthesizing the reducing end tetrasaccharide (Lee et al., 2007b; York and O’Neill, 2008; Liepman et al., 2010). Mutations in these genes cause an increase in the heterodispersity of xylan degree of polymerization, indicating that theses genes are necessary for controlling chain elongation (Brown et al., 2007; Peña et al., 2007).
The genes GlcA Substitution of Xylan1 (GUX1) and GUX2 have also been implicated in xylan synthesis in coexpression analyses from several groups (Brown et al., 2005; Persson et al., 2005; Ko et al., 2006; Oikawa et al., 2010). These proteins were initially thought to be involved in starch synthesis and were named Plant Glycogenin-like Starch Initiation Proteins (PGSIPs) based on their homology to mammalian glycogenin (Chatterjee et al., 2005). PGSIP1/GUX1 was also predicted to be chloroplast localized, and RNA interference knockdown of GUX1 showed less starch accumulation (Chatterjee et al., 2005). However, both GUX1 and GUX2 have since been shown to localize to the Golgi apparatus (Mortimer et al., 2010; Oikawa et al., 2010). In addition, gux1 and gux2 mutants showed a significant reduction in both xylan GlcA substitutions and xylan GlcA transferase (GlcAT) activity in microsomal fractions (Mortimer et al., 2010; Oikawa et al., 2010; Lee et al., 2012). Here, we provide biochemical evidence that GUX1 is directly responsible for adding GlcA substitutions to xylan. We also show that GUX2 and another related protein, GUX4, have xylan glucuronosyltransferase activity.
RESULTS
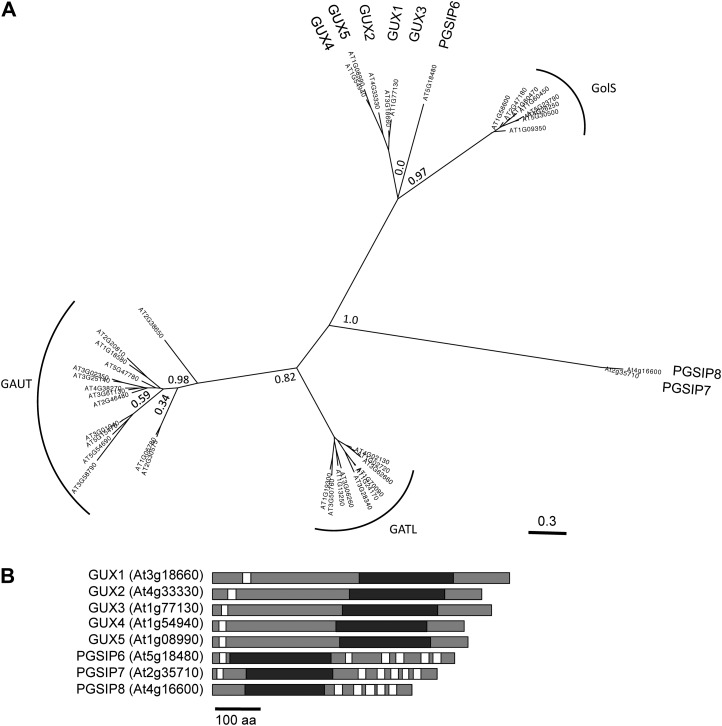
Phylogenetic Analysis of the GUX/PGSIP Family of Proteins
The GUX1 and GUX2 proteins belong to Glycosyltransferase Family 8 (GT8), which is quite diverse but is considered a single glycosyltransferase family according to the CAZy database (www.cazy.org; Cantarel et al., 2009). In plants, GT8 contains the GUX clade, Galactinol Synthase (GolS), Galacturonosyltransferase (GAUT), and GAUT-Like (GATL) clades (Yin et al., 2010). The three GT8 proteins in Arabidopsis that do not belong to these clades have been annotated as PGSIP6, PGSIP7, and PGSIP8 (Yin et al., 2010). The positions of these proteins in the GT8 family tree are shown in Figure 1A. The predicted topologies of the GUX and PGSIP proteins are shown in Figure 1B. All five GUX proteins are predicted to be type II membrane proteins with a single N-terminal transmembrane domain. In contrast, PGSIP6, PGSIP7, and PGSIP8 have between five and seven predicted transmembrane domains with scores above 0.5, according to the Aramemnon plant membrane protein database (http://aramemnon.uni-koeln.de; Schwacke et al., 2003).
Figure 1.
A, Phylogenetic tree of GT8 family proteins in Arabidopsis. Approximate likelihood values are shown at selected nodes. B, Predicted protein structures of the GUX1 to GUX5 and PGISP6 to PGSIP8 proteins. White bars represent transmembrane domains, and black bars represent the GT8 domain. The scale bar at bottom represents 100 amino acids (aa).
Expression and Purification of GUX1
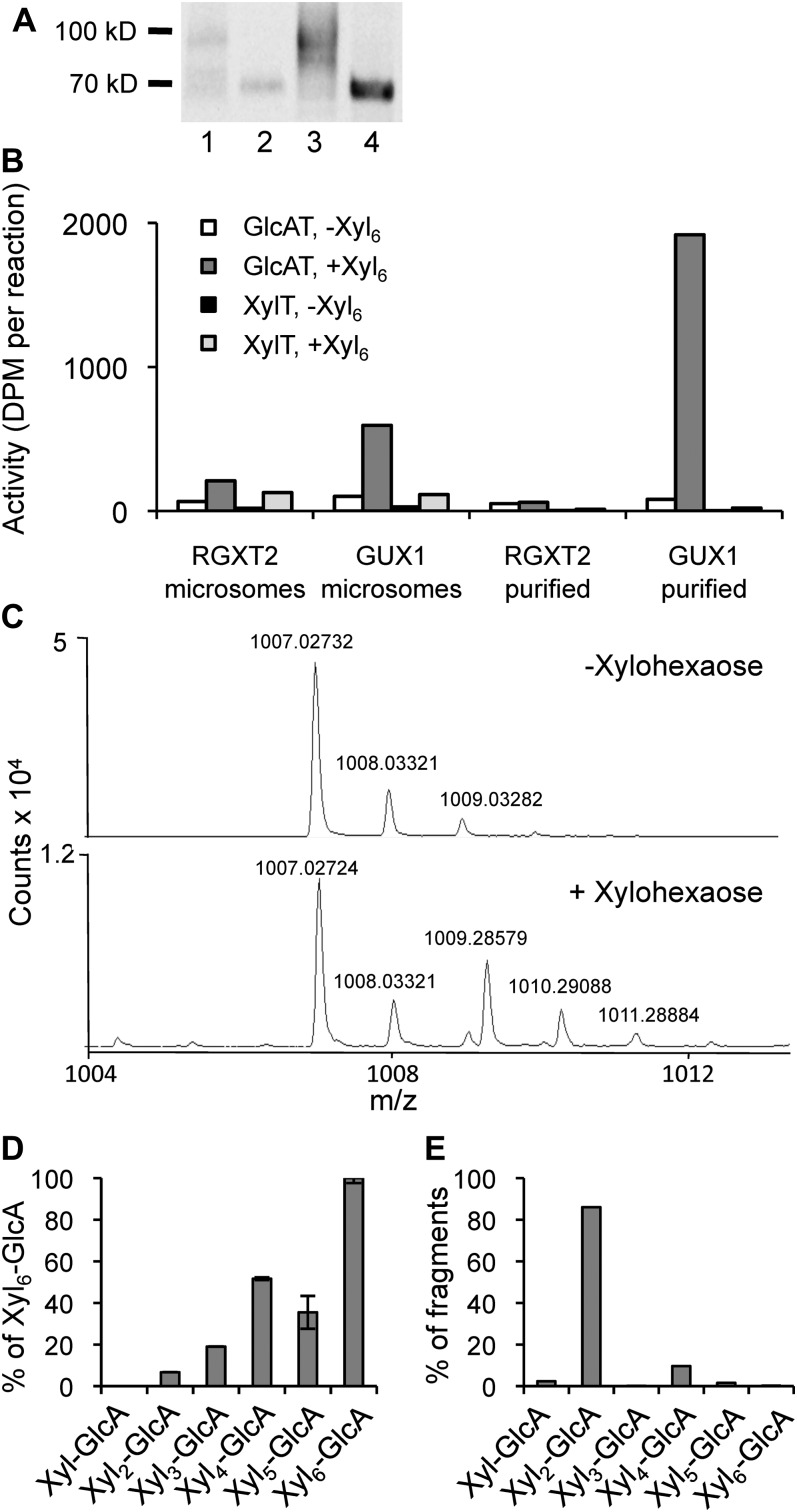
In order to investigate the biochemical function of GUX1, we transiently overexpressed the fusion protein GUX1-yellow fluorescent protein (YFP)-hemagglutinin (HA) by infiltration of Nicotiana benthamiana leaves with Agrobacterium tumefaciens carrying the appropriate construct. We also expressed the Arabidopsis Rhamnogalacturonan Xylosyltransferase2 (RGXT2; Egelund et al., 2006) as a control to ensure that the results of overexpressing GUX1 were not a general effect of overexpressing a Golgi-localized glycosyltransferase. Plants were coinfiltrated with Agrobacterium carrying a construct with the p19 gene from Tomato bushy stunt virus, which encodes a protein that suppresses gene silencing, to ensure that proteins were highly expressed (Voinnet et al., 2003). Microsomes were purified from the infiltrated plants and used directly in GlcAT assays with UDP-[14C]GlcA and xylohexaose as substrates, removal of unincorporated radiolabel by paper chromatography, and quantification of the remaining labeled product by scintillation counting. The microsomes from plants expressing the GUX1 fusion protein had a high xylan GlcAT activity compared with the controls (Fig. 2B). To further investigate that this activity was directly associated with GUX1, the GUX1-YFP-HA protein was isolated from microsomes by affinity purification with anti-HA resin prior to activity assays (Fig. 2A). After purification, the activity was much higher, whereas the activity in control purifications was at a background level (Fig. 2B). XylT activity was assayed in the same samples to confirm that the decarboxylation of UDP-[14C]GlcA to UDP-[14C]Xyl and the addition of Xyl to the xylohexaose acceptor did not contribute to the activity. The amount of XylT activity was negligible, consistent with background RGXT2 activity levels (Fig. 2B).
