Summary
The Muscleblind-like (Mbnl) family of RNA binding proteins plays important roles in muscle and eye development and in Myotonic Dystrophy (DM), where expanded CUG or CCUG repeats functionally deplete Mbnl proteins. We identified transcriptome-wide functional and biophysical targets of Mbnl proteins in brain, heart, muscle, and myoblasts using RNA-Seq and CLIP-Seq approaches. This analysis identified several hundred splicing events whose regulation depended on Mbnl function, in a pattern indicating functional interchangeability between Mbnl1 and Mbnl2. A nucleotide resolution RNA map associated repression or activation of exon splicing with Mbnl binding near either 3' splice site or near the downstream 5' splice site, respectively. Transcriptomic analysis of sub-cellular compartments uncovered a global role for Mbnls in regulating localization of mRNAs in both mouse and Drosophila cells, and Mbnl-dependent translation and protein secretion was observed for a subset of mRNAs with Mbnl-dependent localization. These findings hold several new implications for DM pathogenesis.
Introduction
Muscleblind-like (Mbnl) proteins are a deeply conserved, developmentally regulated family of RNA binding factors which contribute to muscle and eye development and have been implicated in the genetic disease myotonic dystrophy (DM) (Artero et al., 1998; Begemann et al., 1997; Miller et al., 2000). Flies possess a single Mbnl gene, while mammals express 3 closely related Mbnl genes (Fardaei et al., 2002). In mouse and human, Mbnl1 and Mbnl2 are expressed across many tissues, including brain, heart, and muscle, while Mbnl3 is expressed primarily in placenta (Kanadia et al., 2003b; Squillace et al., 2002). Mammalian Mbnl proteins contain two pairs of highly conserved zinc fingers, which bind to pre-mRNA to regulate alternative splicing (Pascual et al., 2006). In DM, Mbnls are sequestered away from their normal RNA targets by interaction with expanded CUG or CCUG repeats (Miller et al., 2000). This reverses the normal developmental accumulation of Mbnls, shifting splicing towards fetal isoforms (Lin et al., 2006).
The hypothesis that Mbnl proteins are responsible for a large fraction of the aberrant splicing patterns observed in DM was supported by a splicing microarray analysis (Du et al., 2010), which found that 80% of ~200 alternative isoform changes observed in a CUG-expressing mouse model of DM – including 55 cassette exons – were reproduced in mice lacking functional Mbnl1 protein. These results suggested that CUG repeat-induced transcriptome changes are largely Mbnl-dependent, but did not identify direct targets of Mbnl.
Mbnl proteins also have significant cytoplasmic expression and have been proposed to contribute to regulation of mRNA stability (Du et al., 2010; Masuda et al., 2012; Osborne et al., 2009) or localization (Adereth et al., 2005). Most mRNAs exhibit specific patterns of localization in the cell, which may be mediated by sequence-specific RBPs that interact with cis-regulatory elements located in 3' UTR regions (Andreassi and Riccio, 2009; Lecuyer et al., 2007; Martin and Ephrussi, 2009). Localized translation of mRNAs is thought to play important roles in proper protein localization and assembly of protein complexes (Besse and Ephrussi, 2008; Lecuyer et al., 2009), and affect cellular properties such as synaptic plasticity (Horne-Badovinac and Bilder, 2008; Kislauskis et al., 1997; Martin and Zukin, 2006). Localization of mRNAs to the rough endoplasmic reticulum (ER) via interaction of the signal recognition particle (SRP) with the nascent signal peptide can also involve recognition of sequences in the mRNA to a substantial extent (Pyhtila et al., 2008). Thus, sequence-specific RBPs like Mbnls that have significant expression in both nucleus and cytoplasm may potentially influence diverse aspects of mRNA biogenesis and function from processing through localization and translation.
To identify direct regulatory targets of Mbnl1, we analyzed the transcriptomes of brain, heart, and muscle tissue isolated from mice lacking functional Mbnl1 protein (Kanadia et al., 2003a), in conjunction with ultraviolet crosslinking/immunoprecipitation/sequencing (CLIP-Seq) to identify binding targets in the same tissues and in mouse myoblasts. These data yielded a transcriptome-wide RNA map for Mbnl-dependent splicing regulation. We observed extensive binding to 3' UTRs of mRNAs encoding membrane and synaptic proteins, and uncovered a widespread, conserved role for Mbnls in regulating localization. This function may contribute to the molecular events responsible for DM pathology.
Results
Identification of hundreds of Mbnl-responsive exons
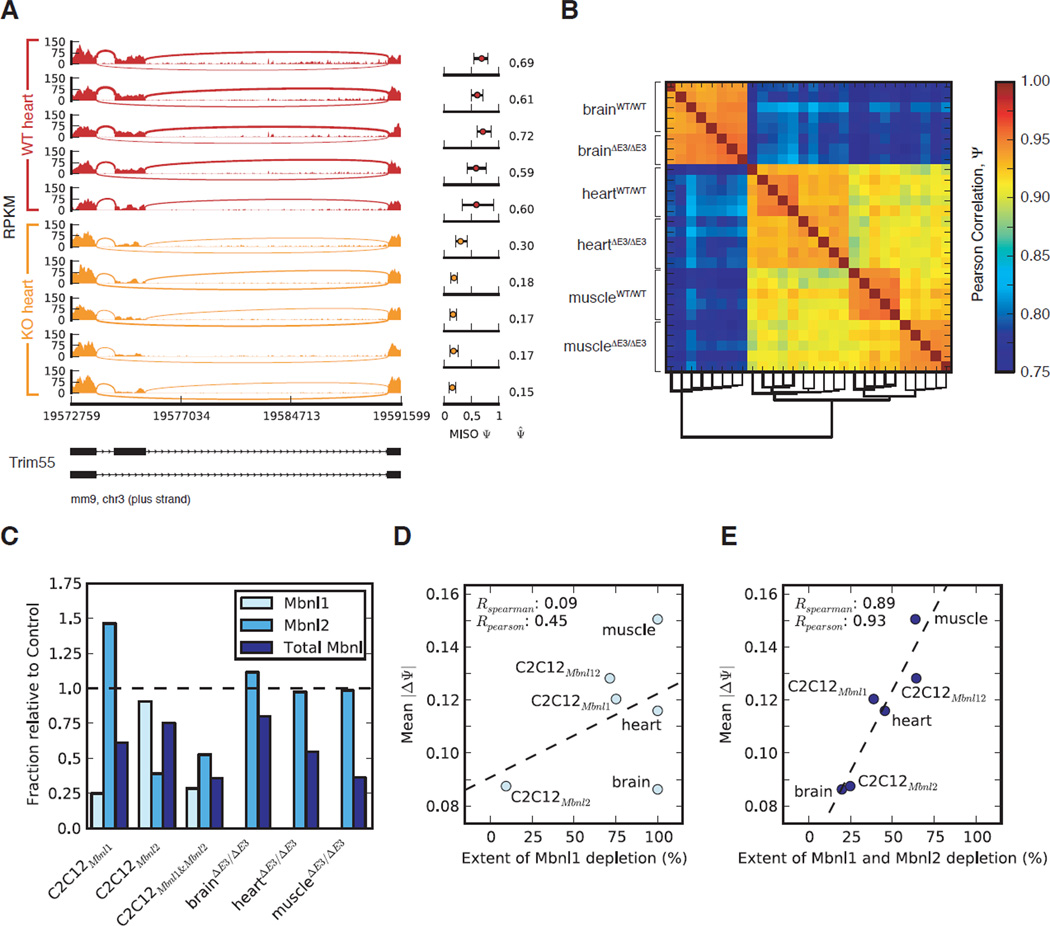
We performed single-end RNA-Seq of poly(A)+ RNA from brain, heart, and muscle from five 4 month-old Mbnl1ΔE3/ΔE3 mice and age-matched controls (129/SvJ), yielding 247M uniquely mapped 48-base reads. Mbnl1ΔE3/ΔE3 mice lack MBNL with RNA-binding activity (Kanadia et al., 2003a) (Figure S1A). MISO was used to compute “percent spliced in” (PSI or Ψ) values for cassette alternative exons, representing the fraction of a gene’s mRNAs that include the exon (Katz et al., 2010). These values showed high concordance with Ψ values assessed by RT-PCR (Figure S1B). We observed hundreds of consistent changes between wild type and knockout animals, with largest changes in muscle, intermediate changes in heart and smallest changes in brain (Figure S1D, Table S1). For example, exon 8 of the Trim55 gene, which encodes a muscle-specific RING finger protein involved in sarcomere assembly (Pizon et al., 2002), had 59–72% inclusion in wild type heart and 15–30% inclusion in knockout heart (Figure 1A). Using Ψ values of widely expressed exons, the samples clustered first by tissue, then by genotype, as expected (Figure 1B). Two knockout brains detected as outliers in this analysis were omitted (data not shown).
Figure 1. Dependence of splicing changes on total MBNL levels by RNA-Seq analysis.
(A) RNA-Seq read coverage across Trim55 exon 8, from wild type and Mbnl1 knockout heart tissue: MISO Ψ values and 95% CI shown at right. (B) Global correlation of Ψ values for cassette exons with dendrogram shown below. (C) The fraction of MBNL1, MBNL2, or total MBNL remaining in the indicated knockout tissues and knockdown myoblasts relative to control tissues/cells. (D & E) Mean splicing change (mean |ΔΨ| correlates much better with total MBNL depletion than with MBNL1 depletion. See also Figures S1, S2 and Table S1.
