Abstract
Pro-apoptotic Bcl-2 family members of the BH3-only subgroup are critical for the establishment and maintenance of tissue homeostasis and can mediate apoptotic cell death in response to developmental cues or exogenously induced forms of cell stress and damage. Based on biochemical experiments as well as genetic studies in mice, the BH3-only proteins Bad and Bmf have been implicated in different proapoptotic events such as those triggered by glucose- or trophic factor-deprivation, glucocorticoids, or histone deacetylase inhibition in lymphocytes as well as suppression of B cell lymphomagenesis upon aberrant expression of c-Myc. To address possible redundancies in cell death regulation and tumor suppression, we generated compound mutant mice lacking both genes. Our studies revealed lack of redundancy in most paradigms of lymphocyte apoptosis tested in tissue culture. Only spontaneous cell death of thymocytes kept in low glucose or that of pre-B cells deprived of cytokines was significantly delayed when both genes were lacking. Of note, despite these minor apoptosis defects we observed compromised lymphocyte homeostasis in vivo that affected mainly the B cell lineage. Long-term follow up revealed significantly reduced latency to spontaneous tumor formation in aged mice when both genes were lacking. Together our study suggests that Bad and Bmf co-regulate lymphocyte homeostasis and limit spontaneous transformation by mechanisms that may not exclusively be limited to the induction of lymphocyte apoptosis.
Keywords: apoptosis, BH3-only proteins, Bad, Bmf, hematopoiesis, cancer
Introduction
Apoptotic cell death can be induced along two distinct signaling cascades, commonly referred to as the “intrinsic” or “extrinsic” apoptosis pathways (1;2). The interplay of members of the Bcl-2 family regulates apoptosis at the level of mitochondria. These proteins can be divided according to their function into proapoptotic and antiapoptotic members that mainly regulate the intrinsic apoptosis pathway. Proapoptotic members again fall into different classes based on the number of Bcl-2 homology (BH)-domains they contain, i.e. in BH123 proteins, such as Bax or Bak, or the so-called “BH3-only” proteins, including Bim, Bid, Bad, Bmf and others that can bind to and neutralize antiapoptotic Bcl-2 or its homologues, such as Bcl-xL or Mcl-1 (1;2).
Activated Bax and/or Bak induce the formation of pores in the outer mitochondrial membrane, allowing the release of apoptogenic factors such as cytochrome c, enabling activation of cell death proteases belonging to the caspase family, triggering apoptotic cell death. Activation of Bax/Bak is discussed to occur via one of two possible routes where either BH3-only proteins, i.e. Bim, Bid or Puma, directly activate Bax and/or Bak while others, e.g. Bad and Bmf, act as sensitizers, inactivating the prosurival Bcl-2 proteins that block apoptosis by sequestering direct activators. In the alternative neutralization model, prosurvival Bcl-2 proteins sequester active Bax/Bak until forced to interact with BH3-only proteins that then can no longer restrain their proapoptotic potential (1;2).
Bad and Bmf are weak inducers of apoptosis in vitro, either because they do not exert direct activator function and may act as redundant sensitizers, or, alternatively because they can only neutralize a limited set of overlapping pro-survival Bcl-2 homologues, i.e. Bcl-xL, Bcl-w and Bcl-2, but not Mcl-1 or A1/Bfl1 (3-5). Of note here, Bad or Bmf have also been implicated in non-apoptotic cellular responses, i.e. autophagy (6), glucose metabolism (7) or necroptosis (8), respectively.
Bad-deficient mice are viable and do not show a gross phenotype in young animals (9). Most hematopoietic cell types develop normally, except B cells that show minor abnormalities in proliferation and IgG production (9). Furthermore, mammary epithelial cultures and primary mouse embryonic fibroblasts (MEF) lacking Bad were less sensitive to stress-induced cell death in the absence of growth factors such as EGF or IGF-1 (9). Aged bad−/− animals were reported to develop diffuse large B-cell lymphoma (DLBCL) with increased frequency and irradiation studies suggested that Bad can also suppress DNA-damage triggered tumorigenesis (9). However, findings on accelerated tumorigenesis were recently challenged (10). Despite this discrepancy, but in line with a tumor suppressor function of Bad, c-Myc-driven B lymphomagenesis was found accelerated in its absence (11). In human cancer, phosphatidylinositol-3 (PI3)-kinase/Akt signaling is frequently hyperactivated, mainly caused by the loss of its negative regulator PTEN (12-14) that results in the inhibition of Bad by phosphorylation-dependent cytoplasmic sequestration via 14-3-3 proteins (15) and may contribute to apoptosis resistance of transforming cells. Genetic alterations of the bad gene have also been observed in rare cases of colon adenocarcinomas (16). Notably, higher expression levels of Bad in prostate and breast cancer have been associated with better outcome and levels of Bad and Bid were reported to predict the outcome of 5-FU treatment (17). Additional studies support a role for Bad in tumor cell killing by tyrosine kinase inhibitors targeting the Bcr-Abl oncogene (18) or the EGF receptor (19).
In hepatocytes, Bad can be part of a holoenzyme complex together with protein kinase A, protein phosphatase-1, WAVE1 and glucokinase. This complex that requires Bad to form, is found in liver mitochondria and affects glucokinase activity, thus mitochondrial respiration is mitigated in the absence of Bad, as it is critical for the other complex partners to assemble (20). In addition, cell death of hepatocytes in low glucose-containing media appeared to be enhanced in wild type (wt) versus Bad-deficient cells (20).