Figure 2.
Activity of recombinant GUX1 protein. A, Immunoblot of recombinant proteins from N. benthamiana. Column 1, microsomal proteins (20 µg) from plants expressing GUX1-YFP-HA; column 2, microsomal proteins from plants expressing RGXT2-YFP-HA (20 µg); column 3, purified GUX1-YFP-HA (corresponding to 100 µg of microsomal protein); column 4, purified RGXT2-YFP-HA (corresponding to 100 µg of microsomal protein). B, Activity assays using microsomal or purified proteins with UDP-[14C]GlcA and xylohexaose as an acceptor. The microsomal GUX1 activity corresponds to 77.5 pmol GlcA h−1 mg−1 protein. C, Mass spectra of products from reactions using purified GUX1 with and without xylohexaose as an acceptor. The sodium adduct of glucuronoxylohexaose is the peak at m/z 1,009.28579 and is found only when xylohexaose is included. D, Products made using purified GUX1 enzyme when different xylooligomers, Xyl to Xyl6, are used as acceptors, detected by LC-MS. Values are shown as percentages relative to the amount of the Xyl6-GlcA product made. Values represent means of two replicates, with error bars showing the highest and lowest values. E, Fragments produced from the digestion of Xyl6-GlcA with xylosidase, detected by LC-MS. Values are shown as percentages of the total fragments produced.
Analysis of the Enzymatic Product
To confirm that the radiolabeled product was in fact xylohexaose with an α-linked [14C]GlcA substitution, we digested the product with a specific α-glucuronidase from Bacteroides ovatus (glycoside hydrolase family 115). Product incubated with the α-glucuronidase contained 7 ± 2 (se) dpm of radiolabel (n = 3), while product incubated with buffer alone contained 430 ± 11 dpm (n = 3). This experiment confirmed that the label was α-linked GlcA, since essentially all of the radiolabel was released by treatment with this enzyme. The product was further characterized by liquid chromatography-time of flight-mass spectrometry (LC-TOF-MS) to confirm that the product from a reaction using xylohexaose and unlabeled UDP-GlcA had the expected molecular mass (Fig. 2D). The compound with measured mass-to-charge ratio (m/z) [M+Na]+ = 1,009.28579 corresponds to within 0.19 ppm of the theoretical m/z of the sodium adduct of glucuronoxylohexaose (1,009.28598), and the compounds with m/z [M+Na]+ of 1,010.29088 and 1,011.28884 represent isotopic peaks of this molecule. The compound with monoisotopic mass 1,007.027 is unidentified, but it was also present in control reactions lacking the xylohexaose acceptor (Fig. 2C) or lacking GUX1 protein (Supplemental Fig. S1) and hence is not related to GUX1 activity. The sodium adduct of glucuronoxylohexaose indicated that it eluted from the HPLC column approximately 10 s later than the unidentified compound (Supplemental Fig. S1), but we were unable to completely separate the peaks. However, we are confident that the sodium adduct of glucuronoxylohexaose is a distinct compound that was not present in the control reaction. No evidence of xylohexaose with the addition of more than one GlcA was observed.
The product was also characterized by digestion with β-xylosidase, which cleaves Xyl residues from the nonreducing end of the xylooligomer but is unable to cleave GlcA-substituted Xyl residues. Digestion fragments were analyzed by LC-TOF-MS and quantified using available xylooligomers as standards. Approximately 85% of digestion fragments were released as glucuronoxylobiose (Fig. 2E), indicating that the GlcA was positioned on the fifth Xyl from the nonreducing end of the molecule. A smaller amount of glucuronoxylotetraose indicated that the third Xyl from the nonreducing end is also a relatively good acceptor site for GUX1.
LC-TOF-MS was also used to characterize products of glucuronosyltransferase reactions using different xylooligomer acceptors. Purified GUX1 supplied with Xyl and xylooligomers from xylobiose to xylohexaose as acceptors was able to transfer GlcA onto xylobiose and larger molecules (Fig. 2D). GUX1 showed a preference for larger acceptors, as the enzyme was most active when xylohexaose was used as the acceptor, and showed less than 10% activity when xylobiose was used as the acceptor.