Changes in splicing of cassette exons (“ΔΨ” values) correlated well with previous estimates by splicing microarray (Du et al., 2010) (Figure S1E). However, our analysis identified 199 cassette exons with significantly altered splicing in muscle (Bayes Factor (BF) > 5 and |ΔΨ| > 0.05), roughly four times the number identified by microarray (Figure SD, SE), and many more Mbnl-dependent cassette exons were identified in heart and brain (Table S1). Alternative 5' and 3' splice sites and other types of events also exhibited Mbnl-dependence, yielding a total of 912 Mbnl-dependent splicing events and 555 Mbnl-dependent alternative 3' UTRs in mouse tissues (Table S1). Together, these exons provide a resource for analyses of the phenotypic consequences of Mbnl depletion.
Splicing regulation by Mbnls is dependent on aggregate levels of Mbnl proteins
To supplement our analysis of mouse tissues, and to assess the roles of Mbnl1 relative to Mbnl2 in splicing regulation, we stably infected C2C12 mouse myoblasts with lentiviral hairpins against Mbnl1, Mbnl2, or both Mbnls (which did not cause gross changes in morphology or viability), and conducted RNA-Seq. Western blotting confirmed reduction in protein levels (Figure S1C) and revealed increased levels of a higher molecular weight isoform of Mbnl2 in Mbnl1-depleted cells, and vice versa, suggesting that these factors cross-regulate each other post-transcriptionally. Expression levels of isoforms were comprehensively assessed (Table S1), and levels of Mbnl1, Mbnl2, and total Mbnl (Mbnl1 + Mbnl2) were determined (Figure 1C). We also calculated the “mean splicing change” as the mean |ΔΨ| for cassette exons that varied in at least one sample relative to control (|ΔΨ| > 0.1, BF > 5, n = 465 cassette exons). The mean splicing change was only modestly correlated to the level of Mbnl1 (Figure 1D) or the level of Mbnl2 (Figure S2A), but showed very strong correlation with total Mbnl levels (Mbnl1 + Mbnl2) (Figure 1E). This trend held not only held for exon skipping, but also for alternative 3' splice sites (178 events), alternative 5' splice sites (107 events) and gene expression changes (Figure S2). Mbnl3 was lowly expressed and did not vary significantly between treatments, so was omitted from this analysis. Together, these data suggested that Mbnl1 and Mbnl2 function interchangeably in regulation of a large set of splicing targets and that the net depletion of both factors will be relevant in DM and genetic models of the disease.
Transcriptome-wide binding locations of Mbnl1 in brain, heart, muscle, and myoblasts
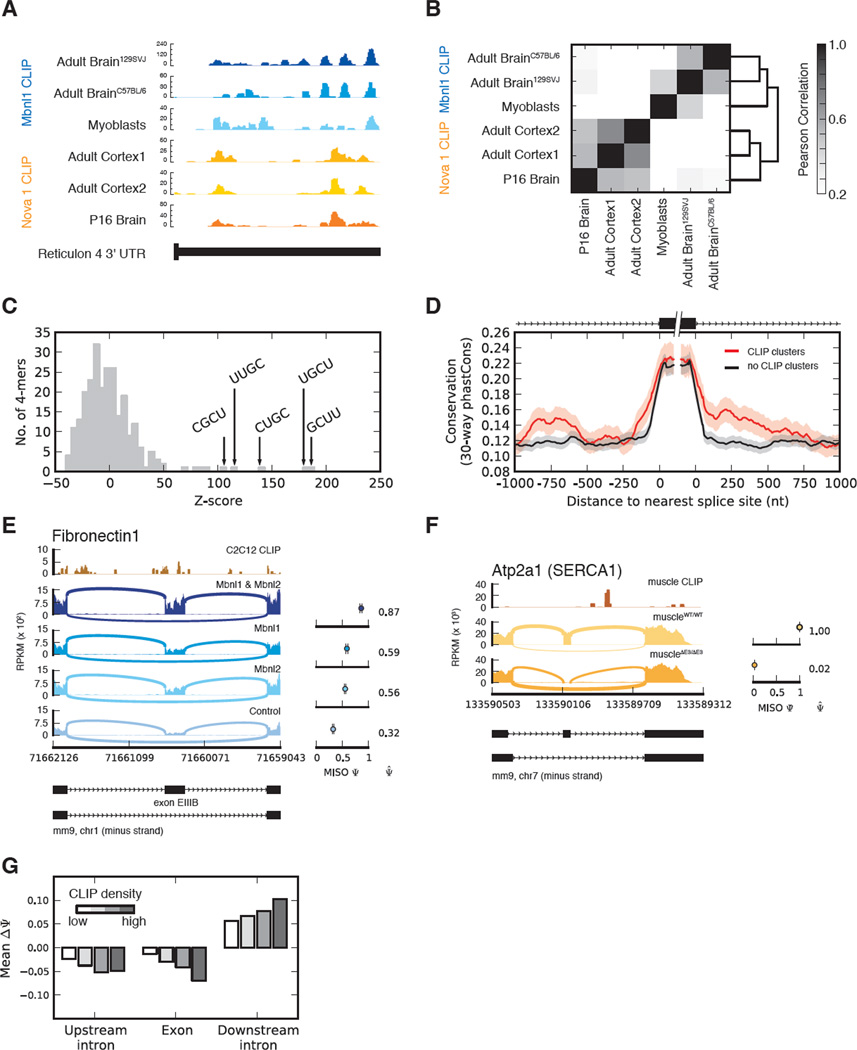
To distinguish between direct and indirect Mbnl targets, and to study context-dependent rules for Mbnl regulation, we performed CLIP-Seq (with random barcodes in some cases) as described (Konig et al., 2010; Ule et al., 2005; Wang et al., 2009), yielding 1.6M, 250K, 120K, 71K, and 2.06M reads uniquely mapping to the genome and splice junctions in the 129/SvJ brain, C57BL/6 brain, heart, muscle, and myoblast samples, respectively (after collapsing identical reads). Mapping occurred predominantly to transcribed regions, with enrichment for introns and/or 3' UTRs (Figure S3).
Mbnl1 CLIP binding locations were consistently observed in different mouse strains, and even when comparing different cell types (e.g., Reticulon 4, Figure 2A). These sites were essentially uncorrelated with CLIP sites for a different RNA binding protein, Nova (Zhang et al., 2010), in this example and overall, supporting the specificity of each CLIP dataset (Figure 2B). Controlling for effects of pre-mRNA length and gene expression (Yeo et al., 2009) CLIP clusters were enriched for UGC- and GCU-containing 4mers in myoblasts (Figure 2C) and other tissues. Alternative exons with CLIP clusters within 1 kilobase were more highly conserved than other alternative exons (Figure 2D, controlling for gene expression), supporting their function.
Figure 2. MBNL binding is conserved and associated with context-dependent splicing activity.
(A) MBNL1 and Nova CLIP tags derived from various tissues and cells in the 3' UTR of Reticulon 4. (B) Correlation of CLIP tag densities in 5 nucleotide windows across all 3' UTRs expressed in brain and C2C12 myoblasts. (C) Histogram of Z-scores of 4mers occurring in MBNL1 CLIP clusters from C2C12 myoblasts, relative to control regions from the same genes. (D) Alternative exons with MBNL1 CLIP clusters upstream or downstream are more conserved than other alternative exons expressed at similar levels. (E & F) Dependence on Mbnl levels of PSI values of Fibronectin 1 exon EIIIB and SercaI exon 22, respectively (CLIP data is shown above). (G) Dependence of change in PSI value (wild type – Mbnl1, Mbnl2 knockdown) on MBNL1 binding in the upstream intron (last 300 bases), alternative exon, or downstream intron (first 300 bases). See also Figures S3, S4 and Table S2.
Context-dependent splicing regulation by Mbnl proteins
Several individual Mbnl-regulated exons have been well characterized, with Mbnl either activating or repressing exon inclusion (Hino et al., 2007; Ho et al., 2004; Warf and Berglund, 2007); (Sen et al., 2010). Binding sites upstream or within the 3' splice site generally repress splicing (Warf and Berglund, 2007) and enrichment of Mbnl motifs downstream of activated exons suggests that Mbnl may activate splicing in this context (Du et al., 2010; Goers et al., 2010). Our CLIP-Seq data confirmed a number of binding sites discovered in previous studies, e.g., sites downstream of SercaI exon 22 (Hino et al., 2007), whose inclusion is highly sensitive to Mbnl levels (Figure 2F). We also uncovered thousands of new locations for Mbnl1 binding associated with splicing activity, including sites in/around exon EIIIB of Fibronectin I (Figure 2E), whose splicing is regulated during development and wound healing (Ffrench-Constant and Hynes, 1989; Ffrench-Constant et al., 1989) and which increased as total Mbnl levels decreased (cf. Figure 1E). Splicing repression was generally associated with binding in the upstream intron and in the alternative exon, with increased binding associated with stronger repression (Figure 2G). Splicing activation was associated with binding in the downstream intron, also increasing with increased binding density (Figure 2G).
