Abstract
We report on a 17-year-old patient with midline defects, ocular hypertelorism, neuropsychomotor development delay, neonatal macrosomy, and dental anomalies. DNA copy number investigations using a Whole Genome TilePath array consisting, of 30K BAC/PAC clones showed a 6.36 Mb deletion in the 9p24.1–p24.3 region and a 14.83 Mb duplication in the 20p12.1–p13 region, which derived from a maternal balanced t(9;20)(p24.1;p12.1) as shown by FISH studies. Monosomy 9p is a well-delineated chromosomal syndrome with characteristic clinical features, while chromosome 20p duplication is a rare genetic condition. Only a handful of cases of monosomy 9/trisomy 20 have been previously described. In this report, we compare the phenotype of our patient with those already reported in the literature, and discuss the role of DMRT, DOCK8, FOXD4, VLDLR, RSPO4, AVP, RASSF2, PROKR2, BMP2, MKKS, and JAG1, all genes mapping to the deleted and duplicated regions.
Keywords: multiple congenital anomalies, midline, chromosomal aberration, array-CGH, monosomy 9p, trisomy 20p
INTRODUCTION
Midline facial defects with hypertelorism (MFDH) constitute a rare and heterogeneous group of craniofacial disorders mainly characterized by ocular hypertelorism and bifid nose [Gil-da-Silva-Lopes, 1997; Guion-Almeida, 2000; Gil-da-Silva-Lopes and Maciel-Guerra, 2007]. These features are part of the frontonasal dysplasia spectrum, which is clinically and etiologically heterogeneous [Gil-da-Silva-Lopes and Maciel-Guerra, 2007; Wu et al., 2007]. Due to its rarity, few reports of MFDH are found in the literature, so the pathogenesis of these conditions is mostly unknown.
Array-based comparative genomic hybridization (Array-CGH) enables a high-resolution screening of DNA copy number abnormalities in the genome, and has become an increasingly important tool to detect copy number losses and gains in patients with an abnormal phenotype.
Very few descriptions of chromosomal abnormalities in MFDH have been described [Chen, 1994; Stevens and Qumsiyeh, 1995], but in those cases reported, chromosomes 2, 3, 7, 11, and 15 have been implicated in this disorder. Beyond these, there are two isolated reports of MFDH in which 22q11.2 deletions were detected [Stratton and Payne, 1997; Kirkpatrick and Pauli, 1998].
In this case report, we characterized a cryptic chromosomal rearrangement in a MFDH patient with partial monosomy 9p and a partial trisomy 20p derived from a maternal balanced translocation t(9;20)(p24.1;p12.1). Although 9p deletion is a well-recognized syndrome with several cases described [Swinkels et al., 2008], very few cases of 20p duplication have been described [Sidwell et al., 2000; Thomas et al., 2004; Venditti et al., 2004; Chaabouni et al., 2007]. Only a few patients with concomitant partial monosomy 9p and partial trisomy 20p have been reported to date [Hoo et al., 1982; Hauge et al., 2008; DECIPHER cases 1578 and 250016].
CLINICAL REPORT
The propositus is the first child of healthy unrelated parents with an unremarkable family history. At the time of delivery the mother was 26-years-old and the father was 29-years-old. The pregnancy was complicated by eclampsia. There was no history of maternal diabetes. The patient was born preterm at 36-week gestation. At birth, weight was 3.55 kg and length was 55 cm (both above the 97th centile). Developmental milestones were delayed. He walked unsupported at 20 months, while first words occurred at 3 years, with poor speech progression. He underwent surgery for unilateral inguinal hernia repair.
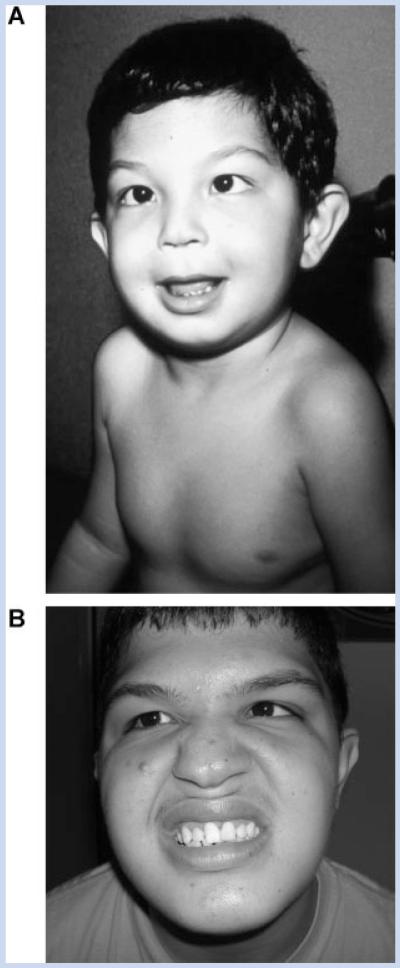
The patient was first evaluated at 3 years of age. Clinical examination disclosed a height of 110 cm (above 97th centile), weight of 22.2 kg (above 97th centile) and OFC 52 cm (97th centile). Physical exam included facial asymmetry, ocular hypertelorism, prominent ears, eyelid ptosis, epicanthal folds, wide nasal bridge, bifid nasal tip, broad columella, bifid uvula, long and flat philtrum, and high palate (Fig. 1A). Dental problems, such as irregular teeth were also noted. Other clinical signs included wide spaced nipples, umbilical hernia, shawl scrotum, wide space between halux and second toes, and hypospadias, which was surgically corrected. Ophthalmological examinations demonstrated hyperopia and convergent strabismus on the left. Audiometry, cardiac, and abdominal ultrasounds were normal. Computerized tomography of the brain was normal.