Characteristics of Purified GUX1 Protein
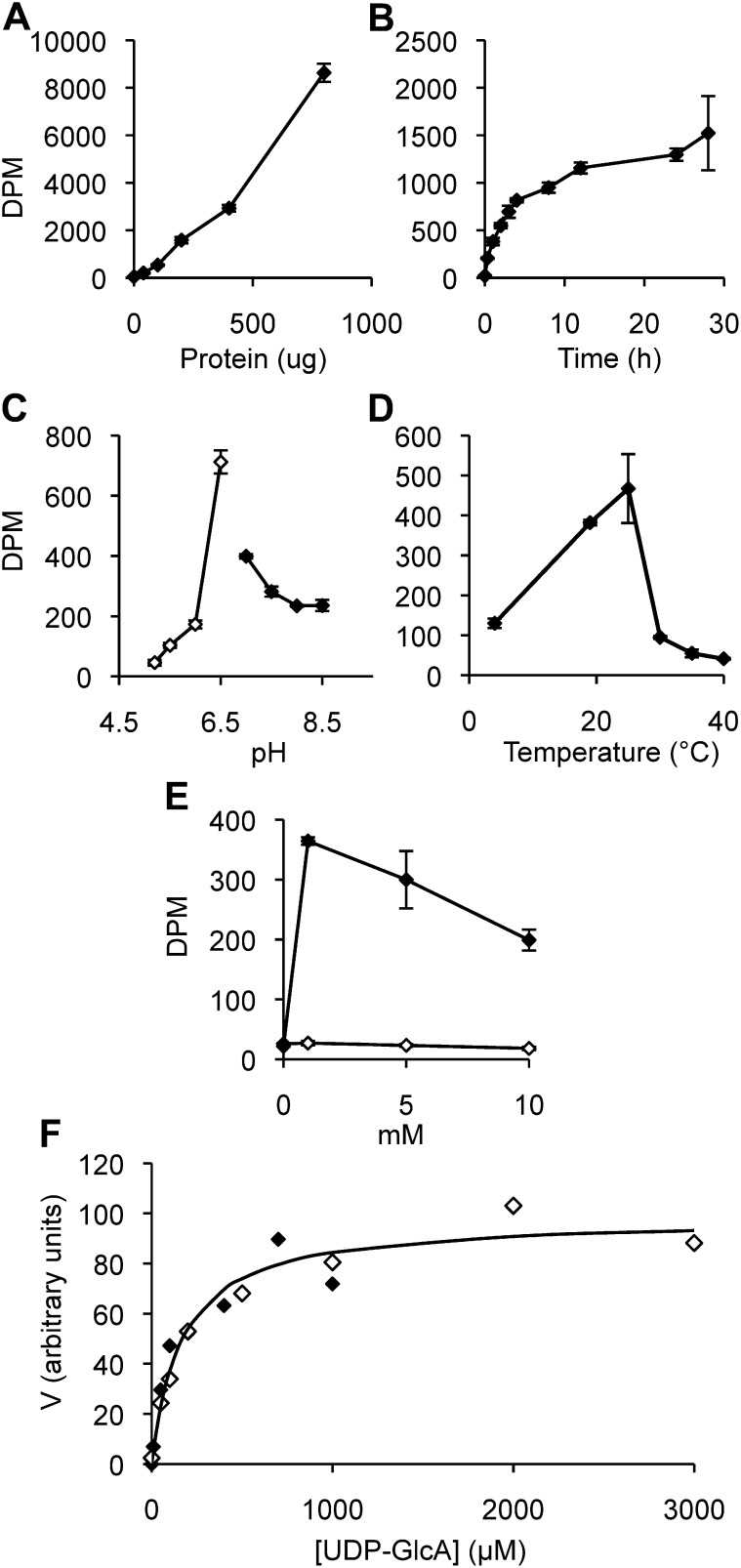
We used purified GUX1-YFP-HA protein to optimize conditions for GlcAT activity and further characterize the enzyme (Fig. 3). GUX1 activity increases with the amount of protein, has a temperature optimum at around 25°C, a pH optimum of 6.5, and requires Mn2+ but not Mg2+. The enzyme is relatively stable, as product increased almost linearly over the first 5 h before it leveled off. The Km value for UDP-GlcA was calculated to be 165 ± 28 μm (n = 2).
Figure 3.
Characteristics of recombinant and purified GUX1 protein. A to E, Enzyme characteristics over amount of protein, where µg of protein corresponds to microsomal proteins used in purification (A); time (B); pH, where white and black symbols designate reactions using MES and HEPES buffer, respectively (C); temperature (D); and divalent cation concentration, where white and black symbols designate Mg2+ and Mn2+, respectively (E). Values represent means of two replicates, with error bars showing the highest and lowest values. F, Kinetic studies of GUX1. Assays were performed with increasing amounts of UDP-GlcA (10–3,000 μm). Each data point is the average of three technical replicates, with the white and black symbols designating data from two independently purified GUX1 preparations. Vmax values are normalized to 100. The solid curve is the Michaelis-Menten curve with Km = 165 µm obtained by nonlinear regression of the experimental data.
Activity of the GUX/PGSIP Family Proteins
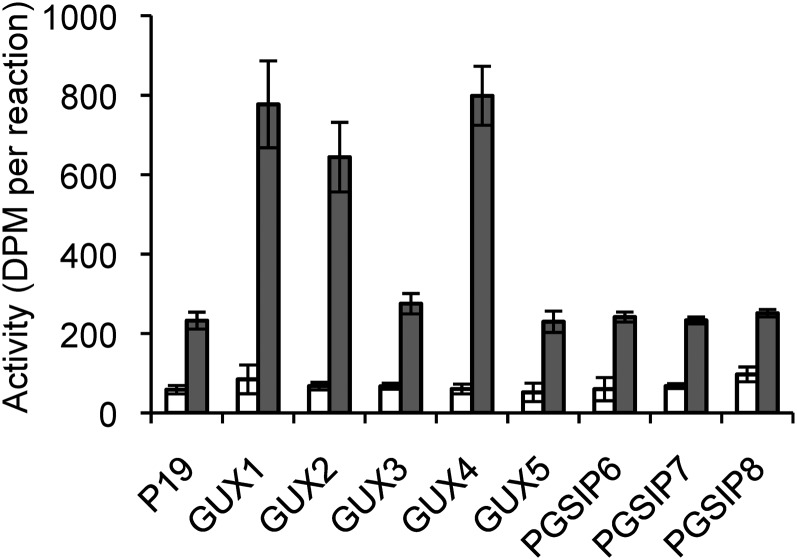
We investigated the activity of the remaining GUX/PGSIP family proteins by transiently overexpressing them in N. benthamiana. Microsomal fractions from these plants were assayed in the same way as GUX1 (Fig. 4). As expected, plants infiltrated with GUX1 showed a large increase in GlcAT activity compared with plants infiltrated with the p19 construct alone. Overexpression of two other proteins, GUX2 and GUX4, caused an increase in GlcAT activity similar to GUX1. Overexpression of the remaining GUX/PGSIP proteins showed no increase in GlcAT activity over background levels.