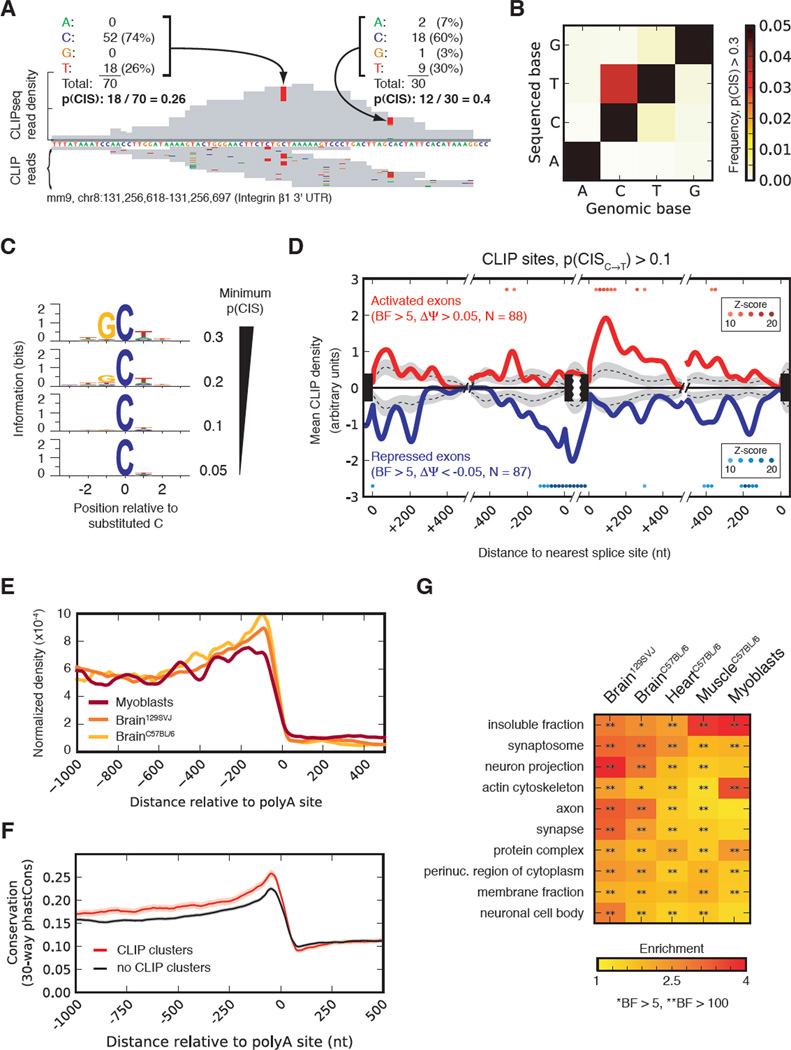
Crosslink-induced substitutions locate precise Mbnl1 binding sites
To define a high-resolution “RNA map” of the regulatory activity associated with binding at specific distances from splice sites, we sought to pinpoint Mbnl binding sites by analysis of crosslink-induced mutations (Kishore et al., 2011). The frequency of crosslink-induced substitutions (CIS) was calculated at each position as the fraction of reads that cover the position but do not match the reference base (Figure 3A). The base with highest CIS frequency was cytosine, and C to T substitution occurred at least 5- to 10-fold more often than any other substitution (Figure 3B). Cytosines with high CIS had biases in flanking bases: the −1 base just preceding the substituted C increasingly biased toward guanine (reaching 94%), and the −2, +1, and +2 bases were preferentially uracil (Figure 3C). The resulting motif closely matched the UGCU motif found in previous studies of Mbnl1 binding affinity and in Figure 2C, suggesting that frequently substituted cytosines represent sites of direct crosslinking to Mbnl1 zinc fingers (Teplova and Patel, 2008). Deletions in CLIP-Seq reads were also observed, but were rarer and less frequently associated with Mbnl motifs, so were not separately analyzed (Figure S3).
Figure 3. Patterns of MBNL binding and a nucleotide-resolution map of splicing activity.
(A) Examples of substitutions observed in MBNL1 CLIP tags and calculation of p(CIS). (B) Cytosine exhibits the greatest p(CIS) within MBNL1 CLIP clusters, and is most often sequenced as thymine. (C) Information (relative entropy) of positions adjacent to frequently substituted cytosines, as a function of substitution frequency. (D) Mean CLIP density near exons activated (red), repressed (blue) by MBNL1, using binding sites with p(CIS)C→T > 0.1. Density around exons unaffected by Mbnl depletion is shown by dotted line, with 90% confidence intervals in gray. Colored dots denote 100 nucleotide-long windows significantly greater density of CLIP sites at regulated than non-regulated exons. (E) Mean MBNL1 CLIP density at positions along 3' UTRs using 3 CLIP datasets. (F) Conservation in 3' UTRs with and without MBNL1 CLIP clusters with similar expression levels and UTR lengths. (G) Significant Gene Ontology categories of genes with 3' UTRs having MBNL1 CLIP. See also Figures S4 and Table S2.
The overlap between MBNL-regulated and MBNL-bound exons was quite extensive (Table S2). In all, 31% of regulated exons (compared to 14% of unregulated exons) were associated with high confidence CLIP sites (P(C→T) > 10%), and this is likely an underestimate as the fraction increased to 44% in genes expressed above median levels, and exceeded 75% when considering all CLIP sites.
We first constructed an RNA map for Mbnl-dependent splicing regulation using the frequently substituted positions (P(C→T) > 10%, Figure 3D). Activation and repression-associated binding were ~4.5-fold and ~9-fold above background, respectively (both Z-scores > 10; Methods), and occurred in almost completely distinct locations. A repression-associated peak was centered on the alternative exon and extended through the 3' splice site ~140 bases into the upstream intron, consistent with repression via direct occlusion of exonic enhancer, 3' splice site or branch site elements. Additional repressive peaks occurred ~100 bases 5' of the downstream 3' splice site, suggesting that repression of splicing may occur by different mechanisms (Figure 3D). An activation-associated peak began just 3' of the regulated exon and extended ~120 bases into the downstream intron. These data establish precise locations where binding of Mbnl is associated with repression or activation of splicing, enabling prediction of regulatory consequences. An RNA map constructed using all CLIP sites yielded qualitatively similar results, but with peaks that were less consistent and pronounced, suggesting lower precision (Figure S4).
An RNA map constructed for alternative 3' splice sites revealed significant enrichment of MBNL binding between the competing splice sites, in cases associated with repression of the intron-proximal site by MBNL (Fig. S4D). This pattern of activity is analogous to that observed for other motifs that function as ESSs (Wang et al., 2006).
3' UTR binding is associated with specific protein functions and localizations
Genes with Mbnl binding in 3' UTRs were most enriched for “cellular component” Gene Ontology categories, as compared to “molecular function” and “biological process” (Figure S4). In all tissues and myoblasts, enriched cellular component categories included synapse, membrane fraction, and insoluble fraction (Figure 3G). Some of these targeted genes included mRNAs encoding proteins involved in synaptic vesicle fusion such as Snap25 and Vamp1, signaling proteins like Camk2α, extracellular matrix (ECM) interacting membrane protein integrin β1, and secreted ECM proteins such as collagens. These data suggested a functional association between Mbnl1 and messages encoding proteins with these localizations. Consistent with this interpretation, the cytoplasmic distribution of Mbnl1 did not appear uniform, with localization at the periphery of the cell or in structures appearing to anchor cellular projections (Figure S4F). Furthermore, as observed previously in human cells (Holt et al., 2009), Mbnl protein partially shifts from the nucleus to cytoplasm following trypsinization and re-plating of mouse myoblasts (Figure S4E). Genes containing MBNL-regulated alternative exons were modestly enriched for some of the same Gene Ontology categories as above, but none reached statistical significance (not shown).
A substantial proportion of Mbnl CLIP reads (sometimes the majority) were located in 3′ UTRs, consistent with a recent study (Masuda et al., 2012), and exhibited enrichment relative to RNA-Seq reads (Figure S3C). Tetramer motifs in CLIP reads mapping to 3' UTRs and introns (based on z-scores, as in Fig. 2C) were highly overlapping, suggesting that MBNL binds similar motifs in 3' UTRs as in introns (not shown). Binding occurred predominantly 5' of the cleavage and polyadenylation site (PAS), consistent with interaction in the cytoplasm, and tended to increase closer to the PAS, suggesting a function other than splicing regulation (Figure 3E). Those 3' UTRs containing CLIP clusters were more highly conserved than other 3' UTRs of similar length and expression level, suggesting enrichment for functional features (Figure 3F).
Mbnl preferentially binds to localized mRNAs
Proteins that bind 3' UTRs are associated with specific cellular and molecular functions, including regulation of mRNA stability, translation, and localization (Moore and Proudfoot, 2009; Zhao et al., 1999). The strong biases for binding to mRNAs associated with particular cellular compartments and a previous study demonstrating that Mbnl2 localizes integrin α3 mRNA to the plasma membrane for translation (Adereth et al., 2005) suggested that Mbnls might play a general role in localization of many mRNAs.