FIG. 1.
Clinical appearance of the present patient at different ages. A: In first evaluation at age of 3 years. B: In second evaluation at age 17 years of age. Note the facial asymmetry, ocular hypertelorism, wide nasal bridge, and the strabismus.
Conventional cytogenetic analysis with G-banded chromosomes, with a resolution of approximately 400 bands level, was normal (46,XY) and FISH was performed on cultured lymphocytes, with the use of a probe for the 22q11.2 region, the HIRA/TUPLE1 gene locus (Vysis®, Downers Groove, IL) and no deletion was detected [Simioni et al., 2010].
At the second examination at 17 years of age (Fig. 1B), height was 176.7 cm (50th–85th centiles), weight 77.2 kg (90th–97th centiles), and OFC 58 cm (relative macrocephaly). He had moderate mental impairment (he was independent in his daily activities, and had some ability to read and write, but had cognitive impairment). The clinical features were unchanged, except for a normal permanent dentition. His puberty started around 12 years. He had normal male genitalia, with right testicle at 15 cm3 (normal) and the left one was reduced in volume (10 cm3); pubic hair stage according to Tanner was P4.
The results of endocrinological investigations were: pretreatment value of TSH was 2.88 mUI/L (3.8–4.5 mUI/L); FT3 was not performed; thyroid autoantibodies were not present. Thyroid ultrasound was normal. He was diagnosed with subclinical hypothyroidism and started therapy with sodic levothyroxin. Follow-up values were: TSH was 3.52 mUI/L (3.8–4.5 mUI/L); Free T4: 0.21 ng/dl (0.8–2.3 ng/dl); FT3 was not performed. Testosterone: 440 ng/dl (270–1070 ng/dl); basal cortisol: 11.1 mcg/dl (5–25 mcg/dl); LH: 2.04 mUI/L (2–12 mUI/L); FSH: 5.49 mUI/ml (0.2–12.8 mUI/ml). LHRH stimulation test was normal. Despite of the volumetric reduction of left testicle, no gonadal dysfunction was detected.
MATERIALS AND METHODS
Genomic DNA was extracted from peripheral blood according to previously described protocols [Araújo et al., 1996]. Array-CGH investigation was carried out using the Sanger 30K Whole Genome TilePath (WGTP) array (NCBI build 36) as described by Fiegler et al. [2006]. All copy number changes observed were compared to copy number variants (CNVs) reported in previous studies of normal populations documented on the Database of Genomic Variants (DGV). Chromosome regions not previously reported as CNVs and comprising at least two clones were singled out for further investigations, which included confirmatory studies by FISH and parent-of-origin analysis. Routine G-banding chromosome and FISH analyses were performed on metaphase preparations of peripheral blood lymphocytes from the patient and his parents using standard techniques. BAC probes for FISH were selected according to mapping data from the Ensembl browser (NCBI build 36, HG 18). BAC DNAs were extracted using Wizard® Plus SV Minipreps DNA Purification System (Promega Corporation, Madison, WI) and labeled by Nick Translation Mix (Roche Applied Science, Indianapolis, IN) with Fluorescein-12-dUTP or Tetramethil-rhodamine-6-dUTP.