Bmf was discovered in a yeast-two-hybrid screen using Mcl-1 as a bait. In healthy cells, a fraction of Bmf can bind to the myosinV motor complex via interaction with dynein light chain 2 (DLC2) molecules. Upon apoptotic signaling, such as the one triggered by UV-radiation or loss of contact to the extracellular matrix (anoikis) it can bind to prosurvival Bcl-2 proteins and was thought to contribute to cell death initiation under such conditions (21;22).
Analysis of Bmf-deficient mice revealed that it is dispensable for embryonic development and most types of stress-induced cell death, including anoikis or UV-mediated killing of gastrointestinal epithelial cells or fibroblasts, respectively (23). It has been shown, however, that Bmf is involved in apoptosis induced by glucocorticoids or HDAC inhibition in lymphocytes (23;24). Of note, Bmf knockout mice showed an excessive amount of B cells but in contrast to mice lacking Bad (9) these cells appeared functional and an increased rate in spontaneous B lymphomagenesis was not observed within their first year of life (11;23). Similar to bad−/− mice, gamma-irradiation studies showed that thymic lymphoma development was accelerated in Bmf-deficient mice (23), as was c-Myc driven B lymphomagenesis. Notably, in this model, animals that lacked Bad and Bmf simultaneously did not contract disease earlier than single-mutant animals expressing the transgene, suggesting that they most likely modulate B cell homeostasis at different and sequential developmental stages (11).
To address possible redundancies between Bad and Bmf in tissue homeostasis and tumor suppression, we monitored cohorts of mice for consequences of simultaneous loss of these two proteins over time. Therefore, we intercrossed single deficient mice to generate double knockout animals and analysed hematopoietic organ composition, lymphocyte survival in response to drug-treatment as well as spontaneous development of malignant disease.
Results
Characterization of Bad and Bmf double-deficient (DKO) mice
DKO mice were generated from intercrosses of double-heterozygous mice and colonies were then maintained by interbreeding DKO mutants, demonstrating that both proteins are dispensable for embryonic development and their absence failed to cause gross phenotypic abnormalities, perturb fertility or cause a gender bias (not shown).
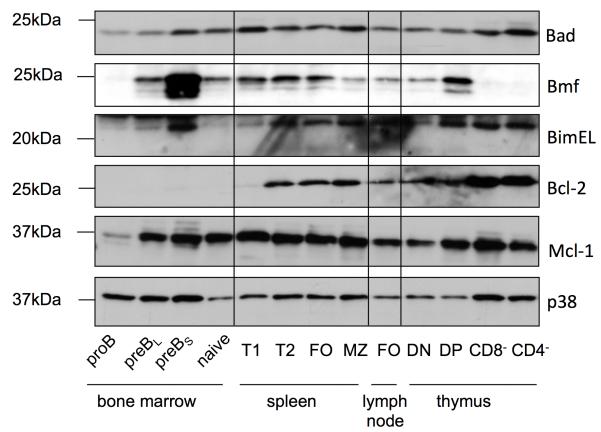
As Bad and Bmf were reported to regulate lymphocyte homeostasis or function (9;23), we first analyzed levels of BH3-only protein expression in immature thymocytes as well as mature T cells and all major B cell subsets present in spleen and isolated by cell sorting. This analysis revealed co-expression of Bmf and Bad in most B cell subsets. While Bad was found expressed throughout B cell maturation, with levels gradually increasing in small resting pre-B cells, Bmf expression was undetectable in pro-B cells, but found in all other subsets with maximal expression in small resting pre-B cells (Figure 1). In T cells, Bmf expression was highest in immature CD4+8+ double positive thymocytes, but poorly detectable in mature CD4+ or CD8+ T cells. In contrast, all these T cell subsets expressed Bad, with highest levels in mature T cells, but also the related protein Bim, that also showed similar expression as Bmf in the B cell lineage. Expression of BH3-only proteins was counter-balanced by expression of anti-apoptotic Mcl-1 and, in more mature stages of T and B lymphocyte development, Bcl-2 (Figure 1).
Figure 1. Analysis of protein expression of Bad and Bmf in sorted B- and T-cell subsets.
Cells were isolated by cell sorting from bone marrow, thymus or spleen, based on cell surface marker expression and lysed with RIPA buffer. The following cell surface markers were stained for subset identification: pro-B cells (B220+IgM−c-Kit+), large/small pre-B cells (B220+IgM−c-Kit−), mature B cells (B220+IgM+D+), immature B cells (B220+IgM+D−), T1 (IgM+CD21−CD23−), T2 (IgM+CD21+CD23+), marginal zone (B220+CD21+CD23−), follicular (B220+CD21+CD23+). Equal amounts of proteins (40 μg/lane) were size fractioned by SDS-PAGE and transferred onto nitrocellulose membranes. Membranes were sequentially probed, or when necessary, stripped and reprobed with antibodies specific for the indicted Bcl-2 family proteins. An antibody recognizing p38 MAP kinase served as a loading control.