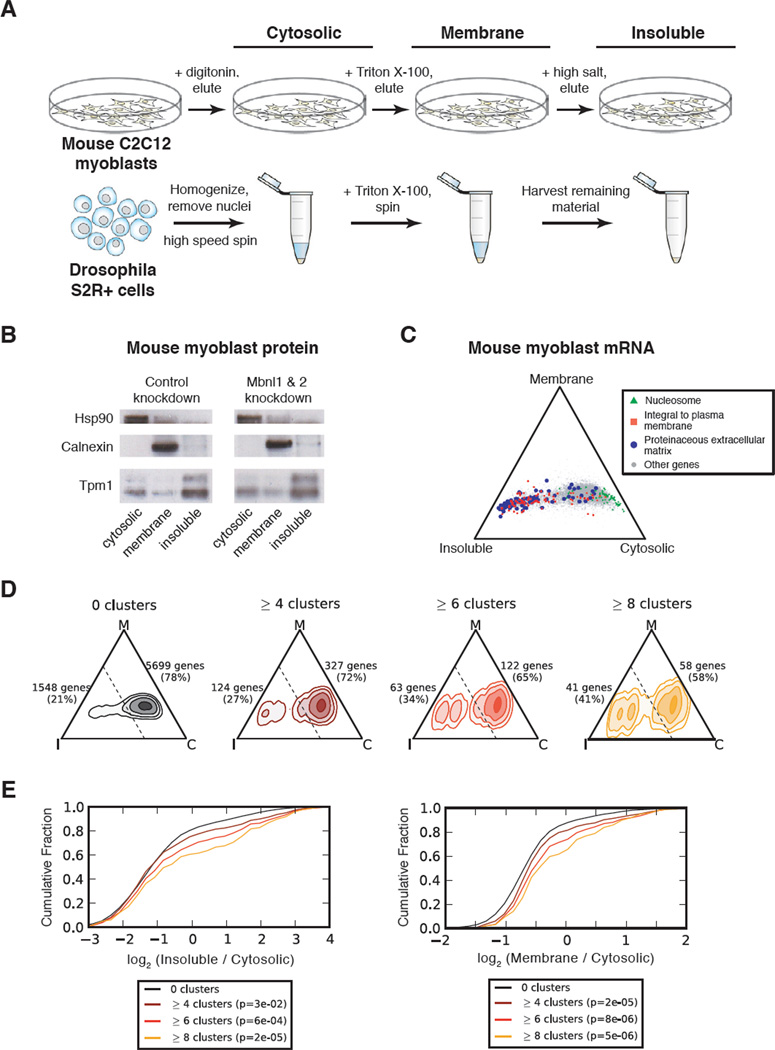
To assess mRNA localization in mouse myoblasts, we used cellular fractionation followed by RNA-Seq. Sequential detergent extractions yielded cytosolic, membrane (including organelles such as rough endoplasmic reticulum (ER)), and insoluble fractions (Jagannathan et al., 2011; Vedeler et al., 1991) (Figure 4A). Western analysis of protein markers Hsp90, calnexin, and tropomyosin 1 in each of these fractions confirmed separation of the corresponding cellular compartments (Figure 4B). Hsp90 was exclusively cytosolic (Csermely et al., 1998); calnexin, an ER resident protein (Bergeron et al., 1994), was almost exclusively associated with the membrane fraction; and tropomyosin, which decorates F-actin filaments (Gunning et al., 2005), was substantially enriched in the insoluble fraction. In parallel, we performed similar analysis in Drosophila S-2R+ cells to assess potential phylogenetic conservation of effects on mRNA localization. Semi-adherent S-2R+ cells were processed by sequential homogenization/centrifugation to obtain analogous cellular compartments (Figure 4A). Assessment of several marker proteins in S-2R+ cells confirmed successful fractionation (Figure S5B), and the mRNA content of each fraction was analyzed by RNA-Seq (Table S3).
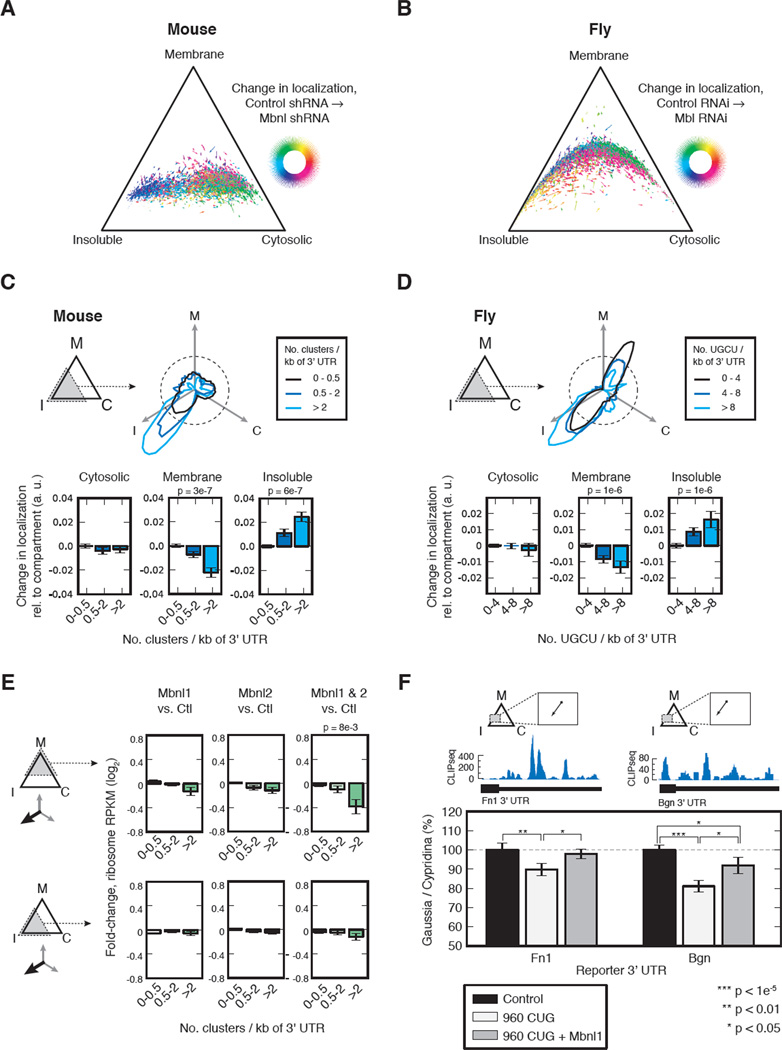
Figure 4. mRNA localization is associated with functional binding by MBNL.
(A) Cellular fractionation scheme in C2C12 mouse myoblasts and fly S2R+ cells. Adherent myoblasts were fractionated on tissue culture plates, while semi-adherent S2R+ cells were fractionated using centrifugation, yielding “cytosolic”, “membrane”, and “insoluble” fractions. (B) Western blot of Hsp90, Calnexin, and Tropomyosin I proteins for each compartment in control and MBNL knockdown myoblasts. (C) Simplex representation of mRNA expression across compartments in control myoblasts, with mRNAs belonging to specific functional categories colored. (D) Density map of MBNL1 CLIP targets in control myoblasts. The relative abundance of genes with 0, ≥4, ≥6, or ≥8 CLIP clusters is illustrated within the simplex, and quantified for areas of the simplex on either side of the dashed line. (E) Cumulative distribution functions of relative enrichment in Insoluble/Cytosolic (left) or Membrane/Cytosolic (right) of genes grouped by number of 3' UTR CLIP clusters, with significance determined by rank-sum test. See also Figures S5 and Table S3.
To visualize differences in mRNA abundance across the three fractions, we displayed mRNAs as points in a triangular simplex, where the proximity to each corner of the triangle represents the relative expression bias towards the corresponding cellular compartment (cytosolic, membrane, or insoluble). While this approach does not provide absolute mRNA abundance in each compartment, it allows the comparison of relative mRNA levels across compartments and conditions. mRNAs belonging to particular functional categories showed compartment-biased expression consistent with the localization patterns of their encoded proteins (Figure 4C), similarly to what has been established for some mRNAs (Besse and Ephrussi, 2008). In myoblasts, mRNAs encoding nucleosomal proteins were enriched in the cytosolic compartment, while integral plasma membrane and proteinaceous ECM proteins were biased towards the membrane and especially insoluble compartments relative to the cytosol (Figure 4C). These observations are consistent with previous studies showing that mRNAs encoding secreted proteins are biased towards membranous cellular compartments such as rough ER and nuclear envelope (Pyhtila et al., 2008), and with studies showing that mRNAs encoding cytoskeletal or ECM proteins are actively translated on cytoskeletally-anchored ribosomes (Hesketh, 1994; Pachter, 1992; Sundell and Singer, 1991). As expected, mRNAs encoding signal peptides were highly enriched in the membrane and especially insoluble compartments (Figure S5G). Similar associations between mRNA localization and protein function were observed in Drosophila cells (Figure S5D).
To assess the relationship between Mbnl binding and mRNA localization, we subdivided genes by the number of 3' UTR CLIP clusters, and plotted their relative density within the simplex. We observed that the proportion of mRNAs biased toward the “insoluble” corner increased significantly with the number of CLIP clusters (Figure 4D, E). These observations support the association of Mbnl with cytoskeletally trafficked mRNAs and suggest a connection to the secretory pathway.
Mbnl depletion alters localization of bound mRNAs
To assess whether Mbnl directly impacts localization of its binding targets, we performed analogous fractionation/RNA-Seq analyses of mouse myoblasts depleted of Mbnl1 and Mbnl2. Comparable separation of cellular compartments in knockdown cells was confirmed by Western analysis (Figure 5B) and similar associations between protein function and mRNA localization were observed (Figure S5C), suggesting that depletion of Mbnls did not grossly alter cellular RNA trafficking. Assessment of mRNA abundance in each compartment in control and Mbnl-depleted cells enabled analysis of changes in localization for individual mRNAs. These changes were displayed as vectors in the triangular simplex, and colored to highlight patterns in direction of change following knockdown (Figure 5A). We focused our analyses on the subset of genes that were biased toward the insoluble corner (in which Mbnl binding was most enriched), and further subdivided these genes according to CLIP cluster density. Following Mbnl depletion, these genes tended to have reduced membrane association and increased bias towards the insoluble compartment (Figure 5C). The extent of re-localization correlated with increasing CLIP density (Figure 5C), supporting a model in which Mbnl binding directly affects mRNA trafficking and localization.
Figure 5. MBNL binding is associated with regulation of mRNA localization in mouse and fly, and may contribute to protein secretion.
(A & B) Change in mRNA localization following depletion of MBNLs in mouse myoblasts and fly S2R+ cells. Arrows represent changes in mRNA localization, where the direction change is encoded by color. (C & D) The aggregate behavior following MBNL depletion of the subset of genes biased toward the insoluble compartment in control cells is shown in a polar plot, where the radial amplitude is the number of genes that relocalized in the given direction following MBNL depletion, stratified by density of CLIP clusters (for mouse) or UGCU 4mers (for fly). The aggregate change in localization towards each compartment is shown bar plots. P-values denote significance when comparing change in localization of MBNL targets versus non-targets (e.g., >2 versus 0–0.5 for mouse myoblasts). (E) The mean fold-change in ribosome footprint density following MBNL1, MBNL2, or double MBNL depletion relative to control was quantitated for mRNAs which were initially biased towards the membrane compartment (top) or the insoluble compartment (bottom), and whose localization shifted towards the insoluble compartment following MBNL depletion. Genes were further stratified by density of MBNL CLIP sites in 3' UTR. The P-value denotes the significance of the change in footprint density when comparing MBNL targets versus non-targets (rank-sum test). (F) Change in mRNA localization of Fibronectin 1 (Fn1) and Biglycan (Bgn) following MBNL depletion in mouse myoblasts (top), MBNL1 CLIP tag density in the 3' UTR of each gene (middle), results of secreted luciferase assay using luciferases containing the Bgn 3' UTR or the Fn1 last intron plus 3' UTR (bottom). See also Figure S6 and Table S4.