RESULTS
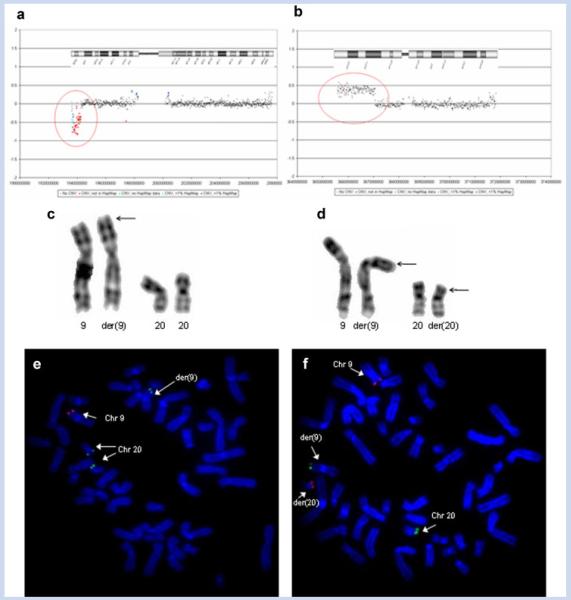
We performed array-CGH experiments using a 30K WGTP array from the Sanger Institute. The analysis identified a chromosome 9p terminal deletion of 6.36 Mb, and a chromosome 20p terminal duplication of 14.8 Mb (Fig. 2a,b). On chromosome 9 the breakpoint mapped to 9p24.1. The chromosome 20 duplication breakpoint mapped to 20p12.1. At the time of first evaluation, the karyotype at the resolution of 400 bands, was considered normal. In view of the large deleted and duplicated segments disclosed by array-CGH, we performed a new chromosome analysis that revealed an abnormal chromosome 9 in the patient, with additional material on the short arm (Fig. 2c). The chromosome analyses of the parents showed the abnormality to be maternally derived [46,XX,t(9;20)(p24;p12)] (Fig. 2d). The patient inherited his mother’s derived copy of chromosome 9 and her normal copy of chromosome 20. Thus, his karyotype was established as 46,XY,der-(9)t(9;20)(p24;p12)mat (Fig. 2c). Subsequent FISH analyses using the RP11-48M17 clone mapping to chromosome 9 (9: 2147378–2305356) and RP5-852M4 mapping to chromosome 20 (20: 379137–437563) confirmed the rearrangement in the patient and his mother (Fig. 2e,f).
FIG. 2.
a: WGTP array profiles of the present patient. Chromosome 9 profile: The WGTP array identified a deletion on chromosome 9. The position of the deletion is depicted on the chromosome 9 ideogram. b: Chromosome 20 profile. The WGTP array identified a duplication on chromosome 20. The position of the duplication is depicted on chromosome 20 ideogram. c: Partial karyotype of the patient showing the normal and the derivative chromosomes 9 (arrow), and the normal chromosomes 20. d: Partial karyotype of the patient’s mother showing the normal and the derivative chromosomes 9 and 20. Arrows point to the derivative chromosomes. Note a reciprocal translocation between the terminal portion of the p arms of chromosomes 9 and 20. e: FISH analysis on metaphase spreads of the patient with BAC RP11-48M17 (labeled in red; 9p24.2-24.3) and PAC RP5-852M4 (labeled in green; 20p13) probes, confirming the aberration. f: FISH analysis on metaphase spreads of the patient’s mother with the same probes, confirming the balanced translocation between chromosomes 9 and 20.
DISCUSSION
Array-CGH investigation in the patient identified a deletion of chromosome 9p24.1–p24.3 and a duplication of chromosome 20p12.1–p13. To our knowledge, at least five other patients with concomitant partial monosomy 9p and partial trisomy 20p have been reported [Hoo et al., 1982; Hauge et al., 2008; DECIPHER cases 1578 and 250016 https://decipher.sanger.ac.uk]. However, no clear genotype–phenotype correlation could be established because for most reported patients, their clinical features were not described in sufficient detail. We have accessed the detailed clinical data from only patient 1578 reported in the DECIPHER database. This patient shares some dysmorphic features and clinical phenotypic characteristics with the patient presented here, such as developmental, speech and motor delay, low-set/malformed ears, ocular hypertelorism, eyelid ptosis, and long philtrum (Table I).
TABLE I. Clinical Features of the Present Case and Other Patients With Similar Chromosome Alterations.