Figure 4.
Glucuronosyltransferase activity of microsomal proteins from N. benthamiana overexpressing GUX and PGSIP proteins. White bars, reaction with no acceptor; gray bars, reaction with xylohexaose as the acceptor. Error bars show se for three biological replicates.
Subcellular Localization of the GUX/PGSIP Proteins
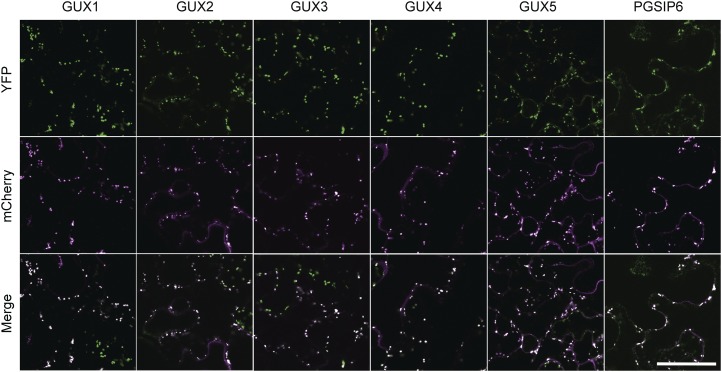
GUX1 and GUX2 have previously been shown to localize to the Golgi (Mortimer et al., 2010; Oikawa et al., 2010). We expressed the remaining members of the family in N. benthamiana with the Golgi marker α-mannosidase-mCherry (Nelson et al., 2007). All five GUX proteins and PGSIP6 colocalized with Golgi markers (Fig. 5). None of the GUX proteins or PGSIP6 colocalized with the plasma membrane marker AtPIP2A-mCherry (Supplemental Fig. S2).
Figure 5.
Subcellular localizations of the GUX1 to GUX5 and PGSIP6 proteins. Single-plane confocal micrographs of the proteins fused with C-terminal YFP (top row), the Golgi marker α-mannosidase-mCherry (middle row), and merged YFP and mCherry channels (bottom row) are shown. Bar = 100 μm.
DISCUSSION
Although the GUX1 and GUX2 proteins were originally identified as PGSIP1 and PGSIP3, they have been implicated in secondary cell wall deposition in coexpression analyses by several groups (Persson et al., 2005; Ko et al., 2006; Mortimer et al., 2010; Oikawa et al., 2010). In addition, gux1 and gux2 mutants have reduced xylan GlcA content and xylan GlcAT activity (Mortimer et al., 2010; Oikawa et al., 2010). However, the previous studies did not provide direct evidence for the catalytic function of GUX proteins. Here, we provide definitive biochemical evidence that GUX1 and GUX2, as well as a related protein, GUX4, have xylan glucuronosyltransferase activity.
Although both GUX1 and GUX2 have been implicated in xylan synthesis, we chose GUX1 for purification and characterization because GUX1 is most highly coexpressed with other xylan biosynthetic genes and the gux1 phenotype is the most severe (Mortimer et al., 2010). GUX1 protein expressed in and purified from N. benthamiana showed a large increase in GlcAT activity that was not present in another purified Golgi-localized glycosyltransferase, RGXT2, indicating that GUX1 is responsible for this activity. The lack of XylT activity indicates that the conversion of UDP-[14C]GlcA to UDP-[14C]Xyl was not a significant factor in the assay, and analysis of the product confirmed that it was in fact α-glucuronoxylohexaose. Therefore, we are confident that our assay shows the formation of an α-linkage between xylohexaose and GlcA. The linkage has not been determined in our experiments, but the data strongly suggest that it is the α(1,2)-linkage characteristic for glucuronosyl substitutions on xylan.
We further analyzed the glucuronoxylohexaose product produced by purified GUX1 by digesting it with β-xylosidase. LC-MS analysis of the digestion products showed that approximately 85% of the GlcA-containing fragments released were glucuronoxylobiose (Fig. 2E). This result indicates that the majority of the glucuronoxylohexaose molecules had GlcA positioned on the fifth Xyl from the nonreducing end. A smaller number of molecules (about 10%) were cleaved to glucuronoxylotetraose, indicating that GUX1 was also able to transfer GlcA onto the third Xyl from the nonreducing end, although at lower efficiency. We also analyzed the preference of GUX1 for differently sized acceptors. GUX1 was able to transfer GlcA to acceptors as small as xylobiose, although it showed higher activity with larger xylooligomer acceptors (Fig. 2D). The highest activity was detected when xylohexaose was used as the acceptor. Taken together, these results indicate that although GUX1 does show preferences regarding acceptor size and the placement of GlcA on the acceptor, its activity is somewhat flexible. This finding is interesting given that xylan extracted from Arabidopsis invariably shows a ratio of one GlcA for every eight Xyl residues even in mutants with large decreases in xylan content, indicating that the proportion of GlcA is controlled by a robust mechanism (Brown et al., 2007). It is likely that mechanisms in the Golgi, such as interactions between GUX1 and other xylan biosynthetic enzymes or properties of the xylan polymer, control GlcA addition in ways that were not evident in our assays using purified protein and xylooligomer acceptors.
GUX1 is a relatively stable protein and has typical properties in terms of pH and temperature optima. The Km for UDP-GlcA was determined to be 165 µm, which is similar to that of other glycosyltransferases in the Golgi, such as RGXT2 (140 μm; Egelund et al., 2008). UDP-Xyl synthase is another Golgi-localized enzyme that uses UDP-GlcA as a substrate, and for this enzyme a Km of 190 μm was determined (Harper and Bar-Peled, 2002).
The GUX proteins belong to GT8, which in Arabidopsis also contains the GolS, GAUT, and GATL clades (Fig. 1; Cantarel et al., 2009). The GAUT and GATL clades have been designated as putative cell wall biosynthesis-related genes, while the remaining genes were grouped into a putative non-cell wall biosynthesis-related class (Yin et al., 2010). The latter class includes the GolS proteins and eight proteins that were annotated as PGSIPs. Three of the PGSIP proteins have previously been reannotated as GUX1 to GUX3 after GUX1 and GUX2 were implicated in xylan synthesis (Mortimer et al., 2010; Oikawa et al., 2010). Based on the results presented here, we have reannotated PGSIP4 and PGSIP5 as GUX4 and GUX5, respectively. While GUX1 to GUX5 are closely related to one another, the placement of PGSIP6 is less clear. Although PGSIP6 appears to be more closely related to the GUX clade than to the GolS clade, the support for this node is very low, indicating that the placement of PGSIP6 within the GUX and GolS clades cannot be determined definitively (Fig. 1A). PGSIP7 and PGSIP8 appear to be only distantly related to both the GUX and GolS clades.