Muscleblind proteins are highly conserved across species, and have been shown to be functionally interchangeable between mammal and fly in certain cases (Goers et al., 2010). To assess whether the localization activity of mouse Mbnl1 is conserved to fly, we depleted mbl mRNA from Drosophila S-2R+ cells (Figure S5A) and performed fractionation/RNA-Seq analysis as above (Figure 5B, Table S4). Again, mbl depletion did not cause gross changes in cellular morphology, viability or bulk mRNA trafficking (Figure S5D). To assess effects of mbl depletion, we analyzed mRNAs biased towards the insoluble compartment, using UGCU motif density as a proxy for mbl binding (Goers et al., 2010). As in mouse, we observed a bias for re-localization of putative mbl targets away from membrane and towards the insoluble compartment, and again, the strength of this trend increased with motif density (Figure 5D). Thus, depletion of muscleblind proteins results in similar alterations in the localization of putative mRNA binding targets (Tables S2 and S3) in both mammalian and fly cells.
Mbnls have also been implicated in regulating mRNA stability (Du et al., 2010; Masuda et al., 2012; Osborne et al., 2009). To investigate a potential link between message stability and localization, we analyzed total gene expression of Mbnl 3' UTR binding targets following Mbnl depletion. Modest increases (< 5%) in mean expression of Mbnl targets following Mbnl depletion were observed overall, and in mRNAs localized to particular subcellular compartments (Figure S6).
Mbnl function contributes to mRNA translation
Because mRNA localization is often coupled to translational regulation, we sought to assess effects of Mbnl at the translational level by performing ribosome footprint profiling (Ingolia et al., 2011) using control C2C12 mouse myoblasts, and myoblasts depleted of MBNL1, MBNL2, or both MBNL1 and MBNL2. These data, representing millions of ribosome-protected mRNA fragments mapping primarily to coding and 5' UTR regions (Figure S6), were used to obtain genome-wide ribosome RPKM measurements that are analogous to RNA-Seq-based RPKMs, but reflect the ribosomal association of mRNAs rather than mRNA abundance. We observed that subsets of MBNL 3' UTR targets, in particular those messages that had membrane-biased localization and moved toward the insoluble compartment following MBNL depletion, had substantially reduced ribosome RPKMs in myoblasts depleted of both MBNLs relative to controls or cells depleted of MBNL1 or MBNL2 alone (Figure 5E). By contrast, the overall abundance of these mRNAs (based on RNA-Seq) actually increased slightly following depletion of MBNLs (Figure S6), implying that Mbnl depletion results in reduced translation of this subset of mRNAs. Alternatively, these effects could possibly result from changes in mRNA stability dependent on the translational status of the mRNA. The analogous subset of messages biased to the insoluble compartment did not exhibit significantly decreased ribosome footprint RPKMs (Figure 5E).
Mbnl function contributes to protein secretion
Many of the mRNAs biased towards the insoluble compartment that re-localized following Mbnl depletion encode secreted proteins (Figure S5G). While, classically, recognition of the signal peptide by the signal recognition particle (SRP) targets mRNAs to the secretory pathway (Blobel and Dobberstein, 1975a, b), more recent studies have demonstrated that RNA elements at the 5' ends of ORFs also contribute to targeting of mRNAs to the rough ER or to efficiency of nuclear export (Cenik et al., 2011; Palazzo et al., 2007; Pyhtila et al., 2008). We therefore hypothesized that effects of Mbnl on mRNA localization might play a role in targeting mRNAs to the secretory pathway.
To test this hypothesis, we performed a dual luciferase assay using two efficiently secreted luciferases, Gaussia and Cypridina. We cloned the 3' UTRs of Biglycan and Fibronectin 1 onto the 3' end of Gaussia luciferase; each gene has abundant Mbnl binding in the 3' UTR and substantially changes in localization following Mbnl depletion (Figure 5F). Upon co-transfection with a plasmid encoding 960 CUG repeats, which mimics DM via functional inactivation of Mbnls (Cooper et al., 2009), we observed a 10–20% reduction in secretion of Gaussia with the Fn1 or Bgn 3' UTR relative to control (Figure 5F). Over-expression of Mbnl1 coding sequence led to a complete rescue of the Fn1 3' UTR-mediated secretion defect and a partial rescue of Bgn 3' UTR-mediated secretion (Figure 5F), suggesting that the effects of CUG repeats on secretion are attributable to the depletion of Mbnls. Taken together, these data and the footprinting results support a model in which Mbnl binding to the 3' UTR of an mRNA promotes productive translation and/or secretion of the encoded protein.
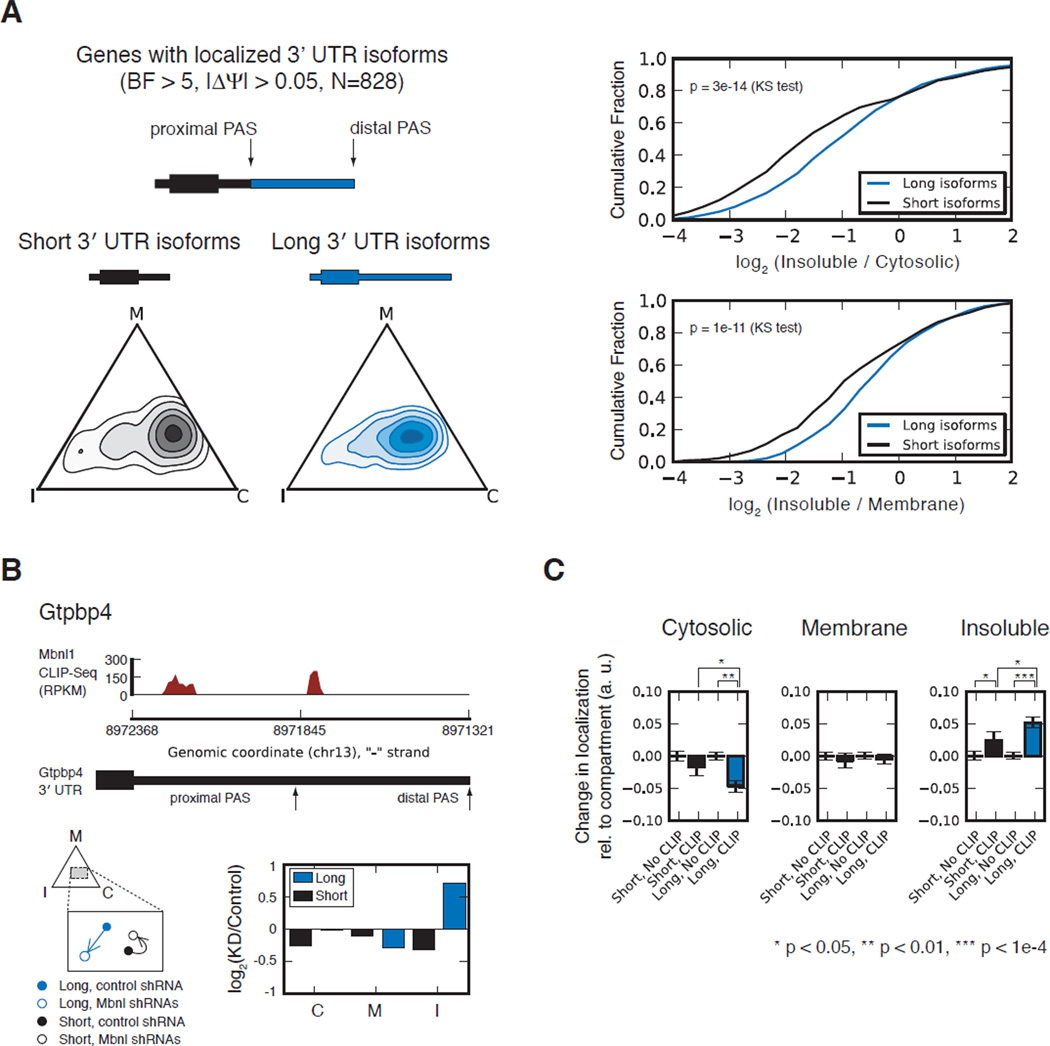
Mbnl function is associated with localization of alternative 3' UTR isoforms
Thousands of mammalian genes express mRNA isoforms with different 3' ends as a result of alternative PAS usage, which can be quantitated based on RNA-Seq data (Katz et al., 2010; Wang et al., 2008). Alternative 3' UTR isoforms provide natural “reporters” for the function of regulatory elements located in the distal (long isoform-specific) region. We found hundreds of genes whose alternative 3' UTR isoforms had differing localization patterns in myoblasts (Table S4). These longer isoforms tended to be localized towards the insoluble compartment relative to the shorter isoform (Figure 6A). This effect is likely attributable to RNA sequence elements uniquely present in the distal regions, which were frequently bound by Mbnl1 (Figure 6B).
Figure 6. MBNL regulates Isoform-specific mRNA localization via binding to alternative 3′ UTRs.