In light of these observations, we were interested to see if combined absence of Bad and Bmf would reveal new phenotypic abnormalities in the hematopoietic compartment of bad−/−bmf−/− mice, maybe similar to the ones reported for mice lacking Bim (25), that should depend on the presence of Bad and/or Bmf for function, at least according to the “direct activation” model (1;2). Comparing hematopoiesis in 8 to 10 week old wt, single-deficient and DKO animals by determining organ weight (suppl. Fig. 1), cell counting and flow cytometric analysis revealed largely normal leukocyte development (Figure 2) but also saddle differences in mice lacking both genes, some not noted before in single-deficient animals (Table 1). Loss of Bmf lead to mild but significantly increased thymic weight, but this effect was not enhanced by additional loss of Bad (suppl. Fig. 1). Thymic cellularity was also mildly increased in the absence of Bmf but this did not reach statistical significance. Spleen weight was only modestly increased in the absence of Bad, increased in the absence of Bmf but this phenomenon was most pronounced in DKO mice (suppl. Fig. 1). These minor increases in weight, however, manifested in significantly increased cell numbers in spleens of Bad-deficient animals that were mainly due to elevated B220+ B cell numbers. Of note, the number of Ter119+ nucleated erythroid progenitors also appeared elevated in bone marrow and spleen of bad−/− and DKO mice, but not in those lacking Bmf. Platelets were found increased in the peripheral blood of Bad-deficient mice but this was no longer detected in DKO mice, possibly due to reduced platelet production caused by loss of Bmf (Table 1).
Figure 2. Flow cytometric analysis of the hematopoietic compartment in mice lacking Bad, Bmf or both genes.
Representative dot blots of the flow cytometric analysis performed on major hematopoietic organs are shown. A quantification of this analysis can be found in Table 1. (a) wt, bmf−/−, bad−/−, and bmf−/−bad−/− thymocyte suspensions stained with antibodies recognizing the T cell markers CD4 or CD8. (b) Spleen cell suspensions stained as in (a). (c) Spleen cell suspensions stained with antibodies recognizing the B cell markers surface, IgM or IgD, or (d) antibodies recognizing the myeloid cell markers Mac1 and Gr-1, to identify Mac1+Gr1low monocytes and Mac1+Gr1high granulocytes. (e) Bone marrow stained as in (d). (f) Bone marrow cell suspensions stained with antibodies recognizing the B cell markers B220, IgM and CD43, in order to identify B220+IgM-CD43+ pro-B cells and B220+IgM−CD43− pre-B cells as well as B220+IgMlow naïve and B220+IgMhigh recirculating mature B cells.
Table 1. Composition of primary and secondary lymphatic organs of wild type, Bad-, Bmf- and double-deficient mice.
Total numbers of hematopoietic cells from 8-10-week-old animals were calculated by counting single-cell suspensions derived from the indicated organs and tissues that were subsequently analyzed by flow cytometry after staining for cell surface markers: total T cells (Thy1+), granulocytes (Gr1+), macrophages (Mac1+Gr1−), total myeloid cells (Mac1+), total nucleated erythroid progenitor cells (Ter119+), total B cells (B220+), pro–B cells (B220+CD43+IgM−), pre-B cells (B220+CD43−IgM−), T1 B cells (IgMhighCD21low), immature B cells (IgMhighCD21−) and mature B cells (IgMlowIgD+). Platelet numbers were assessed in an automated blood cell counter. Numbers represent the mean (± the SEM) of ≥ 8 animals per genotype with the exception of thymic cellularity is based on the analysis of 4 male mice per genotoype. Total cellularity of both inguinal lymph nodes and both femura are presented.
| Cell subsets | wild type | bad −/− | bmf−/− | DKO |
|---|---|---|---|---|
| Thymus | ||||
| Total cellularity (×10 7) | 17.70 ± 3.33 | 14.51 ± 2.35 | 19.85 ± 2.43 | 21.72 ± 2.84 |
| CD4+ cells | 1.05 ± 0.11 | 1.20 ± 0.09 | 1.77 ± 0.27 | 2.54 ± 0.66 |
| CD8+ cells | 0.44 ± 0.02 | 0.42 ± 0.05 | 0.91 ± 0.19 | 1.22 ± 0.58 |
| CD4+8+ cells | 12.54 ± 1.61 | 9.91 ± 2.47 | 17.43 ± 3.38 | 14.63 ± 2.61 |
| CD4−8− cells | 0.63 ± 0.14 | 0.54 ± 0.04 | 1.14 ± 0.28 | 1.62 ± 0.83 |
| Bone marrow | ||||