In order to determine the functions of these proteins, we cloned and transiently expressed the five GUX proteins as well as PGSIP6 to PGSIP8 in N. benthamiana. GlcAT assays using microsomal proteins from these N. benthamiana plants indicated that GUX1, GUX2, and GUX4, but none of the other proteins, have xylohexaose-dependent glucuronosyltransferase activity (Fig. 5). Although all five GUX proteins and PGSIP6 were expressed and localized to the Golgi (Fig. 5), it is possible that the YFP-HA tag prevented the proteins from being active as glucuronosyltransferases or that our assay conditions were inappropriate for detecting the activity of these proteins. However, GUX3 and GUX5 are closely related to GUX1, GUX2 and GUX4, which all had activity in our assays. The finding that only some members of a glycosyltransferase clade have activity is not without precedent. For example, only three of five Arabidopsis members of the GT75 family had detectable activity, even though all proteins were expressed in the same way in Escherichia coli (Rautengarten et al., 2011). Furthermore, unlike the GAUT1 homogalacturonan synthase, GAUT7 did not have detectable activity when expressed in the same system (Sterling et al., 2006), and it appears that the GAUT7 protein lacks key amino acid residues predicted to be involved in catalysis (Atmodjo et al., 2011). In the case of GAUT7, the protein appears to have a structural function as an anchor for GAUT1 rather than a catalytic function (Atmodjo et al., 2011). Hence, it may be a common finding that some glycosyltransferase homologs have lost their catalytic function and play a different role (e.g. as subunits in synthase complexes). It is also possible that GUX3, GUX5, and PGSIP6 have functions unrelated to xylan synthesis. GUX3 and PGSIP6 were identified in proteomic analyses of Golgi vesicles isolated from Arabidopsis cell culture (Dunkley et al., 2006; Parsons et al., 2012). Since undifferentiated cell cultures are enriched for primary cell walls, GUX3 and PGSIP6 may be involved in the synthesis of a polymer that is present in higher amounts in primary walls.
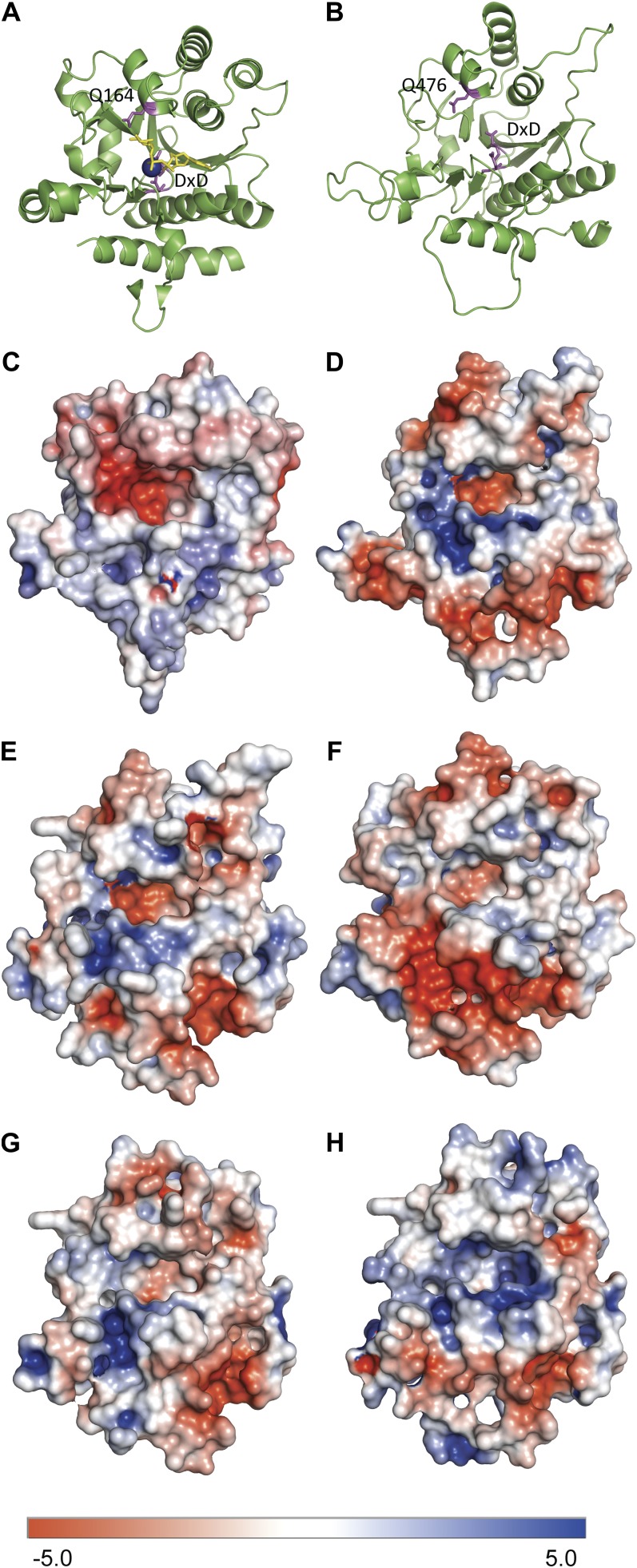
GT8 includes proteins with known functions as glucosyltransferases (glycogenin; Lomako et al., 2004), galactosyltransferases (LgtC; Persson et al., 2001), galacturonosyltransferases (GAUT1; Sterling et al., 2006), and glucuronosyltransferases (this work). Three structures are known from GT8: the LgtC galactosyltransferase from Neisseria meningitidis (Persson et al., 2001) and glycogenin from rabbit (Gibbons et al., 2002) and human (Chaikuad et al., 2011). Structural modeling of GAUT1, which adds GalA onto the negatively charged polymer homogalacturonan, showed that the region where the acceptor is expected to lie contains a patch of positively charged residues, consistent with the accommodation of a negatively charged acceptor (Yin et al., 2010), also observed for modeling of QUA1/GAUT8 (Hansen, 2009). We have analyzed the GUX proteins in a similar way (Fig. 6). GUX and PGSIP proteins contain a DQG motif that is absent in the GAUT and GATL clades. The Gln residue in this motif (Gln-164 in glycogenin) has been suggested to act as a catalytic residue that transiently attaches to the substrate sugar molecule before transferring it to the acceptor (Persson et al., 2001; Gibbons et al., 2002). Surface electrostatic models of GUX1 show a positively charged patch directly adjacent to this Gln, which may help to stabilize the negatively charged GlcA while it is transiently attached to the enzyme (Fig. 6D). This positively charged region is not found near the catalytically active Gln in glycogenin, which is expected to form an intermediate with the neutral sugar Glc (Fig. 6C). In addition, although the electrostatic models show that this region in GUX1 is the most positively charged, similar positively charged regions are found in GUX2, GUX4, and GUX5 but not in GUX3. This observation could explain why GUX3 is not active even though it is the protein most closely related to GUX1. It is noteworthy that modeling of the GUX proteins shows that GUX1, GUX2, and GUX4 have similar electrostatic patterns in the region of substrate binding while GUX3 and GUX5 are clearly more divergent (Fig. 6). The lack of activity and the apparently different structures of GUX3 and GUX5 suggest that these proteins may have a different role. However, given the very close phylogenetic relationship of all five GUX proteins, we suggest that the most likely role of GUX3 and GUX5 is as noncatalytic subunits in a GUX complex. This explanation could also account for why Lee et al. (2012) found that overexpression of GUX3 in Nicotiana tabacum BY2 cells led to an increase in xylan GlcAT activity in the cells even though our results and structural modeling suggest that GUX3 is not catalytically active. Further studies of protein-protein interactions among the GUX proteins and other proteins involved in xylan biosynthesis are needed to clarify the possible interactions between GUX proteins and other xylan biosynthetic proteins.