(A) The localization of long and short alternative 3' UTR isoforms is shown at left (as in Fig. 4B). Relative enrichment in pairs of cellular compartments is shown for long and short isoforms at right (as in Fig. 4E). (B) CLIP data for the 3' UTR of Gtpbp4. Localization of the long and short isoforms in normal myoblasts is displayed in solid blue and black circles, respectively, and in myoblasts depleted of Mbnl in outlined blue and black circles, respectively, with arrows indicating change in localization. Bar plot shows change in RPKM (Mbnl-depleted versus control myoblasts) for the long and short 3' UTR isoforms of Gtpbp4. (C) Genes were separated into those with and without CLIP clusters in the core or extended 3' UTR region, for short and long isoforms, respectively, and assessed for direction of re-localization following Mbnl depletion. Bar plot shows the extent of re-localization towards each compartment, where the magnitude of re-localization for CLIP targets is normalized relative to non-targets. See also Figure S7 and Table S5.
Depletion of Mbnls led to relocalization of specific subsets of Mbnl-bound mRNA isoforms. Focusing on mRNA isoforms biased towards the insoluble compartment as in Figure 5, we observed that Mbnl1-bound long isoforms shifted strongly away from cytosol and towards the insoluble compartment following depletion of Mbnl1 and Mbnl2. More modest shifts were observed for Mbnl1-bound short isoforms (Figure 6C). These effects persisted when restricting the analysis to 3' UTRs whose overall isoform abundance did not change (Figure S7A), suggesting that they resulted from Mbnl activity outside the nucleus rather than from altered PAS choice in the nucleus. This analysis suggests that Mbnl may play a role in regulating isoform-specific subcellular localization of long UTR isoforms, which are particularly abundant in mammalian brain and muscle (Ramsköld et al., 2009).
Discussion
Because of the extreme heterogeneity of clinical symptoms, DM has been described as one of the most variable disorders known in medicine (Harper, 1979). Some of the heterogeneity in DM1 undoubtedly results from the variability in inherited triplet repeat length, and from differences in the extent of somatic expansion in different tissues (Ashizawa et al., 1993). This genomic variability interacts with the diverse tissue expression of DMPK mRNA and of Mbnl and CELF family proteins. Thus, even between different patients with the same inherited repeat length, Mbnl proteins may be depleted to different extents in skeletal muscle, brain or other tissues. A comprehensive understanding of the molecular basis of DM will therefore require not only an understanding of the normal functions of Mbnl proteins in affected cell and tissue types, but also an appreciation of the extent to which different functions are impacted by particular degrees of Mbnl depletion.
Our global analyses yielded several insights into Mbnl function and established genomic resources for future functional, modeling and clinical studies, identifying hundreds of Mbnl-regulated exons and thousands of Mbnl1 binding sites across the transcriptome, many of which are likely to be conserved to human (Figures 2D, 4B). Knowledge of direct regulatory targets should aid in reconstructing the order of events that occur during disease pathogenesis as Mbnl levels are depleted, and provide insights into mechanism.
The transcriptome data indicate that Mbnl1 and Mbnl2 co-regulate a set of shared targets and that their splicing functions may be largely interchangeable. The effects of interventions that alter the levels of Mbnl1 or Mbnl2 are therefore likely to depend strongly on the level of the other factor, which will vary substantially depending on the affected tissue. For example, Mbnl2 depletion might produce greater changes than Mbnl1 depletion in neurons, and vice versa in muscle, while the effects of simultaneous depletion of both factors by CUG repeats should depend more on the size of the total pool of Mbnls than on the level of either factor alone.
We present several lines of evidence for extra-nuclear functions of Mbnl1 in regulation of mRNA localization and protein expression. A previous study demonstrated that Mbnl2 localizes integrin α3 to the plasma membrane (Adereth et al., 2005). In this and many other cases, membrane localization is associated with efficient mRNA translation. Here, we show that Mbnls contribute to targeting of hundreds of mRNAs to membranes. Further, we show that the efficient translation of these mRNAs – particularly those that are membrane-biased – is dependent on Mbnl. We also observed that Mbnl-bound 3' UTRs contribute to protein secretion (Figure 5F), perhaps by facilitating targeting to rough ER.
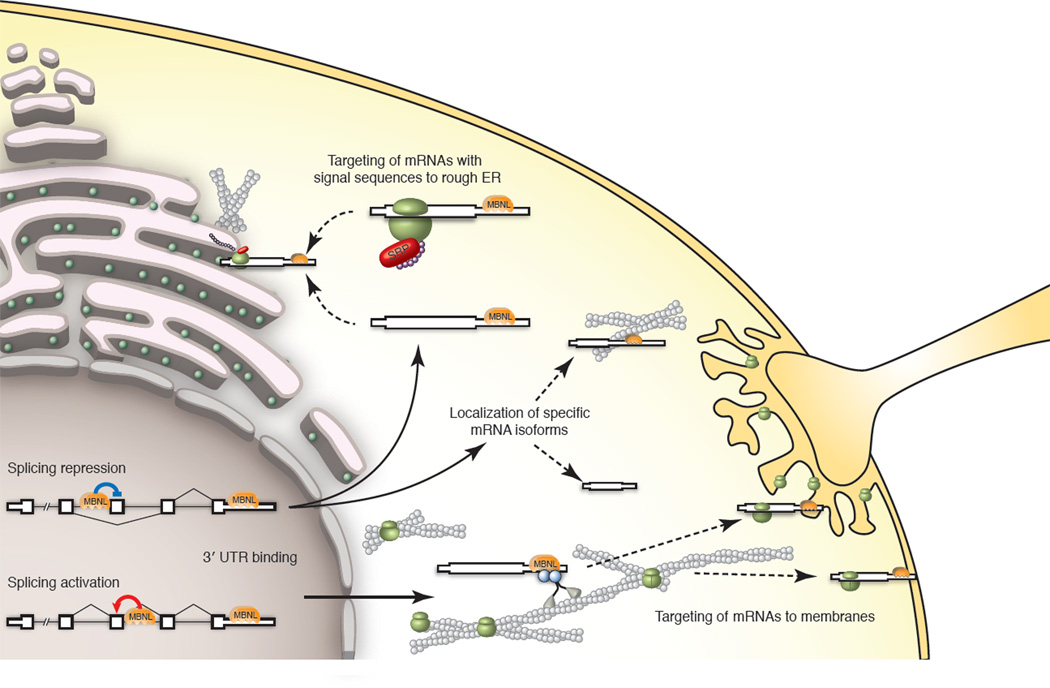
Our results led to a model of the activities of Mbnl proteins in nucleus and cytoplasm that is illustrated in Figure 7. In this model, nuclear Mbnls bind introns or exons to activate or repress splicing while cytoplasmic Mbnls bind 3' UTRs and facilitate transport along the cytoskeleton to the rough ER or other membranes, presumably through interaction with cellular transport machineries. Mbnl’s nuclear and cytoplasmic functions may be related. Indeed, splicing regulation by Mbnls was associated with 3' UTR binding: genes with evidence of direct splicing regulation by Mbnl1 were twice as likely to have CLIP clusters in their 3' UTR (p = 7e-6, Fisher exact test, Figure S7B, C). We observed a positive correlation between the density of 3' UTR CLIP clusters and proximity to MBNL-regulated exons, but whether this correlation reflects a functional relationship or simply results from the correlation between high C+G content and short intron length is not clear (Fig. S7D).
Figure 7. A model for nuclear and cytoplasmic functions of MBNL.
MBNLs repress or activate splicing depending on binding location. In the cytoplasm, MBNL binding in 3' UTRs may facilitate targeting of mRNAs with signal sequences to the rough ER. Alternatively, transcripts may be targeted to membrane-rich organelles for localized translation via actin-, microtubule-, or intermediate filament-based molecular motors. These organelles may include synapses, NMJs, or the plasma membrane, depending on cell type. MBNL may mediate isoform-specific mRNA localization, in which MBNL binding sites within distal 3' UTRs are required for targeting to particular compartments. (Model based on data from previous figures. See also Figure S7.)
Both cytoplasmic and nuclear pools of Mbnl are affected in DM (Miller et al., 2000), suggesting that functions of Mbnl outside the nucleus may play a role in DM pathogenesis. Cellular structures particularly sensitive to alterations in mRNA localization, such as synapses and neuromuscular junctions (NMJs), may be perturbed in DM as a result of Mbnl sequestration. Indeed, NMJ degeneration has been observed in diaphragm muscles in mouse models of DM (Panaite et al., 2008). Additionally, motor neurons derived from human DM ES cells fail to form mature neuromuscular connections with normal muscle (Marteyn et al., 2011), and worms deficient in neuronal muscleblind expression fail to form distal NMJs, implicating pre-synaptic dysregulation (Spilker et al., 2012). Secretion of proteins with important functions in muscle may also be affected in DM. Biglycan, whose 3'UTR confers Mbnl-dependent secretion, is of particular interest because its loss leads to NMJ degeneration, and because it has been proposed as a therapy for Duchenne’s muscular dystrophy (Amenta, 2012). Other relevant secreted factors include fibronectin 1 and thymosin-beta 4 (Figure 6E and Table S2). Our findings suggest that downstream consequences of Mbnl’s cytoplasmic functions, such as altered levels of extracellular proteins or circulating factors, may play important roles in disease pathogenesis and could serve as easily accessible diagnostics for monitoring disease progression and assessing response to therapy.
Experimental Procedures
RNA-Seq library preparation and sequencing
Five 16 week-old 129/SvJ Mbnl1ΔE3/ΔE3 mice and littermate WT mice were sacrificed for isolation of whole brain, heart, and skeletal muscle (vastus lateralis). Poly(A)+ RNA was prepared for single end RNA sequencing (50 cycles by Illumina GAII). RNAseq libraries for C2C12 mouse myoblasts were performed similarly (paired-end 2×36 cycles by Illumina GAIIx). For the cellular fractionation experiments, Ribo-Zero columns (Epicentre Biotechnologies) were used to reduce ribosomal RNA abundance instead of oligo dT beads, and barcodes were used to multiplex 3 samples per lane (paired-end 2 × 36 by Illumina HiSeq 2000).