| Total cellularity (×10 6) | 22.90 ± 1.35 | 28.82 ± 2.51 | 28.99 ± 0.23a | 29.94 ± 4.12 |
| T cells | 1.16 ± 0.15 | 1.91 ± 0.55 | 1.31 ± 0.40 | 2.47 ± 0.84 |
| Granulocytes | 0.76 ± 0.11 | 0.67 ± 0.09 | 0.87 ± 0.13 | 0.92 ± 0.16 |
| Macrophages | 0.46 ± 0.04 | 0.44 ± 0.04 | 0.58 ± 0.07 | 0.63 ± 0.06a |
| Myeloid cells | 12.20 ± 1.41 | 11.20 ± 1.13 | 14.22 ± 2.49 | 16.00 ± 2.58 |
| Erythroid progenitors | 3.52 ± 1.07 | 6.68 ± 1.69 | 3.74 ± 1.82 | 8.22 ± 2.39 |
| B cells | 4.65 ± 0.64 | 7.02 ± 1.65 | 6.75 ± 0.78a | 7.38 ± 0.88a |
| Pro Bcells | 0.75 ± 0.21 | 1.04 ± 0.42 | 0.88 ± 0.20 | 1.22 ± 0.29 |
| Pre B cells | 2.03 ± 0.23 | 2.87 ± 0.57 | 2.86 ± 0.52 | 3.14 ± 0.36a |
| T1 B cells | 0.69 ± 0.12 | 1.13 ± 0.27 | 0.92 ± 0.23 | 1.08 ± 0.16 |
| Mature B cells | 1.06 ± 0.15 | 1.34 ± 0.22 | 1.52 ± 0.21 | 1.98 ± 0.25a |
| Spleen | ||||
| Total cellularity (×10 7) | 10.33 ± 1.00 | 16.11 ± 1.59a | 16.04 ± 2.23a | 20.12 ± 1.85b |
| CD4+ T cells | 2.04 ± 0.17 | 2.38 ± 0.25 | 3.87 ± 1.20 | 2.85 ± 0.37a |
| CD8+ T cells | 1.35 ± 0.13 | 1.34 ± 0.14 | 2.57 ± 1.03 | 2.00 ± 0.27a |
| Myeloid cells | 1.67 ± 0.25 | 1.42 ± 0.18 | 1.48 ± 0.20 | 2.12 ± 0.49 |
| Erythroid progenitors | 1.03 ± 0.13 | 2.25 ± 0.68a | 2.29 ± 0.83 | 3.41 ± 1.02a |
| B cells | 4.97 ± 0.45 | 7.83 ± 1.29a | 9.81 ± 2.23a | 13.02 ± 2.47a |
| Mature B cells | 3.79 ± 0.32 | 4.97 ± 0.78 | 6.86 ± 1.32a | 9.04 ± 0.86b |
| immature B cells | 1.09 ± 0.17 | 1.42 ± 0.35 | 1.93 ± 0.46 | 1.61 ± 0.23 |
| Peipheral blood | ||||
| Total cellularity (×10 6/ml) | 12.68 ± 1.96 | 11.86 ± 1.04 | 14.28 ± 2.92 | 18.7 ± 1.51a |
| CD4+ T cells | 0.91 ± 0.12 | 1.11 ± 0.11 | 1.43 ± 0.18a | 1.00 ± 0.14 |
| CD8+ T cells | 0.78 ± 0.08 | 0.85 ± 0.11 | 1.27 ± 0.19a | 1.00 ± 0.10 |
| Myeloid cells | 3.61 ± 0.93 | 2.19 ± 0.38 | 3.28 ± 0.59 | 3.15 ± 0.46 |
| B cells | 4.49 ± 0.60 | 6.40 ± 1.16 | 11.02 ± 1.82a | 11.31 ± 1.31b |
| T1 B cells | 0.63 ± 0.11 | 0.91 ± 0.21 | 1.17 ± 0.28 | 1.51 ± 0.32a |
| Platelets | 614 ± 68 | 1021 ± 116a | 689 ± 156 | 620 ± 89 |
| Inguinal lymph nodes | ||||
| Total cellularity (×10 6) | 6.42 ± 0.69 | 7.21 ± 0.69 | 8.85 ± 1.02a | 11.60 ± 0.72b |
| CD4+ T cells | 2.40 ± 0.30 | 2.52 ± 0.22 | 3.22 ± 0.39 | 3.40 ± 0.34 |
| CD8+ T cells | 1.67 ± 0.19 | 1.74 ± 0.19 | 2.31 ± 0.23a | 2.86 ± 0.29a |
| B cells | 1.49 ± 0.21 | 2.35 ± 0.45 | 2.34 ± 0.39 | 3.02 ± 0.60a |
p < 0.05 compared to wt mice.
p < 0.0001 compared to wt mice.
Bmf-deficient mice are known to present with higher B cell numbers in their spleen (23), reconfirmed here, as well as mildly elevated total cellularities in bone marrow, inguinal lymph nodes and significantly elevated numbers in peripheral blood (Table 1). DKO mice presented with even further increased splenic cellularity and erythroid progenitor and B cell numbers were most strongly elevated, the latter also noted in the peripheral blood and inguinal lymph nodes of these mice (Table 1). Furthermore, DKO mice showed significant changes in the number of pre-B cells, recirculating mature B cells but also Mac1+Gr1− monocyte/macrophages in the bone marrow when compared to wt controls, but not single knockout mice. Together, these observations indicate that loss of both BH3-only proteins causes minor changes in cell survival that preferentially lead to the accumulation of mature B lymphocyte and nucleated erythroid progenitors, but does not impact on the developmentally regulated death of hematopoietic cells.
We wondered whether the minor phenotypes observed in the hematopoietic compartment of young DKO mice might be due to the compensatory upregulation of other BH3-only proteins or the downregulation of antiapoptotic Bcl-2 family members. Therefore, we assessed protein levels of Bim, Bid, Bcl-2, Bcl-xL and Mcl-1 by Western Blot in all major lymphatic tissues, but this analysis failed to reveal prominent changes in expression between wt and double knockout mice (Figure 3). We conclude that the absence of both BH3-only proteins does not cause altered expression of some of the most abundant Bcl-2 family members to compensate for their absence, at least not under steady-state conditions.
Figure 3. Assessment of potential compensatory expression of other Bcl-2 family proteins in hematopoietic organs of DKO mice.