Figure 6.
Structural modeling of glycogenin and GUX1 to GUX5 proteins. A, Cartoon structure of glycogenin showing the active site with Mn2+ ion (blue), UDP-Glc (yellow), and DxD and Gln-164 (Q164) residues (magenta). B, Homology model of GUX1 showing the DxD and Gln-164 residues (magenta). C to H, Surface electrostatic models of glycogenin (C), GUX1 (D), GUX2 (E), GUX3 (F), GUX4 (G), and GUX5 (H). Positive and negative charges are shown as blue and red, respectively, as shown on the bar at bottom.
The GUX1 to GUX5 proteins all have one predicted N-terminal transmembrane domain, which is expected for classical type II Golgi-localized glycosyltransferases. In contrast, PGSIP6, PGSIP7, and PGSIP8 are predicted to have between five and seven transmembrane domains distributed along the length of the protein. This topology, along with their relatively distant relationship to the GUX proteins, indicates that these proteins are unlikely to function in transferring GlcA onto xylan. Analysis of knockout mutants may provide indications of such functions, although our preliminary studies of such mutants have not given any obvious phenotypes or suggestions of function.
MATERIALS AND METHODS
Phylogenetic Analysis
Protein sequences obtained from The Arabidopsis Information Resource (www.arabidopsis.org) were aligned using MUSCLE 3.7 (Edgar, 2004), and the phylogenetic tree was built using PhyML 3.0 aLRT (Guindon et al., 2010) and viewed using FigTree version 1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/).
Plant Material and Transient Expression
Three- to 4-week old Nicotiana benthamiana ‘Domin’ plants were used for infiltration with Agrobacterium tumefaciens. Agrobacterium strain C58-1 pGV3850 carrying the appropriate vectors was grown to log phase, pelleted at 3,500g, and resuspended in 10 mm MES-KOH, pH 5.6, 10 mm MgCl2, and 200 μm acetosyringone before being infiltrated into the abaxial surfaces of leaves. Agrobacteria were resuspended to an optical density at 600 nm of 1.0 for protein purification or an optical density at 600 nm of 0.1 for confocal microscopy. Plants used for protein purification and activity assays were coinfiltrated with a strain carrying a plasmid with the p19 gene from Tomato bushy stunt virus (Voinnet et al., 2003). As much of each leaf as possible (approximately 95%) was infiltrated. The expression of genes fused to YFP was verified by monitoring YFP fluorescence with an epifluorescence microscope, and all cells in infiltrated areas were shown to express protein.
Microsome Preparation
Entire infiltrated leaves were harvested on day 3 after infiltration. All microsome preparation steps took place at 4°C. Leaf tissue was ground in buffer containing 50 mm HEPES-KOH, pH 7.0, 400 mm Suc, 1 mm phenylmethanesulfonyl fluoride, 1% (w/v) polyvinylpolypyrrolidone, and protease inhibitors (Roche Complete protease inhibitor tablets). The homogenate was filtered through two layers of Miracloth (EMD Millipore) and centrifuged at 3,000g for 10 min, then the supernatant was centrifuged at 50,000g for 1 h. The pellet was resuspended in 50 mm HEPES-KOH, pH 7.0, and 400 mm Suc. Microsomes were used immediately or frozen in liquid nitrogen and stored at −80°C. No significant loss of activity was detected after freezing.
Cloning and Construction of Expression Vectors
All clones used in this study were constructed using Gateway technology (Invitrogen). The entry clones were obtained via BP (attB × attP recombination) reaction in pDONR-Zeo. The genes were cloned using cDNA from Arabidopsis (Arabidopsis thaliana) stems or leaves as template. The reverse primers contained no stop codon to enable C-terminal fusions. Sequences of forward and reverse primers can be provided on request. All entry clones were verified by restriction analysis and sequencing. Gateway expression vectors were constructed via LR (attL × attR recombination) reaction with the corresponding entry clone and the binary vector pEarleyGate101, which contains a 2× 35S promoter and a C-terminal YFP-HA tag (Earley et al., 2006).
Protein Purification
All purification steps took place at 4°C. Microsomal proteins were first solubilized by incubating with 1% Triton X-100 for 10 min and subsequently centrifuging at 100,000g for 30 min. The supernatant was incubated with EZview Red anti-HA resin (Sigma-Aldrich) for 3 h, washed three times with 1% Triton X-100, 400 mm Suc, 50 mm HEPES-KOH, pH 7.0, and 200 mm NaCl, and then washed three times with 400 mm Suc and 50 mm HEPES-KOH, pH 7.0. Resin-bound protein was used directly in GlcAT assays. For immunoblot analysis, proteins were resolved by SDS-PAGE on 7% to 15% gradient gels and blotted onto nitrocellulose membranes (GE Healthcare). Blots were probed with a 1:10,000 dilution of rabbit anti-HA antibody (Sigma-Aldrich), followed by a 1:20,000 dilution of goat anti-rabbit IgG conjugated to horseradish peroxidase (Sigma-Aldrich), before applying ECL Plus detection reagent (GE Healthcare). Blots were imaged using a ChemiDoc-It 600 Imaging System (UVP).
Glucuronosyltransferase and Xylosyltransferase Assays
Xylan glucuronosyltransferase activity in microsomes or purified proteins was determined essentially as described (Lee et al., 2007a) using 3.7 µm UDP-[14C]d-GlcA (740 Bq per reaction; MP Biomedicals), 50 µm unlabeled UDP-d-GlcA, and 400 μm xylohexaose (Megazyme) as acceptor in a 30-µL reaction volume. Products were separated by paper chromatography and analyzed by liquid scintillation counting according to Lee et al. (2007a). Microsomes corresponding to 100 µg of protein were used. The reaction took place at 20°C (except in Fig. 3D), and reaction time was 2 h (except in Fig. 3B). For determining the optimal Mn2+ and Mg2+ concentrations, protein was incubated with 10 mm EDTA in a total volume of 5 μL on ice for 10 min before being added to the 30-μL reaction. For determining the optimal pH, MES buffer was used for pH 5.0, 5.5, 6.0, and 6.5 and HEPES buffer was used for pH 7.0, 7.5, 8.0, and 8.5. For reactions with purified GUX1, an amount of protein corresponding to 100 µg of microsomal protein was used in each assay (except in Fig. 3A). The concentration of the purified protein was too low to be determined. The Km for UDP-GlcA was determined by varying the UDP-GlcA concentration and determining the amount of product made by the purified GUX1 in a 1-h reaction. Xylosyltransferase assays were performed as described (Lee et al., 2007a).