CLIP-Seq library preparation and sequencing
CLIP was performed using 254 nm UV irradation as previously described (Wang et al., 2009) using whole brain, heart, and muscle, or C2C12 myoblasts. Minor modifications to the protocol can be found in Extended Experimental Procedures.
Immunoprecipitation was performed using a polyclonal antibody which does not exhibit cross-reactivity with Mbnl2 (personal communication, Tom Cooper).
Cellular fractionation for C2C12 mouse myoblasts
Extraction of cellular compartments from C2C12 monolayers was performed using cytosolic lysis buffer with 0.015% digitonin, membrane lysis buffer with 0.5% Triton X-100, and cytoskeletal lysis buffer with 1 M NaCl. Cells were washed with PBS between each extraction. Protein and RNA from each compartment was prepared by heating in SDS page buffer or by guanidum isothiocyanate, phenol/chloroform, and RNeasy mini columns, respectively.
Cellular fractionation for fly S-2R+ cells
S-2R+ cells were fractionated using hypotonic lysis and ultracentrifugation. Cells were lysed in hypotonic buffer (20mM Tris HCl pH 7.5, 20mM NaCl, 5mM EGTA, 1mM EDTA, 1mM DTT, 1mM PMSF, 0.15 units/mL aprotinin, 20µM leupeptin, and 40 units/mL RNAse-Out (Invitrogen)), homogenized by dounce, and spun for 10 minutes at 1000g at 4 degrees to clear nuclear and cell debris. The lysate was spun to obtain the cytosolic fraction as the supernatant. The remaining pellet was resuspended in hypotonic buffer containing 1% Triton X-100, homogenized, and spun again to obtain a membrane fraction as the supernatant, and a insoluble fraction as the pellet. RNA was extracted using Trizol (Invitrogen).
Ribosome footprinting
Ribosome footprinting of C2C12 mouse myoblasts was performed as previously described (Ingolia et al., 2011), with several modifications. Instead of isolating ribosomes from polysome gradients, ribosomes were isolated by pelleting through a sucrose cushion. Further details can be found in Extended Experimental Procedures.
Immunoblotting
Bis-tris SDS-PAGE gels (Invitrogen) were used to separate proteins by molecular weight, and protein was transferred to nitrocellulose using the iBlot transfer apparatus (Invitrogen). Nitrocellulose blots were blocked in 5% milk-PBS with 0.1% Tween-20. Mouse antibodies against Mbnl1 and Mbnl2, MB1a and MB2a, respectively, were kind gifts from the Wolffson Center for Inherited Neuromuscular Disease, and used at 1:300 dilutions. Rabbit Upf1 antibody (Bethyl Laboratories) was used at 1:10K. Antibodies against Hsp90 (mouse), Calnexin (rabbit), and Tropomyosin I (mouse) were used at dilutions of 1:1000. SuperSignal West Femto ECL reagent (ThermoScientific) was used to detect activity of HRP-conjugated anti-mouse and anti-rabbit secondary antibodies.
Cell culture, cloning, and luciferase assays
C2C12 mouse myoblasts were cultured in high glucose DMEM supplemented with 20% FBS and penicillin/streptomycin at 37 degrees and 5% CO2. Knockdowns of Mbnl1 and Mbnl2 in mouse myoblasts were performed via polybrene-assisted infection of lentivirus derived from the pLKO.1 vector, followed by puromycin selection starting 24 hours after infection. RNA and/or protein were isolated from C2C12 mouse myoblasts 7 to 10 days after infection.
Fly S-2R+ cells were cultured in 1X Schneider’s Drosophila media (Lonza) supplemented with 10% FBS, 50 units/mL penicillin, and 50 µg/mL streptomycin at room temperature. DsRNA synthesis and knockdown in S-2R+ cells were performed as described previously (Clemens et al., 2000). DsRNAs were used to knock down cells on days 1, 3, and 4, and cells were harvested on day 5.
The Infusion system (Clontech) was used to clone fragments of Bgn and Fn1 into the 3′ of Gaussia luciferase (pCMV-Gluc, New England Biolabs N8081). The Fn1 fragment contained the last intron of Fn1 in addition to the 3′ UTR. Liposome mixtures of 400 ng plasmid encoding Gaussia luciferase, 100 ng plasmid encoding Cypridina luciferase, and either 100 ng of a control (pLKO.1) plasmid, 100 ng of a plasmid containing 960 CUG repeats (DT960), or 100 ng DT960 plus 100ng of a plasmid encoding the Mbnl1 coding sequence were prepared using Trans-IT (Mirus). The liposome mixtures were added to C2C12 cells in 12-well plates, and luciferase assays were performed 24 hours later. Luciferase assays (Targeting Systems) were used to assay Gaussia and Cypridina activity. Significance between means was computed using the Mann-Whitney U test.
Computational analysis of RNA-Seq, CLIP-Seq, and ribosome footprinting data
RNA-Seq, CLIP-Seq and ribosome footpringint data have been submitted to GEO (accession number pending). Computational and statistical methods used to analyze short read data are described in Extended Experimental Procedures.
Supplementary Material
Highlights.
A nucleotide-resolution RNA map of the splicing regulatory activity of Mbnl1
Functional interchangeability of Mbnl1 and Mbnl2 in splicing regulation
A global role for Mbnls in regulating mRNA localization in mouse and fly
Mbnl-dependent regulation of translation and protein secretion of Mbnl-bound mRNAs
Acknowledgements
We thank Tom Cooper for the anti-Mbnl1 antibody used to perform CLIP. We thank Andy Berglund and David Brook for insightful conversations about Muscleblinds, and Phil Sharp and Bob Brown, Jr. for their comments and suggestions to perform cellular fractionation experiments. We thank the MIT Biomicro Center and the IRCM molecular biology core facility assistance with deep sequencing, and the Koch Institute Swanson Biotechnology Center for microscopy assistance. This work was supported by a Poitras Fellowship (E. T. W.), by grant no. 0821391 from the National Science Foundation, and by grants from the National Institutes of Health to C. B. B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
E.T.W. performed CLIP-Seq, cellular fractionation of mouse cells, ribosome footprint profiling and luciferase assays, and analyzed RNA-Seq, CLIP-Seq and footprint data. N.C. performed cellular fractionation of fly cells. S.J., M.B., and S.R. isolated RNA from mouse tissues and performed splicing RT-PCR assays. T.T.W. performed lentiviral knockdown of Mbnls in mouse cells. D.T. and S.L. prepared RNA-Seq libraries. N.C., E.L., S.R., D.H., and G.S. contributed to study design. E.T.W. and C.B.B. designed the study and wrote the paper with contributions from E.L. and N.C.