Indicated organs were harvested from wt and DKO mice and single cell suspensions lysed with RIPA buffer. Equal amounts of proteins (40 μg/lane) were size fractioned by SDS-PAGE and transferred onto nitrocellulose membranes. Membranes were sequentially probed, or when necessary, stripped and reprobed with antibodies specific for the indicted Bcl-2 family proteins. An antibody recognizing p38 MAP kinase served as a loading control. Two individual animals were assessed per genotype
Minor effects of Bad and/or Bmf-deficiency on lymphocyte apoptosis
Thymocytes express Bad and Bmf at significant levels and to elicit possible redundant roles in stress-induced apoptosis cells were put in culture and analyzed for their apoptotic response. Spontaneous cell death was monitored under normal culture conditions or in response to glucose-limitation or deprivation. In addition, cell death was also assessed after treatment with the histone deacetylase inhibitor SAHA (also known as Vorinostat), etoposide or dexamethasone. Serum withdrawal was also combined with glucose-deprivation.
Under normal culture conditions (25mM glucose) no differences in spontaneous cell death rates were seen, but when the level of glucose was reduced to 5mM only wt cells were sensitized to spontaneous death in culture. This difference was no longer detectable in glucose-free medium where cells of all genotypes died with similar kinetics (Figure 4a). In the absence of serum no significant protection was observed across genotypes due to loss of Bad, Bmf or both, regardless of the glucose content in the medium. Reducing the glucose concentration to 5mM did not change the response to serum withdrawal, while under conditions of serum- plus glucose-deprivation thymocytes of all genotypes seemed most vulnerable, but indistinguishable in their death kinetics (Figure 4b). To rule out that under such conditions thymocytes die by non-apoptotic mechanisms we assessed if thymocyte death in culture was inhibited in the presence of the pan-caspase inhibitor Q-VD-OPh, or the necroptosis inhibitor Nec-1. In addition, we also monitored necrosis-associated lactate-dehydrogenase (LDH)-release into the culture supernatant, cellular PI-uptake as well as changes in intracellular ATP-levels. These experiments revealed that caspase-inhibition delayed the rise of Annexin-V+ apoptotic cells at early time points in culture, while addition of Nec-1 had no effect on AnnexinV-exposure (suppl. Fig. 2) or cellular PI-uptake (suppl. Fig. 3), independent of glucose levels present in the media (not shown). This suggests that apoptosis is the key-mechanism of thymocyte death under these conditions while programmed necroptosis is not involved and cellular PI-uptake is due to secondary passive necrosis in cell culture. Consistently, significant necrosis-associated LDH-release into the supernatant was only observed at later time-points and this did not differ between the genotypes (suppl. Fig. 4) while ATP-levels were well maintained over the first 24h in culture, followed by a rapid drop in the next 24h (suppl. Fig. 4). This drop in ATP-levels was delayed in DKO cells, also under glucose limiting conditions, indicative for prolonged mitochondrial integrity in the absence of Bad and Bmf (suppl. Fig. 5a). Finally, we also monitored accumulation of lactate in thymocytes that were cultured in the presence of graded doses of glucose. Levels of lactate did not differ significantly between genotypes in freshly isolated cells and accumulated over time in culture to similar levels, suggesting normal glycolytic flux in resting as well as stressed thymocytes lacking Bad and/or Bmf (suppl. Fig. 5b).
Figure 4. Survival analysis of thymocytes lacking Bad and/or Bmf.
Thymocytes were isolated from mice of the indicated genotypes. Induction of cell death was triggered by (a) glucose- and/or (b) serum-deprivation, (c) treatment with the glucocorticoid dexamethasone (10−7M), the topoisomerase II inhibitor VP16 (1μg/ml) or the histone deacetylase inhibitor SAHA (0.5 μM). (d) Large and small pre-B cells were sorted based on cell surface marker expression (B220+IgM−c-kit−) and additional subdivision according to size. Cell death was monitored by Annexin-V/7-AAD staining and flow cytometric analysis at the indicated time points. Specific apoptosis was calculated by the following equation: [(spontaneous cell death – induced apoptosis)/spontaneous cell death]*100. Means ± the SEM from four independent experiments using n ≥ 4 animals per genotype are shown. (e) Representative dot blots of thymocytes stained with antibodies recognizing CD4 and CD8 after gamma-irradiation in vivo (1.75 Gy). Means ± the SEM from two independent experiments using n = 4 animals per genotype are shown.
Treating thymocytes with different drugs reported to depend in part on Bmf for killing, including SAHA or dexamethasone, confirmed these findings but double-deficiency did not lead to a further increase in apoptosis resistance and all cells of all genotypes died equally fast in response to etoposide (Figure 4c).
In addition to immature T cell precursors, we also analyzed spontaneous and glucose-deprivation-mediated killing of pre-B cells. These cells can be subdivided morphologically into small and large pre-B cells based on their forward-side-scattering profile in flow cytometric analysis. Bmf expression is highest in small resting pre-B cells where Bad protein can also be found (Figure 1).
Therefore, we assessed the in vitro viability of large and small pre-B cells, isolated by cell sorting from bone marrow over time in order to document possible differences between the genotypes. Large pre-B cells lacking both genes were less susceptible to spontaneous cell death compared to wt controls. Interestingly, double-deficient pre-B cells were less sensitive to spontaneous death compared to wt cells under normal culture conditions (Figure 4d). No significant differences in cell death responses were noted upon glucose deprivation (not shown).
Of note, both Bad and Bmf have been implicated in the suppression of thymic lymphomagenesis in response to gamma-irradiation. Hence we also assessed thymocyte depletion after whole body-irradiation in vivo. Albeit death triggered by the DNA-damaging drug etoposide occurred normally in single and DKO thymocytes in vitro, significantly more thymocytes survived after whole body irradiation in vivo (Figure 4e).