| Present case |
DECIPHER case 1578 |
DECIPHER case 250016 |
Case 8 from Hauge et al. [2008] |
Case 9 from Hauge et al. [2008] |
Five affected individuals from a family [Hoo et al., 1982] |
9p Deletiona (1, 2, 3, 4) |
20p Duplicationb (4, 5) |
|
|---|---|---|---|---|---|---|---|---|
| Deletion sizes at 9p | ~6.3 Mb | ~11.34 Mb | ~6.36 Mb | ~ 10.4 Mb | ~ 10.5Mb | Breakpoint at 9p23 |
||
| Duplication sizes at 20p | ~14.8 Mb | ~19.14 Mb | ~4.73 Mb | ~4.3 Mb | ~14Mb | Unkown | ||
| Sex | Male | Female | Male | Male | Male | |||
| Developmental delay | + | + | + | + | + | + | + | + |
| Speech delay | + | + | + | + | - | - | - | - |
| Motor delay | + | + | - | + | + | + | + | + |
| Trigonocephaly | - | + | - | - | + | + | + | - |
| Facial asymmetry | + | - | - | - | - | - | + | nd |
| Prominent ears | + | - | - | - | - | - | + | + |
| Low-set/malformed ears | + | + | - | - | + | - | + | + |
| Ocular hypertelorism | + | + | - | - | - | - | + | + |
| Strabismus | + | - | - | - | - | - | + | + |
| Eyelid ptosis | + | + | - | - | - | - | + | + |
| Epicanthal folds | + | - | - | - | - | - | + | + |
| Wide/flat nasal bridge | + | + | - | - | - | + | + | + |
| Bifid nasal tip | + | - | - | - | - | - | nd | nd |
| Broad columella | + | - | - | - | - | - | nd | nd |
| Long/flat philtrum | + | + | - | + | + | - | + | + |
| High/narrow palate | + | - | - | - | - | + | + | + |
| Bifid uvula | + | - | - | - | - | - | + | + |
| Hypothyroidism | + | - | - | - | - | - | nd | nd |
| Wide spaced nipples | + | - | - | - | - | - | + | + |
| Congenital heart defects | - | + | - | na | na | - | + | + |
| Umbilical hernia/onphalocele | + | - | - | - | - | - | + | + |
| Inguinal hernia | + | - | - | - | - | - | + | + |
| External genital abnormalities | + | - | - | + | - | - | + | + |
Includes multiple cases described by Swinkels et al. [2008] (1), Huret et al. [1988] (2), Alfi et al. [1973] (3), and registered at the ECARUCA (4).
Includes multiple cases described by Oppenheimer et al. [2000] (5) and registered at the ECARUCA (4); nd, not described; na, not assessed.
The present case and four others described before have been detected or characterized by array-CGH [Hauge et al., 2008; DECIPHER]. We have accessed the breakpoint data and found variability in the extent of deleted and duplicated segments on chromosomes 9 and 20, respectively (Table 1). Although the number of patients presenting the same chromosome rearrangement between 9p and 20p could suggest a shared underlying mechanism of formation, this is not supported by the breakpoint heterogeneity observed among the cases. Monosomy 9p syndrome (OMIM 158170) is a well-delineated chromosomal syndrome with cardinal features including psychomotor retardation, trigonocephaly, flat nasal bridge, long philtrum, short, broad and webbed neck, genital anomalies, and sex reversal [Alfi et al., 1973; Hoo et al., 1982; Swinkels et al., 2008].
Previous studies have aimed to characterize the critical deleted region in 9p deletion syndrome [Hauge et al., 2008; Swinkels et al., 2008]. Swinkels et al. [2008] have distinguished two different groups of patients; the first group encompassing those patients that have the clinically recognizable phenotype of deletion 9p (consensus phenotype consisting of trigonocephaly, small palpebral fissures, flat nasal bridge with anteverted nostrils, long philtrum, and micro/retrognathia), and the second group encompassing those patients who, although they have a deletion 9p, do not meet the criteria for the consensus phenotype, in particular, lacking the trigonocephaly. They mapped a critical region for the consensus phenotype to the 9p22.3 region.
However, Hauge et al. [2008] described trigonocephaly in a patient with the deletion of the distal 4 Mb of 9pter. They concluded that the minimal deleted region shared by the patients with clinically relevant phenotypes includes the distal 2 Mb of 9pter (p24.3), a critical region even more telomeric to the previously discussed region. Similarly, Barbaro et al. [2009] suggested that the mild cranial dysmorphism in patients with deletions distal to the Swinkels’ critical region could be caused either by misregulation of the candidate gene for trigonocephaly or by the deletion of other gene(s) involved in craniofacial development.
Our patient does not present with trigonocephaly, but presents with other facial dysmorphisms considered characteristic for the 9p deletion phenotype, such as flat nasal bridge and long philtrum. Considering the extent of the 9p deletion in this patient (6.3 Mb from telomere), this case supports the observations made by Hauge et al. [2008] and Barbaro et al. [2009], suggesting a more distal critical region for the cranial dysmorphisms observed in the 9p deletion syndrome.