Analysis of Radiolabeled Glucuronoxylohexaose
After the GlcAT reaction, labeled product was separated from unincorporated UDP-GlcA by spotting the reaction onto Whatman 3M chromatography paper and developing it in 95% ethanol:1.0 m ammonium acetate (2:1, v/v). The origin was cut out, and the product was eluted from the paper in 1 mL of water, dried down, and resuspended in 20 μL of water. α-Glucuronidase (0.5 μg) from Bacteroides ovatus was used to digest the product in 100 mm ammonium acetate, pH 5.5, in a total volume of 50 μL for 1 h at 21°C. Digested product and product incubated with buffer alone were then analyzed by paper chromatography and scintillation counting as above.
Xylosidase Digestion of Glucuronoxylohexaose
Purified GUX1 corresponding to 500 µg of microsomal proteins was incubated with 50 mm HEPES-KOH, pH 6.8, 5 mm MnCl2, 400 μm Xyl6, and 500 µm UDP-GlcA to produce glucuronoxylohexaose. This product was then digested with β-xylosidase (Sigma) in 100 mm HEPES-NaOH, pH 7.0, for 1 h at 70°C. Digested fragments were quantified using LC-TOF-MS.
LC-TOF-MS Analysis
Products from reactions using purified GUX1 both with and without xylohexaose were analyzed. Enzyme reactions were centrifuged in a 3.5-kD dialyzer (Novagen) before analysis. The solvents used were of HPLC grade or better (Honeywell Burdick & Jackson). Chemical standards were purchased from Megazyme and were made up to 20 µm, as the stock solution, in methanol:water (50:50, v/v). The separation of metabolites was conducted on a Fermentation Monitoring HPX-87H column with 8% cross-linkage (150 mm length, 7.8 mm internal diameter, and 9 µm particle size; Bio-Rad) using an Agilent Technologies 1100 Series HPLC system. A sample injection volume of 10 µL was used throughout. The temperature of the sample tray was maintained at 4°C by an Agilent FC/ALS Thermostat. The column compartment was set to 50°C. Metabolites were eluted isocratically with a mobile phase composition of 0.1% formic acid in water. A flow rate of 0.5 mL min−1 was used throughout. The HPLC system was coupled to an Agilent Technologies 6210 TOF mass spectrometer using a one-to-five postcolumn split. Contact between both instruments was established by a local area network card in order to trigger the mass spectrometer into operation upon the initiation of a run cycle from the MassHunter workstation (Agilent Technologies). Nitrogen gas was used as both the nebulizing and drying gases to facilitate the production of gas-phase ions. The drying and nebulizing gases were set to 12 L min−1 and 30 ψ, respectively, and a drying gas temperature of 330°C was used throughout. Electrospray ionization was conducted in the positive ion mode, and a capillary voltage of 3,500 V was utilized. MS experiments were carried out in the full scan mode, at 0.86 spectra s−1, for the detection of [M+Na]+ ions. The instrument was tuned for a range of 50 to 1,700 m/z. Prior to LC-TOF-MS analysis, the TOF mass spectrometer was calibrated via an electrospray ionization low-concentration tuning mix (Agilent Technologies). Data acquisition and processing were performed with the MassHunter software package. Xylooligomer standards ranging from Xyl to xylohexaose were used to estimate the amounts of glucuronoxylooligomers.
Fluorescence Confocal Microscopy
Abaxial epidermal sections from infiltrated leaves were used for microscopy. A Zeiss LSM 710 confocal microscope equipped with Argon and InTune lasers was used for confocal laser-scanning microscopy. All images were obtained with a 1.30 numerical aperture oil 40× objective. YFP and mCherry channels were imaged by simultaneous scanning using excitation/emission of 514/519 to 560 nm for YFP and 587/600 to 634 nm for mCherry. The Zen software package (Carl Zeiss) was used for image acquisition and processing.
Structural Modeling
The SWISS-MODEL server was used to predict protein structures (Arnold et al., 2006; Kiefer et al., 2009). Aligned regions were selected by the SWISS-MODEL server with an E-value cutoff of 10−6, and amino acids 317 to 577 (GUX1), 296 to 555 (GUX2), 281 to 541 (GUX3), 266 to 517 (GUX4), and 274 to 526 (GUX5) were used to make homology models with the glycogenin structure (ILL2; Gibbons et al., 2002). Electrostatic outputs for these models were generated using PDB2PQR (Dolinsky et al., 2004, 2007), and surface electrostatics were calculated using the Adaptive Poisson-Boltzmann Solver software (Baker et al., 2001).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. LC-MS analysis of the GUX1 reaction product.
Supplemental Figure S2. Subcellular localizations of the GUX1 to GUX5 and PGSIP6 proteins.
Supplementary Material
Acknowledgments
We thank Sherry Chan for assistance with plant growth, Drs. David Bolam, Paul Dupree, and Artur Rogowski for providing α-glucuronidase, Dr. Michelle Smith for assistance with confocal microscopy, and Dr. Vaishali Sharma for assistance with mass spectrometry.
Glossary
- GlcAT
GlcA transferase
- GT8
Glycosyltransferase Family 8
- YFP
yellow fluorescent protein
- HA
hemagglutinin
- LC
liquid chromatography
- TOF
time of flight
- MS
mass spectrometry
- m/z
mass-to-charge ratio
References
- Andersson SI, Samuelson O, Ishihara M, Shimizu K. (1983) Structure of the reducing end-groups in spruce xylan. Carbohydr Res 111: 283–288 [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. (2006) The SWISS-MODEL workspace: a Web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 [DOI] [PubMed] [Google Scholar]
- Atmodjo MA, Sakuragi Y, Zhu X, Burrell AJ, Mohanty SS, Atwood JA, III, Orlando R, Scheller HV, Mohnen D. (2011) Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan:galacturonosyltransferase complex. Proc Natl Acad Sci USA 108: 20225–20230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA 98: 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DM, Goubet F, Wong VW, Goodacre R, Stephens E, Dupree P, Turner SR. (2007) Comparison of five xylan synthesis mutants reveals new insight into the mechanisms of xylan synthesis. Plant J 52: 1154–1168 [DOI] [PubMed] [Google Scholar]
- Brown DM, Zeef LA, Ellis J, Goodacre R, Turner SR. (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37: D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC. (1996) Structure and biogenesis of the cell walls of grasses. Annu Rev Plant Physiol Plant Mol Biol 47: 445–476 [DOI] [PubMed] [Google Scholar]
- Chaikuad A, Froese DS, Berridge G, von Delft F, Oppermann U, Yue WW. (2011) Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis. Proc Natl Acad Sci USA 108: 21028–21033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee M, Berbezy P, Vyas D, Coates S, Barsby T. (2005) Reduced expression of a protein homologous to glycogenin leads to reduction of starch content in Arabidopsis leaves. Plant Sci 168: 501–509 [Google Scholar]
- Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. (2007) PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res 35: W522–W525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. (2004) PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res 32: W665–W667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkley TPJ, Hester S, Shadforth IP, Runions J, Weimar T, Hanton SL, Griffin JL, Bessant C, Brandizzi F, Hawes C, et al. (2006) Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci USA 103: 6518–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Ebringerová A, Heinze T. (2000) Xylan and xylan derivatives: biopolymers with valuable properties. 1. Naturally occurring xylan structures, isolation procedures and properties. Macromol Rapid Commun 21: 542–556 [Google Scholar]
- Ebringerová A, Hromádková Z, Heinze T. (2005) Hemicellulose. Adv Polym Sci 186: 1–67 [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelund J, Damager I, Faber K, Olsen C-E, Ulvskov P, Petersen BL. (2008) Functional characterisation of a putative rhamnogalacturonan II specific xylosyltransferase. FEBS Lett 582: 3217–3222 [DOI] [PubMed] [Google Scholar]
- Egelund J, Petersen BL, Motawia MS, Damager I, Faik A, Olsen CE, Ishii T, Clausen H, Ulvskov P, Geshi N. (2006) Arabidopsis thaliana RGXT1 and RGXT2 encode Golgi-localized (1,3)-α-d-xylosyltransferases involved in the synthesis of pectic rhamnogalacturonan-II. Plant Cell 18: 2593–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC. (1986) Cross-linking of matrix polymers in the growing cell walls of angiosperms. Annu Rev Plant Physiol 37: 165–186 [Google Scholar]
- Gibbons BJ, Roach PJ, Hurley TD. (2002) Crystal structure of the autocatalytic initiator of glycogen biosynthesis, glycogenin. J Mol Biol 319: 463–477 [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Hansen SF. (2009) Identification et caractérisation de genes de glycosyltransférases impliqués dans la biosynthèse des polysaccharides pariétaux. PhD thesis. Joseph Fourier University, Grenoble, France
- Harper AD, Bar-Peled M. (2002) Biosynthesis of UDP-xylose: cloning and characterization of a novel Arabidopsis gene family, UXS, encoding soluble and putative membrane-bound UDP-glucuronic acid decarboxylase isoforms. Plant Physiol 130: 2188–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MH, Samuelson O. (1977) Reducing end groups in birch xylan and their alkaline degradation. Wood Sci Technol 11: 251–263 [Google Scholar]
- Keppler BD, Showalter AM. (2010) IRX14 and IRX14-LIKE, two glycosyl transferases involved in glucuronoxylan biosynthesis and drought tolerance in Arabidopsis. Mol Plant 3: 834–841 [DOI] [PubMed] [Google Scholar]
- Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T. (2009) The SWISS-MODEL repository and associated resources. Nucleic Acids Res 37: D387–D392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JH, Beers EP, Han KH. (2006) Global comparative transcriptome analysis identifies gene network regulating secondary xylem development in Arabidopsis thaliana. Mol Genet Genomics 276: 517–531 [DOI] [PubMed] [Google Scholar]
- Lee C, O’Neill MA, Tsumuraya Y, Darvill AG, Ye Z-H. (2007a) The irregular xylem9 mutant is deficient in xylan xylosyltransferase activity. Plant Cell Physiol 48: 1624–1634 [DOI] [PubMed] [Google Scholar]
- Lee C, Teng Q, Zhong R, Ye Z-H. (2012) Arabidopsis GUX proteins are glucuronyltransferases responsible for the addition of glucuronic acid side chains onto xylan. Plant Cell Physiol (in press) [DOI] [PubMed] [Google Scholar]
- Lee C, Zhong R, Richardson EA, Himmelsbach DS, McPhail BT, Ye ZH. (2007b) The PARVUS gene is expressed in cells undergoing secondary wall thickening and is essential for glucuronoxylan biosynthesis. Plant Cell Physiol 48: 1659–1672 [DOI] [PubMed] [Google Scholar]
- Liepman AH, Wightman R, Geshi N, Turner SR, Scheller HV. (2010) Arabidopsis: a powerful model system for plant cell wall research. Plant J 61: 1107–1121 [DOI] [PubMed] [Google Scholar]
- Lomako J, Lomako WM, Whelan WJ. (2004) Glycogenin: the primer for mammalian and yeast glycogen synthesis. Biochim Biophys Acta 1673: 45–55 [DOI] [PubMed] [Google Scholar]
- Mortimer JC, Miles GP, Brown DM, Zhang Z, Segura MP, Weimar T, Yu X, Seffen KA, Stephens E, Turner SR, et al. (2010) Absence of branches from xylan in Arabidopsis gux mutants reveals potential for simplification of lignocellulosic biomass. Proc Natl Acad Sci USA 107: 17409–17414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Oikawa A, Joshi HJ, Rennie EA, Ebert B, Manisseri C, Heazlewood JL, Scheller HV. (2010) An integrative approach to the identification of Arabidopsis and rice genes involved in xylan and secondary wall development. PLoS ONE 5: e15481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons HT, Christiansen K, Knierim B, Carroll A, Ito J, Batth TS, Smith-Moritz AM, Morrison S, McInerney P, Hadi MZ, et al. (2012) Isolation and proteomic characterization of the Arabidopsis Golgi defines functional and novel components involved in plant cell wall biosynthesis. Plant Physiol 159: 12–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña MJ, Zhong R, Zhou G-K, Richardson EA, O’Neill MA, Darvill AG, York WS, Ye Z-H. (2007) Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19: 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson K, Ly HD, Dieckelmann M, Wakarchuk WW, Withers SG, Strynadka NCJ. (2001) Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs. Nat Struct Biol 8: 166–175 [DOI] [PubMed] [Google Scholar]
- Persson S, Wei H, Milne J, Page GP, Somerville CR. (2005) Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA 102: 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C, Ebert B, Herter T, Petzold CJ, Ishii T, Mukhopadhyay A, Usadel B, Scheller HV. (2011) The interconversion of UDP-arabinopyranose and UDP-arabinofuranose is indispensable for plant development in Arabidopsis. Plant Cell 23: 1373–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P. (2010) Hemicelluloses. Annu Rev Plant Biol 61: 263–289 [DOI] [PubMed] [Google Scholar]
- Schwacke R, Schneider A, van der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flügge U-I, Kunze R. (2003) ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol 131: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterling JD, Atmodjo MA, Inwood SE, Kumar Kolli VS, Quigley HF, Hahn MG, Mohnen D. (2006) Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proc Natl Acad Sci USA 103: 5236–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Koshijima T. (1988) Evidence for an ester linkage between lignin and glucuronic acid in lignin-carbohydrate complexes by DDQ-oxidation. Agric Chem Soc Jpn 52: 2953–2955 [Google Scholar]
- Wu AM, Hörnblad E, Voxeur A, Gerber L, Rihouey C, Lerouge P, Marchant A. (2010) Analysis of the Arabidopsis IRX9/IRX9-L and IRX14/IRX14-L pairs of glycosyltransferase genes reveals critical contributions to biosynthesis of the hemicellulose glucuronoxylan. Plant Physiol 153: 542–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Chen H, Hahn MG, Mohnen D, Xu Y. (2010) Evolution and function of the plant cell wall synthesis-related glycosyltransferase family 8. Plant Physiol 153: 1729–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- York WS, O’Neill MA. (2008) Biochemical control of xylan biosynthesis: which end is up? Curr Opin Plant Biol 11: 258–265 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.