Supplementary Information
Figures S1–S7
Tables S1–S5
Extended Experimental Procedures
Supplementary Figure Legends
References
- Adereth Y, Dammai V, Kose N, Li R, Hsu T. RNA-dependent integrin alpha3 protein localization regulated by the Muscleblind-like protein MLP1. Nat Cell Biol. 2005;7:1240–1247. doi: 10.1038/ncb1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amenta AR, Creely HE, Mercado MT, Hagiwara H, McKechnie BA, et al. Biglycan is an extracellular MuSK binding protein important for synapse stability. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:2324–2334. doi: 10.1523/JNEUROSCI.4610-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3'UTR ends. Trends Cell Biol. 2009;19:465–474. doi: 10.1016/j.tcb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Artero R, Prokop A, Paricio N, Begemann G, Pueyo I, Mlodzik M, Perez-Alonso M, Baylies MK. The muscleblind gene participates in the organization of Z-bands and epidermal attachments of Drosophila muscles and is regulated by Dmef2. Dev Biol. 1998;195:131–143. doi: 10.1006/dbio.1997.8833. [DOI] [PubMed] [Google Scholar]
- Ashizawa T, Dubel JR, Harati Y. Somatic instability of CTG repeat in myotonic dystrophy. Neurology. 1993;43:2674–2678. doi: 10.1212/wnl.43.12.2674. [DOI] [PubMed] [Google Scholar]
- Begemann G, Paricio N, Artero R, Kiss I, Pérez-Alonso M, Mlodzik M. muscleblind, a gene required for photoreceptor differentiation in Drosophila, encodes novel nuclear Cys3His-type zinc-finger-containing proteins. Development. 1997;124:4321–4331. doi: 10.1242/dev.124.21.4321. [DOI] [PubMed] [Google Scholar]
- Bergeron JJ, Brenner MB, Thomas DY, Williams DB. Calnexin: a membrane-bound chaperone of the endoplasmic reticulum. Trends Biochem Sci. 1994;19:124–128. doi: 10.1016/0968-0004(94)90205-4. [DOI] [PubMed] [Google Scholar]
- Besse F, Ephrussi A. Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat Rev Mol Cell Biol. 2008;9:971–980. doi: 10.1038/nrm2548. [DOI] [PubMed] [Google Scholar]
- Blobel G, Dobberstein B. Transfer of proteins across membranesIPresence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol. 1975a;67:835–851. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G, Dobberstein B. Transfer of proteins across membranes II. Reconstitution of functional rough microsomes from heterologous components. J Cell Biol. 1975b;67:852–862. doi: 10.1083/jcb.67.3.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenik C, Chua HN, Zhang H, Tarnawsky SP, Akef A, Derti A, Tasan M, Moore MJ, Palazzo AF, Roth FP. Genome analysis reveals interplay between 5'UTR introns and nuclear mRNA export for secretory and mitochondrial genes. PLoS Genet. 2011;7:e1001366. doi: 10.1371/journal.pgen.1001366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci U S A. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csermely P, Schnaider T, Soti C, Prohászka Z, Nardai G. The 90-kDa molecular chaperone family: structure, function, and clinical applications. A comprehensive review. Pharmacol Ther. 1998;79:129–168. doi: 10.1016/s0163-7258(98)00013-8. [DOI] [PubMed] [Google Scholar]
- Du H, Cline MS, Osborne RJ, Tuttle DL, Clark TA, Donohue JP, Hall MP, Shiue L, Swanson MS, Thornton CA, et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol. 2010;17:187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS, Brook JD. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum Mol Genet. 2002;11:805–814. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- Ffrench-Constant C, Hynes RO. Alternative splicing of fibronectin is temporally and spatially regulated in the chicken embryo. Development. 1989;106:375–388. doi: 10.1242/dev.106.2.375. [DOI] [PubMed] [Google Scholar]
- Ffrench-Constant C, Van de Water L, Dvorak HF, Hynes RO. Reappearance of an embryonic pattern of fibronectin splicing during wound healing in the adult rat. J Cell Biol. 1989;109:903–914. doi: 10.1083/jcb.109.2.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goers ES, Purcell J, Voelker RB, Gates DP, Berglund JA. MBNL1 binds GC motifs embedded in pyrimidines to regulate alternative splicing. Nucleic Acids Res. 2010;38:2467–2484. doi: 10.1093/nar/gkp1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning PW, Schevzov G, Kee AJ, Hardeman EC. Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol. 2005;15:333–341. doi: 10.1016/j.tcb.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Harper PS. Myotonic dystrophy. Vol 9. Philadelphia: Saunders; 1979. [Google Scholar]
- Hesketh J. Translation and the cytoskeleton: a mechanism for targeted protein synthesis. Mol Biol Rep. 1994;19:233–243. doi: 10.1007/BF00986965. [DOI] [PubMed] [Google Scholar]
- Hino S-I, Kondo S, Sekiya H, Saito A, Kanemoto S, Murakami T, Chihara K, Aoki Y, Nakamori M, Takahashi MP, et al. Molecular mechanisms responsible for aberrant splicing of SERCA1 in myotonic dystrophy type 1. Hum Mol Genet. 2007;16:2834–2843. doi: 10.1093/hmg/ddm239. [DOI] [PubMed] [Google Scholar]
- Ho TH, Charlet BN, Poulos MG, Singh G, Swanson MS, Cooper TA. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt I, Jacquemin V, Fardaei M, Sewry CA, Butler-Browne GS, Furling D, Brook JD, Morris GE. Muscleblind-like proteins: similarities and differences in normal and myotonic dystrophy muscle. Am J Pathol. 2009;174:216–227. doi: 10.2353/ajpath.2009.080520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne-Badovinac S, Bilder D. Dynein regulates epithelial polarity and the apical localization of stardust A mRNA. PLoS Genet. 2008;4:e8. doi: 10.1371/journal.pgen.0040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan S, Nwosu C, Nicchitta CV. Analyzing mRNA localization to the endoplasmic reticulum via cell fractionation. Methods Mol Biol. 2011;714:301–321. doi: 10.1007/978-1-61779-005-8_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscleblind knockout model for myotonic dystrophy. Science. 2003a;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- Kanadia RN, Urbinati CR, Crusselle VJ, Luo D, Lee Y-J, Harrison JK, Oh SP, Swanson MS. Developmental expression of mouse muscleblind genes Mbnl1, Mbnl2 and Mbnl3. Gene Expr Patterns. 2003b;3:459–462. doi: 10.1016/s1567-133x(03)00064-4. [DOI] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore S, Jaskiewicz L, Burger L, Hausser J, Khorshid M, Zavolan M. A quantitative analysis of CLIP methods for identifying binding sites of RNAbinding proteins. Nat Methods. 2011;8:559–564. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
- Kislauskis EH, Zhu X, Singer RH. beta-Actin messenger RNA localization and protein synthesis augment cell motility. J Cell Biol. 1997;136:1263–1270. doi: 10.1083/jcb.136.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Krause HM. Global implications of mRNA localization pathways in cellular organization. Curr Opin Cell Biol. 2009;21:409–415. doi: 10.1016/j.ceb.2009.01.027. [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Lin X, Miller JW, Mankodi A, Kanadia RN, Yuan Y, Moxley RT, Swanson MS, Thornton CA. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum Mol Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- Marteyn A, Maury Y, Gauthier MM, Lecuyer C, Vernet R, Denis JA, Pietu G, Peschanski M, Martinat C. Mutant human embryonic stem cells reveal neurite and synapse formation defects in type 1 myotonic dystrophy. Cell stem cell. 2011;8:434–444. doi: 10.1016/j.stem.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KC, Zukin RS. RNA trafficking and local protein synthesis in dendrites: an overview. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2006;26:7131–7134. doi: 10.1523/JNEUROSCI.1801-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda A, Andersen HS, Doktor TK, Okamoto T, Ito M, Andresen BS, Ohno K. CUGBP1 and MBNL1 preferentially bind to 3′ UTRs and facilitate mRNA decay. Sci Rep. 2012;2 doi: 10.1038/srep00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Osborne RJ, Lin X, Welle S, Sobczak K, O'Rourke JR, Swanson MS, Thornton CA. Transcriptional and post-transcriptional impact of toxic RNA in myotonic dystrophy. Hum Mol Genet. 2009;18:1471–1481. doi: 10.1093/hmg/ddp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachter JS. Association of mRNA with the cytoskeletal framework: its role in the regulation of gene expression. Crit Rev Eukaryot Gene Expr. 1992;2:1–18. [PubMed] [Google Scholar]
- Palazzo AF, Springer M, Shibata Y, Lee C-S, Dias AP, Rapoport TA. The signal sequence coding region promotes nuclear export of mRNA. PLoS Biol. 2007;5:e322. doi: 10.1371/journal.pbio.0050322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaite PA, Gantelet E, Kraftsik R, Gourdon G, Kuntzer T, Barakat-Walter I. Myotonic dystrophy transgenic mice exhibit pathologic abnormalities in diaphragm neuromuscular junctions and phrenic nerves. Journal of neuropathology and experimental neurology. 2008;67:763–772. doi: 10.1097/NEN.0b013e318180ec64. [DOI] [PubMed] [Google Scholar]
- Pascual M, Vicente M, Monferrer L, Artero R. The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing. Differentiation; research in biological diversity. 2006;74:65–80. doi: 10.1111/j.1432-0436.2006.00060.x. [DOI] [PubMed] [Google Scholar]
- Pizon V, Iakovenko A, Van Der Ven PF, Kelly R, Fatu C, Furst DO, Karsenti E, Gautel M. Transient association of titin and myosin with microtubules in nascent myofibrils directed by the MURF2 RING-finger protein. Journal of cell science. 2002;115:4469–4482. doi: 10.1242/jcs.00131. [DOI] [PubMed] [Google Scholar]
- Pyhtila B, Zheng T, Lager PJ, Keene JD, Reedy MC, Nicchitta CV. Signal sequence- and translation-independent mRNA localization to the endoplasmic reticulum. RNA. 2008;14:445–453. doi: 10.1261/rna.721108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsköld D, Wang ET, Burge CB, Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol. 2009;5:e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S, Talukdar I, Liu Y, Tam J, Reddy S, Webster NJ. Muscleblindlike 1 (Mbnl1) promotes insulin receptor exon 11 inclusion via binding to a downstream evolutionarily conserved intronic enhancer. J Biol Chem. 2010;285:25426–25437. doi: 10.1074/jbc.M109.095224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilker KA, Wang GJ, Tugizova MS, Shen K. Celegans Muscleblind homolog mbl-1 functions in neurons to regulate synapse formation. Neural development. 2012;7:7. doi: 10.1186/1749-8104-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squillace RM, Chenault DM, Wang EH. Inhibition of muscle differentiation by the novel muscleblind-related protein CHCR. Dev Biol. 2002;250:218–230. doi: 10.1006/dbio.2002.0798. [DOI] [PubMed] [Google Scholar]
- Sundell CL, Singer RH. Requirement of microfilaments in sorting of actin messenger RNA. Science. 1991;253:1275–1277. doi: 10.1126/science.1891715. [DOI] [PubMed] [Google Scholar]
- Teplova M, Patel DJ. Structural insights into RNA recognition by the alternative-splicing regulator muscleblind-like MBNL1. Nat Struct Mol Biol. 2008;15:1343–1351. doi: 10.1038/nsmb.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Vedeler A, Pryme IF, Hesketh JE. The characterization of free, cytoskeletal and membrane-bound polysomes in Krebs II ascites and 3T3 cells. Mol Cell Biochem. 1991;100:183–193. doi: 10.1007/BF00234167. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Tollervey J, Briese M, Turner D, Ule J. CLIP: construction of cDNA libraries for high-throughput sequencing from RNAs cross-linked to proteins in vivo. Methods. 2009;48:287–293. doi: 10.1016/j.ymeth.2009.02.021. [DOI] [PubMed] [Google Scholar]
- Wang Z, Xiao X, Van Nostrand E, Burge CB. General and specific functions of exonic splicing silencers in splicing control. Mol Cell. 2006;23:61–70. doi: 10.1016/j.molcel.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf MB, Berglund JA. MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. RNA. 2007;13:2238–2251. doi: 10.1261/rna.610607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu X-D, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Frias MA, Mele A, Ruggiu M, Eom T, Marney CB, Wang H, Licatalosi DD, Fak JJ, Darnell RB. Integrative modeling defines the Nova splicing-regulatory network and its combinatorial controls. Science. 2010;329:439–443. doi: 10.1126/science.1191150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C. Formation of mRNA 3' ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.