Genes mapping within these two regions and that have been previously associated with disease are depicted in Table II. The 9p deleted region includes the candidate sex-determining genes DMRT1, 2 and 3 (Doublesex- and MAB3-Related Transcription Factor; OMIM 602424, 604935), frequently associated with gonadal dysgenesis and sex reversal [McDonald et al., 1997; Veitia et al., 1997; Smith et al., 1999; Muroya et al., 2000; Shan et al., 2000]. Considerable variability regarding impairment of male sex development in cases of 9p has been reported. Our patient had hypospadias and reduction of the left testicle. As he had a normal anterior pituitary gland axis, these features could be attributed to a preaxis dysfunction/hypofunction related to midline defects. Hypospadias is one of the most frequent genital abnormalities (8 in 1,000 live births) [Roberts and Lloyd, 1973], but the cause of the disorder is unknown in most cases. Several chromosome abnormalities have been reported to be associated with ambiguous or female genitalia in 46,XY subjects, including Xp duplications and monosomy of portions of 9p and 10q [Bennett et al., 1993; Wilkie et al., 1993; Bardoni et al., 1994], suggesting that several loci are involved in normal male sexual differentiation. In a male case with hypertelorism and hypospadias associated with a balanced translocation between 8q22.3–23 and 20p13 described by Tar et al. [1997], the authors suggested that a locus necessary for male sex differentiation was located at distal 8q. However, because our patient has hypertelorism, hypospadias, and the same region duplicated, it is conceivable that the 20p region also contribute to the genital malformation.
TABLE II. Selected Genes at 9p and 20p Previously Associated to Diseases.
| Region | Gene | Expression/subcellular localization |
Gene function | Associated disorders |
|---|---|---|---|---|
| 9p24.1p243 | DOCK8 | Several human tissues, including adult and fetal brain |
Involved in processes that affect the organization of filamentous actin, play roles in regulation of migration, morphology, adhesion, and growth of cells, and could be implicate in human cognitive function |
Mental retardation/developmental delay [OMIM 611432] |
| FOXD4 | Heart, uterus, prostate, lung, muscular tissue |
Embryonic transcriptional regulator | Severe speech and language disorder | |
| VLDLR | Heart, pituitary, testis, respiratory tract, muscular tissue |
Involved in neuroblast migration, required for proper development of the cerebral cortex and cerebellum |
VLDLR-associated cerebellar hypoplasia characterized by non-progressive congenital ataxia that is predominantly truncal and results in delayed ambulation, moderate-to-profound mental retardation, dysarthria, strabismus, and seizures |
|
| 20p12.1p13 | RSPO4 | Nervous system, testis, eye |
Play a major role in activating the Wnt/catenin signaling pathway |
Anonychia congenital in which all the fingernails and toenails are absent without significant bone anomalies [OMIM 206800] |
| AVP | Brain, kidney, epithelial tissue (endocrine) |
Having a direct antidiuretic action on the kidney, contributes to brain physiology and in the neural regulation of various behaviors, especially social behaviors |
Neurohypophyseal diabetes insipidus [OMIM 125700], hypertelorism, decreased bone mineral density |
|
| RASSF2 | Lymphoid/immune system, brain, muscular tissue |
Regulating various biologic processes, most notably cell growth, mitogenesis, and oncogenesis, through interactions with a broad range of effector proteins |
Colorectal tumorigenesis | |
| PROKR2 | Brain, epithelial tissue (endocrine) |
Involved in the mobilization of calcium, essential link for coordination of circadian behavior and physiology |
Kallmann syndrome type 3 (KAL3) [OMIM 607123] |
|
| BMP2 | Heart, nervous system (astrocyte), and skeleton system (osteoclast) |
Induce the formation of both cartilage and bone. Inhibits limb bud expansion and induces the formation of chondrocyte and osteoblast precursors |
Wolff–Parkinson–White syndrome | |
| MKKS | Lymphoid/immune, digestive, endocrine systems, brain, and eye |
Encodes a protein with sequence similarity to the chaperonin family. The encoded protein may have a role in protein processing in limb, cardiac, and reproductive systems development |
McKusick–Kaufmann syndrome (MKKS) [OMIN 236700]; Bardet-Biedl syndrome 6 (BBS6) [OMIM 209900] |
|
| JAG1 | Mouth, pharynx, endocrine and reproductive systems, ear |
Encodes a ligand for the Notch receptor and plays a role in determining cell fates in early embryonic development |
Alagille syndrome [OMIM 118450]; deafness; isolated congenital heart defects |
Other relevant genes included in the deleted region and related to mental retardation or developmental disabilities are: DOCK8 (Dedicator of Cytokinesis 8; OMIM 611432), FOXD4 (Forkhead Box D4; OMIM 601092), and VLDLR (Very Low Density Lipoprotein Receptor; OMIM 192977; Table II).
Chromosome 20p duplication is a rare genetic condition characterized by facial appearance that may include a round face with prominent cheeks, coarse and straight hair, upslanting palpebral fissures, developmental delay, and dental anomalies [Sidwell et al., 2000; Thomas et al., 2004; Venditti et al., 2004; Chaabouni et al., 2007].
