Abstract
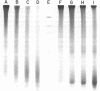
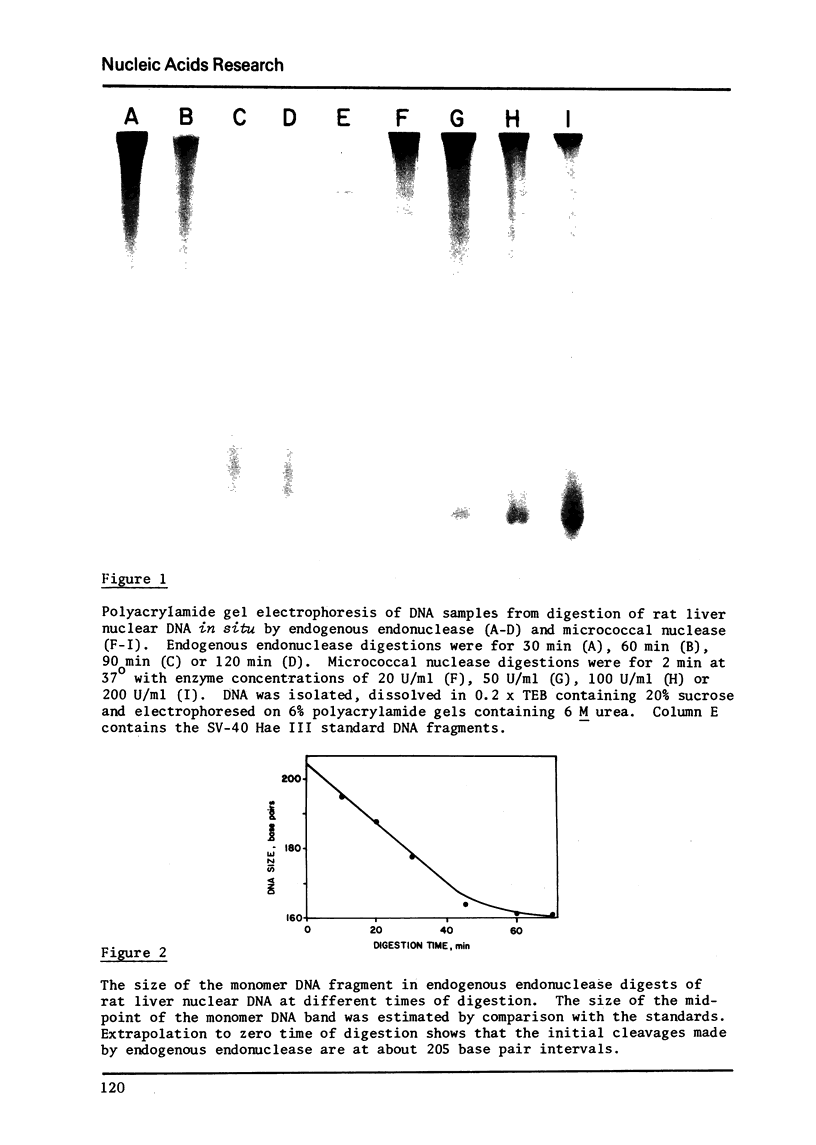
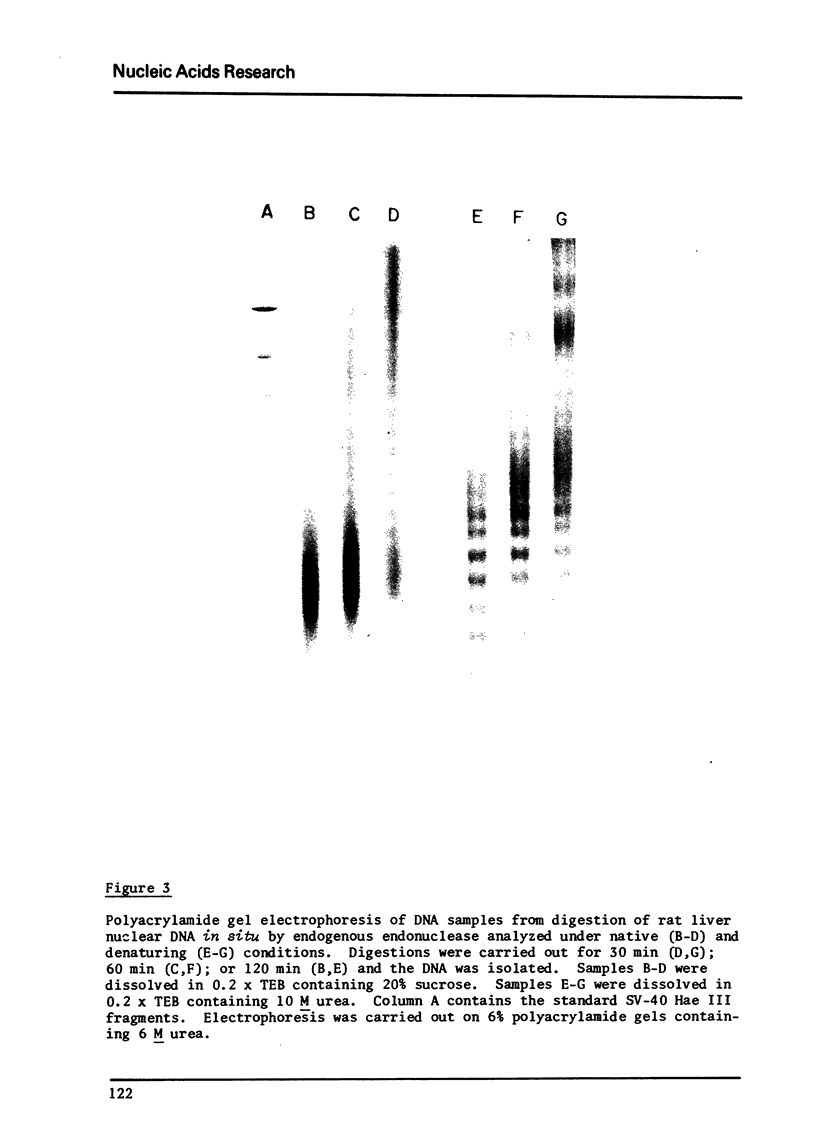
Digestion of rat liver nuclei by an endogenous endonuclease generates double-stranded DNA fragments which are initially about 205 base pairs long, as reported previously by Hewish and Burgoyne. As digestion proceeds, the average size of these fragments is reduced to about 160 base pairs. Electrophoresis under denaturing conditions shows that these DNA fragments contain single strand nicks at ten base intervals. Fifteen bands, 10-150 bases, are clearly resolvable. DNA Fragments of 160 to 200 nucleotides are not resolved as distinct species. The results suggest that the chromosomal subunit contains both a 160 base-pair DNA segment, in a conformation susceptible to single strand nicking at ten base intervals, and a forty base-pair DNA segment in a conformation more uniformly susceptible to endogenous endonuclease activity. This chemical evidence agrees with morphological observations suggesting that chromatin has a "bead and bridge" structure.