Analysis of cell death responses of resting mature T and B cells sorted from spleen of the different genotypes also failed to reveal significant differences between genotypes (data not shown). Hence, Bad and Bmf appear to co-regulate cell death in thymocytes under glucose-limiting but not glucose-free conditions and that of pre-B cells, triggered by factor-withdrawal but not glucose-deprivation. No redundancy exists in response to serum-deprivation, HDAC inhibition, glucocorticoid or etoposide treatment despite the fact that both proteins are co-expressed in thymocytes, as well as mature T and B cells. This suggests that the noted accumulation of B cells in vivo may not necessarily be only due to intrinsic apoptosis defects, but may involve non-cell autonomous mechanisms and/or defects in other cell death modalities.
Long-term analysis of aging cohorts of bad−/−bmf−/− mice
To explore the effect of double-deficiency in spontaneous disease development, cohorts of wt, bad−/−, bmf−/− and DKO mice were monitored for two years. Disease free survival was recorded and statistically evaluated (Figure 5a). Mice that reached the age of 750 days and mice that diseased earlier were sacrificed and major organs were analyzed by histopathological analysis.
Figure 5. Reduced latency to spontaneous tumorigenesis in DKO mice.
(a) Kaplan-Meier analysis of tumor free survival of wt (n=18), bad−/−(n=30), bmf−/− (n=17) and bad−/−bmf−/− (n=28) mice. Animals were monitored weekly until the age of 750 days, sacrificed and organs harvested for histological analysis post mortem. (b) Representative histological images of detected tumors (H&E staining). (A, B) low grade non-Hodgkin lymphoma, (C, D) high grade non-Hodgkin lymphoma, (E, F) hepatocellular carcinoma, (G, H) angiosarcoma, (I) adenocarcinoma, (J) osteosarcoma, (K) undifferentiated sarcoma, (L) fibrosarcoma.
We observed that DKO animals showed increased frequency and reduced latency of spontaneous tumors. Tumors included most frequently high- and low-grade non-Hodgkin lymphomas and occasional hepatocellular- and adeno-carcinomas, as well as different sarcoma entities (Figure 5b and Table 2). It has been reported that Bad-deficient animals show increased spontaneous tumorigenesis and develop diffuse large B cell lymphomas with a frequency of about 20% leading to reduced overall lifespan in Bad-deficient mice (9). Here, we did not observe significantly reduced lifespan and accelerated onset of spontaneous tumors in mice lacking Bad and the overall frequency of lymphatic malignancies was not significantly increased when compared to controls (Table 2). Histopathological assessment revealed that 3/4 of the high-grade non-Hodgkin lymphomas recorded were of diffuse large B cell type in bad−/− animals while the other tumor was a Burkitt or Burkitt-like lymphoma (Table 2). The additional absence of Bmf did not significantly increase the numbers of lymphomas but reduced the overall tumor free survival significantly from a mean of 737.9 to 657.4 days (p=0.0045). In DKO mice 4/6 high-grade non-Hodgkin lymphomas were DLBCL, one of Burkitt or Burkitt-like type while the other represented a follicular lymphoma. Interestingly, we saw occasional cases of hepatocellular carcinomas that appeared to occur more frequently when Bmf function was lost, although overall numbers were too low to draw formal conclusions (Table 2). Loss of Bmf on its own, as loss of Bad, failed to significantly reduce the tumor free survival (Figure 5a).
Table 2. Histopathological assessment of spontaneous tumors.
All major organs of animals that showed signs of disease or that were sacrificed after two years were subjected to histopathological analysis on H&E stained sections in a blinded manner. Number of tumors and relative percentages are given. The χ2-test and Fisher’s exact test were used for comparison of frequency distributions but failed to reveal significant differences. Low- and high-grade lymphomas were pooled for that purpose. NHL = Non-Hodgkin Lymphoma.
| wild type | bad−/− | bmf−/− | DKO | |
|---|---|---|---|---|
| High-grade NHL | 4 (22%) | 4 (18%) | 1 (8%) | 6 (24%) |
| Low-grade NHL | 2 (11%) | 7 (32%) | 3 (25%) | 5 (20%) |
| Hepatocellular carcinoma | 1 (6%) | 1 (5%) | 2 (17%) | 4 (16%) |
| Angiosarcoma | 2 (11%) | - | 1 (8%) | 1 (4%) |
| Osteosarcoma | 1 (6%) | - | - | - |
| Sarcoma | - | 1 (5%) | - | - |
| Fibrosarcoma | - | - | - | 1 (4%) |
| No tumor | 8 (44%) | 9 (41%) | 5 (42%) | 8 (32%) |
| Total | 18 | 22 | 12 | 25 |
Discussion
Meanwhile most members of the Bcl-2 family have been investigated for their physiologic function by gene knockout studies in mice. This has revealed functions in cell death initiation and tissue homeostasis that are sometimes highly specific for a given cell type and frequently also highly stimulus-dependent (1). However, the comparison with gain-of-function studies where individual pro-survival Bcl-2 family members were overexpressed in mice, suggested a large degree of redundancy between individual BH3-only proteins in cell death signaling (25;26). Therefore, different compound mutant animal models have been generated. These studies revealed previously unappreciated overlapping function between the BH3-only proteins Bim and PUMA, e.g. in DNA damage responses (27), or Bim and Blk (28) in spermatogenesis as well as Bim and Bad in the suppression of irradiation-triggered lymphomagenesis (10). Following similar lines, we investigated the consequences of loss of Bad and Bmf, considered to be weak killers due to their limited binding spectrum to anti-apoptotic Bcl-2 family proteins (29) but, according to the direct activation model of BH3-only protein function (4), critical de-repressors that may be required for the efficient activation of direct activators such as Bim.