Similar to our patient, most patients described with partial trisomy 20p have a rearrangement derived from a parental translocation [Taylor et al., 1976; Centerwall and Francke, 1977; Marcus et al., 1979; Funderburk et al., 1983; Zumel et al., 1989; Grammatico et al., 1992; Oppenheimer et al., 2000; Thomas et al., 2004]. Thus, correlation between chromosome 20 duplications and corresponding phenotypes is usually complicated by partial deletion of another chromosome. The genes found in the duplicated 20p12.1p13 interval may act as modifiers in this patient, contributing to the phenotype. It is possible that the genes in the region are only overexpressed, but the breakpoints of the duplication could also conceivably disrupt genes or regulatory regions. Relevant genes included in the 20p duplication region are: RSPO4 (R-Spondin Family Member 4; OMIM 610573), AVP (Arginine Vasopressin; OMIM 192340), RASSF2 (Ras Association Domain Family Protein 2; OMIM 609492), PROKR2 (Prokineticin Receptor 2; OMIM 607123), BMP2 (Bone Morphogenetic Protein 2; OMIM 112261), MKKS (Mckusick-Kaufmann syndrome; OMIM 604896), and JAG1 (Jagged 1; OMIM 601920; Table II).
Although monosomy 9p is a common and well-delineated chromosomal syndrome, trisomy 20p is rare and few cases with both alterations have been reported. The patient shares several but not all clinical features with the previous reported cases. Therefore, it is likely that the phenotypic features presented may be caused by multiple genes on 9p and 20p or other modifying genes in the proximity of the rearrangements.
The present report contributes to the description of unusual chromosomal aberrations affecting chromosomes 9 and 20 in MFDH.
ACKNOWLEDGMENTS
The authors are indebted to the patient and their family. We thank for Dr. Heloísa Marcelina Cunha Palhares for endocrinological evaluation of the patient. Financial support provided in part from: FAPESP (grant number 05/03480-7) and the Wellcome Trust (grant number WT077008).
Grant sponsor: FAPESP; Grant number: 05/03480-7; Grant sponsor: Wellcome Trust; Grant number: WT077008.
REFERENCES
- Alfi O, Donnell GN, Crandall BF, Derencsenyi A, Menon R. Deletion of the short arm of chromosome #9 (46,9p-): A new deletion syndrome. Ann Genet. 1973;16:17–22. [PubMed] [Google Scholar]
- Araújo M, Sanches MR, Suzuki LA, Guerra JRG, Farah SB, Mello MP. Molecular analysis of CYP21 and C4 genes in Brazilian families with the classical form of steroid 21-hydroxylase deficiency. Braz J Med Biol Res. 1996;29:1–13. [PubMed] [Google Scholar]
- Barbaro M, Balsamo A, Anderlid BM, Myhre AG, Gennari M, Nicoletti A, Pittalis MC, Oscarson M, Wedell A. Characterization of deletions at 9p affecting the candidate regions for sex reversal and deletion 9p syndrome by MLPA. Eur J Hum Genet. 2009;17:1439–1447. doi: 10.1038/ejhg.2009.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardoni B, Zanaria E, Guioli S, Floridia G, Worley KC, Tonini G, Ferrante E, Chiumello G, McCabe ER, Fraccaro M, Zuffardi O, Camerino G. A dosage sensitive locus at chromosome Xp21 is involved in male to female sex reversal. Nat Genet. 1994;7:497–501. doi: 10.1038/ng0894-497. [DOI] [PubMed] [Google Scholar]
- Bennett CP, Docherty Z, Robb SA, Ramani P, Hawkins JR, Grant D. Deletion 9p and sex reversal. J Med Genet. 1993;30:518–520. doi: 10.1136/jmg.30.6.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centerwall W, Francke U. Familial trisomy 20p five cases and two carriers in three generations a review. Ann Genet. 1977;20:77–83. [PubMed] [Google Scholar]
- Chaabouni M, Turleau C, Karboul L, Jemaa LB, Maazoul F, Attié-Bitach T, Romana S, Chaabouni H. De novo trisomy 20p of paternal origin. Am J Med Genet Part A. 2007;143A:1100–1103. doi: 10.1002/ajmg.a.31704. [DOI] [PubMed] [Google Scholar]
- Chen H. An approach to work-up of dysmorphic patients: Clinical, cytogenetic, and molecular aspects. Keio J Med. 1994;43:98–107. doi: 10.2302/kjm.43.98. [DOI] [PubMed] [Google Scholar]
- Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER) https://decipher.sanger.ac.uk/ Database of Genomic Variants (DGV): http://projects.tcag.ca/variation/ Ensembl C Genome Browser: http://www.ensembl.org/ European Cytogeneticists Association Register of Unbalanced Chromosome Aberrations (ECARUCA): http://agserver01.azn.nl:8080/ecaruca/ecaruca.jsp.