Full text
PDF



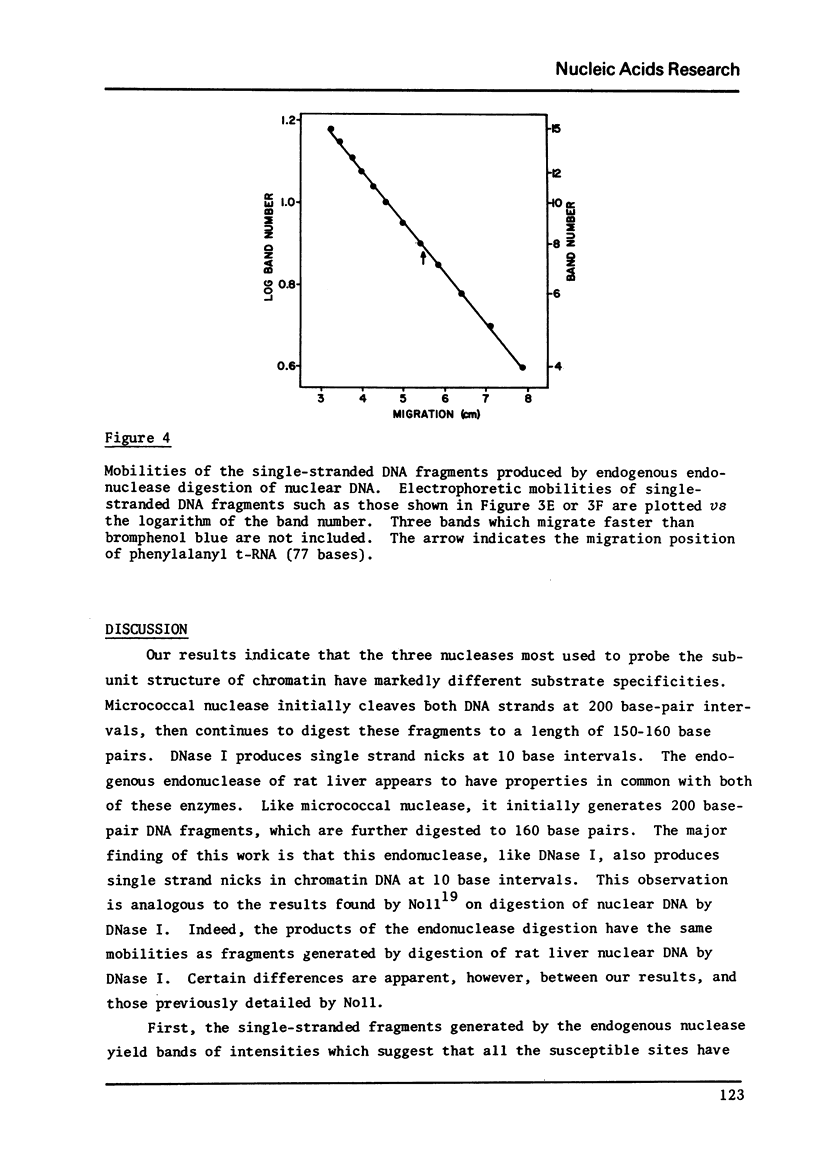
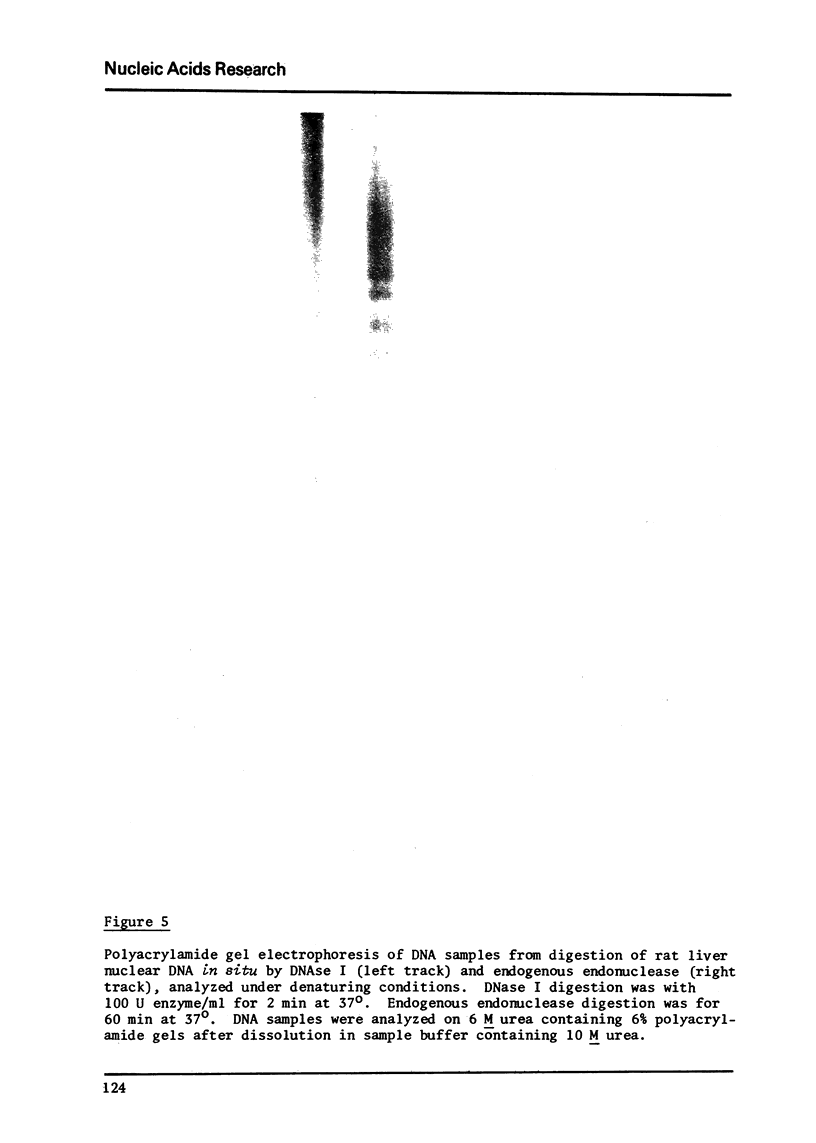
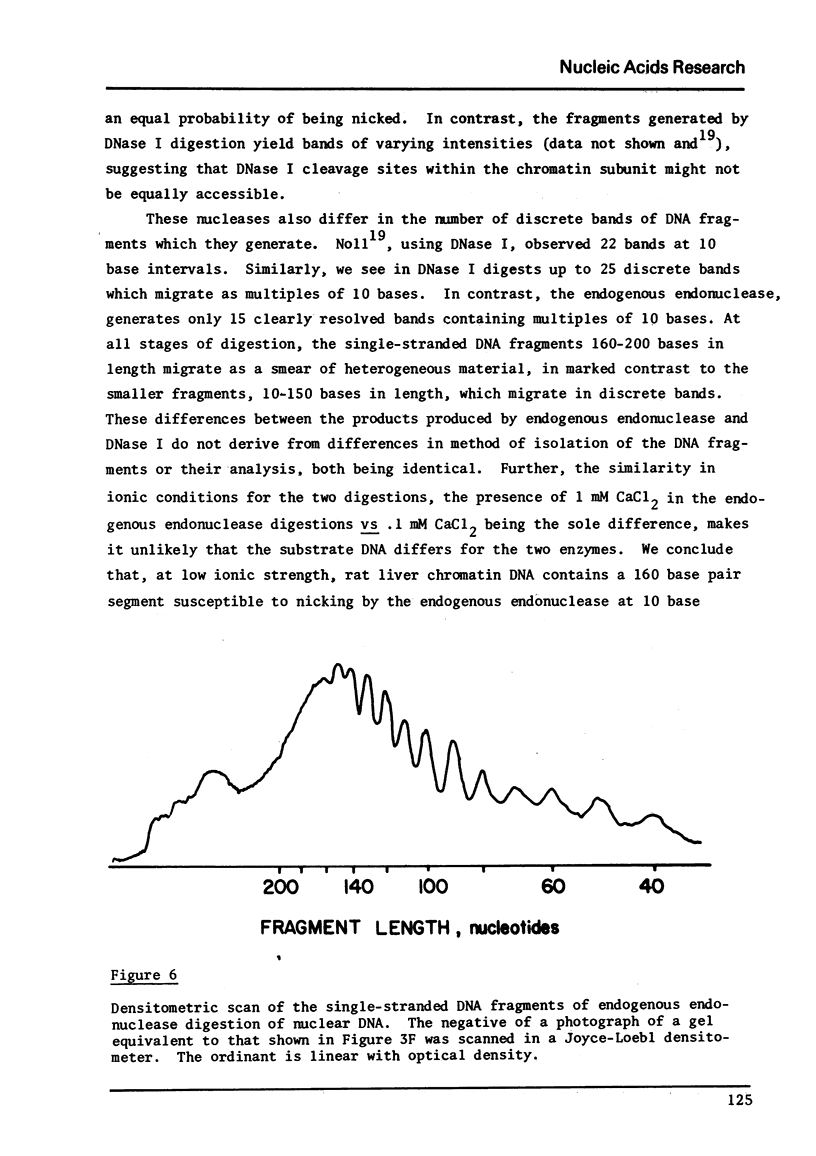


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Axel R. Cleavage of DNA in nuclei and chromatin with staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2921–2925. doi: 10.1021/bi00684a020. [DOI] [PubMed] [Google Scholar]
- Burgoyne L. A., Hewish D. R., Mobbs J. Mammalian chromatin substructure studies with the calcium-magnesium endonuclease and two-dimensional polyacrylamide-gel electrophoresis. Biochem J. 1974 Oct;143(1):67–72. doi: 10.1042/bj1430067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J. D. Chromatin structure: deduced from a minichromosome. Science. 1975 Mar 28;187(4182):1202–1203. doi: 10.1126/science.187.4182.1202. [DOI] [PubMed] [Google Scholar]
- HYMER W. C., KUFF E. L. ISOLATION OF NUCLEI FROM MAMMALIAN TISSUES THROUGH THE USE OF TRITON X-100. J Histochem Cytochem. 1964 May;12:359–363. doi: 10.1177/12.5.359. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Hyde J. E., Walker I. O. A model for chromatin sub-structure incorporating symmetry considerations of histone oligomers. Nucleic Acids Res. 1975 Mar;2(3):405–421. doi: 10.1093/nar/2.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebowitz P., Siegel W., Sklar J. Hemophilus aegyptius restriction edonuclease cleavage map of the simian virus 40 genome and its colinear relation with the hemophilus influenzae cleavage map of SV40. J Mol Biol. 1974 Sep 5;88(1):105–123. doi: 10.1016/0022-2836(74)90297-6. [DOI] [PubMed] [Google Scholar]
- Li H. J. A model for chromatin structure. Nucleic Acids Res. 1975 Aug;2(8):1275–1289. doi: 10.1093/nar/2.8.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M. Internal structure of the chromatin subunit. Nucleic Acids Res. 1974 Nov;1(11):1573–1578. doi: 10.1093/nar/1.11.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M. Subunit structure of chromatin. Nature. 1974 Sep 20;251(5472):249–251. doi: 10.1038/251249a0. [DOI] [PubMed] [Google Scholar]
- Noll M., Thomas J. O., Kornberg R. D. Preparation of native chromatin and damage caused by shearing. Science. 1975 Mar 28;187(4182):1203–1206. doi: 10.1126/science.187.4182.1203. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Spheroid chromatin units (v bodies). Science. 1974 Jan 25;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Resolution of multiple ribonucleic acid species by polyacrylamide gel electrophoresis. Biochemistry. 1967 Jun;6(6):1818–1827. doi: 10.1021/bi00858a033. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Felsenfeld G. A comparison of the digestion of nuclei and chromatin by staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2915–2920. doi: 10.1021/bi00684a019. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Holde K. E., Sahasrabuddhe C. G., Shaw B. R. A model for particulate structure in chromatin. Nucleic Acids Res. 1974 Nov;1(11):1579–1586. doi: 10.1093/nar/1.11.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bruggen E. F., Arnberg A. C., van Holde K. E., Sahasrabuddhe C. G., Shaw B. R. Electron microscopy of chromatin subunit particles. Biochem Biophys Res Commun. 1974 Oct 23;60(4):1365–1370. doi: 10.1016/0006-291x(74)90348-9. [DOI] [PubMed] [Google Scholar]