Bad and Bmf are the two most abundant “de-repressor” BH3-only proteins found constitutively expressed during B- and T-cell development (Figure 1, Labi 2006), and previous reports suggested roles in lymphocyte cell death and function (9;23). Therefore, we reasoned that their combined absence may cause more pronounced phenotypes, maybe similar to those caused by loss of Bim, the key-regulator of lymphocyte homeostasis and development within the Bcl-2 family (25;27). However, in contrast to our anticipation, the hematopoietic system developed largely normal in DKO mice (Table 1), suggesting that neither protein is required for normal leukocyte development. Development did not depend on compensatory upregulation of other BH3-only proteins, such as Bim or Bid, or down-regulation of prosurvival Bcl-2 proteins (Figure 3).
In line with a rate-limiting role in the survival of some hematopoietic cell types, most prominently cells of the B cell lineage, we observed accumulation of mature B cells in the spleen, lymph nodes and peripheral blood of DKO mice. A similar phenotype was already noted in the absence of Bmf (23;30), and was significantly enhanced by additional loss of Bad (Table 1). Similar observations have been made in mice lacking Bim and Bad that represented with increased lymph node cellularity, when compared with Bim-deficient mice (10). Somewhat surprising, we were unable to detect cell death resistance phenotypes in mature B cells derived from single or DKO mice in various cell death paradigms in vitro. This may suggest that accumulation of mature B cells, noted mainly in Bmf and DKO mice, may not entirely be cell autonomous. However, competitive reconstitution experiments using Bmf-deficient and wt bone marrow showed that lymphocytes lacking Bmf accumulate over time in recipient mice (23). However, a non-cell autonomous contribution of Bad in lymphocyte accumulation in DKO mice can presently not be excluded. Of note, radioresistance of thymocytes noted in Bad or Bim/Bad DKO mice, recapitulated in part here (Figure 4e), was ultimately shown to depend on interactions with the Bad-deficient thymic epithelium (10).
Interestingly, we also noted increased numbers of platelets in Bad-deficient mice that was no longer detectable in DKO mice. Again, a similar phenomenon was observed in bad−/−bim−/− DKO mice (10), suggesting that Bim and Bmf contribute to platelet production, while Bad defines their lifespan. However, in contrast to this report we observed increased splenic B cell number in the absence of Bad that reached statistical significance as well as an increased number of nucleated erythroid progenitors in spleen and bone marrow. The latter effect was not modified by simultaneous loss of Bmf while B cell numbers increased further in DKO mice (Table 1). The reason for this discrepancy is currently unclear but cannot be related to genetic background differences, as all mice in both studies were maintained or backcrossed onto C57BL/6. However, differences in the pathogen burden and/or spectrum in the local environments may be involved.
The fact that both proteins are poor agonists of apoptosis, either due to restricted binding patterns to Bcl-2 prosurvival proteins or redundancy with other de-repressors, was confirmed by the in vitro analysis of thymocytes exposed to a number of different cell death stimuli. As it has been reported that Bad is in a complex with glucokinase and acts as a sensor of glucose metabolism and metabolic stress, we also investigated the impact of glucose-free medium on drug-induced cell death. Although glucose-deprivation can trigger and/or sensitize lymphocytes to apoptosis (31), we did not observe resistance caused by lack of Bad in pre-B cells, thymocytes (Figure 4) or resting mature T and B cells from spleen (not shown). This observation is actually in line with the cell type specific expression of glucokinase, restricted to liver and pancreatic β-cells, and that hexokinases, that exerts comparable function in lymphocytes, is presumably not relayed to Bad as a sensor of metabolic state in these cells. Similarly, loss of Bad over Bim failed to protect primary lymphocytes beyond levels caused by loss of Bim (10). A notable exception, recapitulated here, was that thymocyte death triggered by gamma-irradiation that was partially dependent on Bad and cell loss in its absence was mitigated when Bmf was lacking simultaneously (Figure 4). The latter observation is consistent with our previous findings that thymocyte death caused by irradiation appears to depend in part on Bmf but it remains to be investigated whether this effect is cell autonomous. Furthermore, loss of Bad over Bim selectively delayed cytokine-deprivation mediated death of mitogen-activated B cells (10), suggesting that Bad can impact on B cell homeostasis under certain conditions, in line with the observed B cell accumulation in our study. In addition, we noted that pre-B cells showed reduced death in culture when both proteins were lacking, consistent with its expression pattern. However, it remains to be established if Bmf and Bad are involved in apoptosis in response to checkpoint failure during development, and/or upon antigenic stimulation of mature B cells that would explain the noted B cell lymphadenopathy (Table 1).
Despite these mild defects in apoptosis we observed a reduced lifespan of double-deficient mice due to accelerated onset of spontaneous tumorigenesis. Single mutant animals showed no accelerated tumorigenesis, but a skewing of the tumor profile towards lymphoid malignancies whenever Bad was lacking (Table 2). Our own observations support the notion that the shortened lifespan, noted in Bad knockout mice earlier (9), is mainly due to difference in the genetic backgrounds analyzed, as suggested recently also by Kelly et al., who failed to observe spontaneous tumorigenesis over an observation period of 18 month (Kelly et al. 2010) that was also confirmed in our study (Figure 5a).