- Fiegler H, Redon R, Andrews D, Scott C, Andrews R, Carder C, Clark R, Dovey O, Ellis P, Feuk L, French L, Hunt P, Kalaitzopoulos D, Larkin J, Montgomery L, Perry GH, Plumb BW, Porter K, Rigby RE, Rigler D, Valsesia A, Langford C, Humphray SJ, Scherer SW, Lee C, Hurles ME, Carter NP. Accurate and reliable high-throughput detection of copy number variation in the human genome. Genome Res. 2006;16:1566–1574. doi: 10.1101/gr.5630906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funderburk SJ, Sparkes RS, Sparkes MC. Trisomy 20p due to a paternal reciprocal translocation. Ann Genet. 1983;26:94–97. [PubMed] [Google Scholar]
- Gil-da-Silva-Lopes VL. Malformação Frontonasal: estudo genéticoclínico de 31 pacientes ñao portadors de quadros sindrômicos já definidos. Universidade Estadual de Campinas; 1997. Campinas: Tese de Doutorado. [Google Scholar]
- Gil-da-Silva-Lopes VL, Maciel-Guerra AT. A clinical study of 31 individuals with midline facial defects with hypertelorism and a guideline for follow-up. Arq Neuropsiquiatr. 2007;65:396–401. doi: 10.1590/s0004-282x2007000300006. [DOI] [PubMed] [Google Scholar]
- Grammatico P, Cupilari F, Di Rosa C, Falcolini M, Del Porto G. 20p Duplication as a result of parental translocation: Familial case report and a contribution to the clinical delineation of the syndrome. Clin Genet. 1992;41:285–289. doi: 10.1111/j.1399-0004.1992.tb03398.x. [DOI] [PubMed] [Google Scholar]
- Guion-Almeida ML. Hipertelorismo e defeitos de linha média facial: Estudo genético-clínico de uma amostra de pacientes. Universidade Estadual de Campinas; 2000. Campinas: Tese de Doutorado. [Google Scholar]
- Hauge X, Raca G, Cooper S, May K, Spiro R, Adam M, Martin CL. Detailed characterization of, and clinical correlations in, 10 patients with distal deletions of chromosome 9p. Genet Med. 2008;10:599–611. doi: 10.1097/GIM.0b013e31817e2bde. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoo JJ, Fischer A, Fuhrmann W. Familial tiny 9p/20p translocation: 9p24 the critical segment for monosomy 9p syndrome. Ann Genet. 1982;25:249–252. [PubMed] [Google Scholar]
- Huret JL, Leonard C, Forestier B, Rethoré MO, Lejeune J. Eleven new cases of del(9p) and features from 80 cases. Med J Genet. 1988;25:741–749. doi: 10.1136/jmg.25.11.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick SJ, Pauli RM. Frontonasal malformation and deletion of 22q11. Am J Med Genet. 1998;75:443–444. [PubMed] [Google Scholar]
- Marcus ES, Fuller B, Riccardi VM. Triplication of chromosome arm 20p due to inherited translocation and secondary nondisjunction. Am J Med Genet. 1979;4:47–50. doi: 10.1002/ajmg.1320040107. [DOI] [PubMed] [Google Scholar]
- McDonald MT, Flejter W, Sheldon S, Putzi MJ, Gorski JL. XY sex reversal and gonadal dysgenesis due to 9p24 monosomy. Am J Med Genet. 1997;73:321–326. [PubMed] [Google Scholar]
- Muroya K, Okuyama T, Goishi K, Ogiso Y, Fukuda S, Kameyama J, Sato H, Suzuki Y, Terasaki H, Gomyo H, Wakui K, Fukushima Y, Ogata T. Sex-determining gene(s) on distal 9p: Clinical and molecular studies in six cases. J Clin Endocr Metab. 2000;85:3094–3100. doi: 10.1210/jcem.85.9.6771. [DOI] [PubMed] [Google Scholar]
- National Center for Biotechnology Information (NCBI) http://www.ncbi.nlm.nih.gov/ Online Mendelian Inheritance in Man (OMIM): http://www.ncbi.nlm.nih.gov/omim.
- Oppenheimer S, Dignan P, Soukup S. Partial trisomy 20p: Familial occurrence. Am J Med Genet. 2000;95:316–319. [PubMed] [Google Scholar]
- Roberts CJ, Lloyd S. Observations on the epidemiology of simple hypospadiass. Br Med J. 1973;31:768–770. doi: 10.1136/bmj.1.5856.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Z, Zabel B, Trautmann U, Hillig U, Ottolenghi C, Wang Y, Haaf T. FISH mapping of the sex-reversal region on human chromosome 9p in two XY females and in primates. Eur J Hum Genet. 2000;8:167–173. doi: 10.1038/sj.ejhg.5200431. [DOI] [PubMed] [Google Scholar]
- Sidwell RU, Pinson MP, Gibbons B, Byatt SA, Svennevik EC, Hastings RJ, Flynn DM. Pure trisomy 20p resulting from isochromosome formation and whole arm translocation. J Med Genet. 2000;37:454–458. doi: 10.1136/jmg.37.6.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simioni M, Freitas EL, Vieira TAP, Lopes-Cendes I, Gil-da-Silva-Lopes VL. Investigation of the 22q11.2 candidate region in patients with midline facial defects with hypertelorism. J Appl Genet. 2010;51:219–221. doi: 10.1007/BF03195732. [DOI] [PubMed] [Google Scholar]
- Smith CA, McClive PJ, Western PS, Reed KJ, Sinclair AH. Conservation of a sex-determining gene. Nature. 1999;402:601–602. doi: 10.1038/45130. [DOI] [PubMed] [Google Scholar]
- Stevens CA, Qumsiyeh MB. Syndromal frontonasal dysostosis in a child with a complex translocation involving chromosomes 3, 7 and 11. Am J Med Genet. 1995;55:494–497. doi: 10.1002/ajmg.1320550420. [DOI] [PubMed] [Google Scholar]
- Stratton RF, Payne RM. Frontonasal malformation with tetralogy of Fallot associated with a submicroscopic deletion of 22q11. Am J Med Genet. 1997;69:287–289. doi: 10.1002/(sici)1096-8628(19970331)69:3<287::aid-ajmg13>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Swinkels MEM, Simons A, Smeets DF, Vissers LE, Veltman JA, Pfundt R, de Vries BBA, Faas BHW, Schrander-Stumpel CTRM, McCann E, Sweeney E, May P, Draaisma JM, Knoers NV, van Kessel AG, van Ravenswaaij-Arts CMA. Clinical and cytogenetic characterization of 13 Dutch patients with deletion 9p syndrome: Delineation of the critical region for a consensus phenotype. Am J Med Genet Part A. 2008;146A:1430–1438. doi: 10.1002/ajmg.a.32310. [DOI] [PubMed] [Google Scholar]
- Tar A, Ion A, Sólyom J, Györvári B, Stephenson C, Barbaux S, Nunes M, Fellous M, McElreavey K. Hypertelorism and hypospadiass associated with a de novo apparently balanced translocation between 8q22.3-23 and 20p13. Am J Med Genet. 1997;68:231–235. doi: 10.1002/(sici)1096-8628(19970120)68:2<231::aid-ajmg22>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Taylor KM, Wolfinger HL, Brown MG, Chadwick DL, Francke U. Partial trisomy 20p derived from a t(18;20) translocation. Hum Genet. 1976;28:155–162. doi: 10.1007/BF00278884. [DOI] [PubMed] [Google Scholar]
- Thomas MA, Duncan AM, Bardin C, Kaloustian VM. Lissencephaly with der(17)t(17;20)(p13.3;p12.2)mat. Am J Med Genet Part A. 2004;124A:292–295. doi: 10.1002/ajmg.a.20373. [DOI] [PubMed] [Google Scholar]
- Veitia R, Nunes M, Brauner R, Doco-Fenzy M, Joanny-Flinois O, Jaubert F, Lortat-Jacob S, Fellous M, McElreavey K. Deletions of distal 9p associated with 46,X Y male to female sex reversal: Definition of the breakpoints at 9p23.3–p24.1. Genomics. 1997;41:271–274. doi: 10.1006/geno.1997.4648. [DOI] [PubMed] [Google Scholar]
- Venditti CP, Hunt P, Donnenfeld A, Zackai E, Spinner NB. Mosaic paternal uniparental (iso)disomy for chromosome 20 associated with multiple anomalies. Am J Med Genet Part A. 2004;124A:274–279. doi: 10.1002/ajmg.a.20430. [DOI] [PubMed] [Google Scholar]
- Wilkie AO, Campbell FM, Daubeney P, Grant DB, Daniels RJ, Mullarkey M, Affara NA, Fitchett M, Huson SM. Complete and partial XY sex reversal associated with terminal deletion of 10q: Report of 2 cases and literature review. Am J Med Genet. 1993;46:597–600. doi: 10.1002/ajmg.1320460527. [DOI] [PubMed] [Google Scholar]
- Wu E, Vargevik K, Slavotinek AM. Subtypes of frontonasal dysplasia are useful in determining clinical prognosis. Am J Med Genet Part A. 2007;143A:3069–3078. doi: 10.1002/ajmg.a.31963. [DOI] [PubMed] [Google Scholar]
- Zumel RM, Darnaude MT, Delicado A, Diaz de Bustamante A, de Torres ML, López Pajares I. Trisomy 20p from maternal translocation and anencephaly. Case report and genetic review. Ann Genet. 1989;32:247–249. [PubMed] [Google Scholar]