Remarkably, although Bad-deficient mice were reported to show defects in glucose tolerance (20) and diabetes associates with increased risk for liver cancer (32), we observed a trend to HCC development whenever Bmf was lost (Figure 5), albeit tumor numbers are admittedly to low to draw formal conclusions. However, our observation is consistent with a possible role of Bmf in human hepatocellular carcinoma that is regulated by the micro RNA (miR-221) in hepatocellular carcinoma cell lines. Of note, high levels of miR221 were found to correlate with tumor multifocality and reduced time to recurrence after surgery (33).
Collectively, we have found that the simultaneous absence of Bad and Bmf is dispensable for embryogenesis and reproduction. It also has only minor effects on immune homeostasis and causes only minor changes that go beyond those observed in single-deficient mice, thereby affecting mainly the B cell lineage. Redundancy in apoptotic cell death initiation appears restricted to the spontaneous death of thymocytes under glucose limitation and that of pre-B cells. However, our investigations revealed that the tumor-suppressive potential of Bad and Bmf is not limited to oncogene- or irradiation-driven forms of lymphoma (11;23;34) but also contributes to the suppression of spontaneous transformation with a possible role for Bmf in the inhibition of hepatocellular carcinoma development, warranting further investigations into the role of Bmf in liver cancer and treatment.
Material and methods
Mice
All used mice were maintained on a C57BL/6 genetic background. The generation and genotyping of the Bad- and Bmf-deficient mice has been previously described (9;23).
Cell culture and reagents
Thymocytes and sorted B- and T-cells were cultured in DMEM (Dulbecco modified Eagle medium, PAA) supplemented with 10% FCS (fetal calf serum, PAA), 250μM l-glutamine (Gibco), nonessential amino acids (Invitrogen), penicillin/streptomycin (Sigma-Aldrich) and 50μM 2-mercaptoethanol.
Antibodies for flow cytometry
The monoclonal antibodies used, and their specificities, are as follows: CD4-PE (RM4-5), CD4-Alexa647 (RM4-5), TCR-β-FITC (H57-597), Mac1-APC (M1/70), Gr1-bio (RB6-8C5), B220-FITC (RA3-6B 2), IgG-bio (polyclonal), IgD-FITC (11-26c.2a), CD5-PE (53-7.3), CD43-bio (R2/60), CD19-PE (MB19-1), CD44-bio (IM7), CD127-PE-Cy7 (A7R34), Sca1-FITC (D7), Lin-bio Flow Panel (B220-bio, Gr1-bio, CD3e-bio, Ter119-bio, CD11b-bio) all Ebioscience and, CD138-PE (281-2), CD62L-APC (MEL-14), CD25-APC (PC61), B220-PE (RA3-6B2) all Becton Dickinson, CD19-bio (6D5), IgM-APC (RMM-1), IgM-PE (RMM-1), CD23-PE (B3B4), c-kit-APC (2B8), CD8a-Alexa647 (53-6.7), CD21-bio (7E9), CD8a-FITC (53-6.7), Ter119-bio (TER-119), Annexin V-FITC from Biolegend and Thy1-bio was self labeled. Biotinylated antibodies were detected by the use of streptavidin-R-phycoerythrin (DAKO) or streptavidin-phycoerythrin-Cy7 (Becton Dickinson).
Cell sorting
Single-cell suspensions from peripheral blood, bone marrow, lymph nodes, spleen, and thymus were surface stained with conjugated monoclonal antibodies. Sorting of cells was performed with the usage of a FACSVantage cell sorter (Becton Dickinson). Cells were sorted with the use of B220-PE, CD4-Alexa647 and CD8a-Alexa647. From the living cells that were stained with 7-AAD (Sigma-Aldrich), B cells were selected as B220 positive, and T-cells were taken from the Alexa647 (CD4 and CD8) positive population. Pre-B cells were sorted as living (7-AAD negative), B220-FITC, c-Kit-APC positive and IgM-PE negative. Further they were separated by size as small and large pre-B cells based on forward-side scattering.
Cell lysis and Immunoblotting
The single cell suspensions of organs and sorted cell substes were lysed in RIPA buffer (150mM NaCl, 50mM Tris, 1% (v/v) NP40, 0.5% (v/v) sodium deoxycholate), 0.1% (v/v) SDS) with phosphatase inhibitors (PhosSTOP, Roche). Western blotting was performed as previously described (35). The membranes were probed with the subsequent primary antibodies: Bad CS9292, Mcl1, Bim CS2819, Bcl-xl CS2764, BID CS2003 and p38 MAPK CS9212 from Cell Signaling, Bmf 17A9 from Enzo Life Sciences and Bcl2 3F11 from Becton Dickinson. The proper loading was confirmed by probing the membranes with p38 MAPK. HRP (horseradish peroxidise) labeled secondary antibodies were used in combination with ECL (Enhanced Chemiluminescence) Western blotting detection reagents (GE Healthcare).
Cell death assays
For the induction of cell death, the following reagents were used: dexamethasone, etoposide, suberoylanilide hydroxamic acid (SAHA), ionomycin (Sigma), staurosporine (Sigma). The percentage of viable cells was quantified by flow cytometric analysis in a FACScan (Becton Dickinson) using 7-AAD (1μg/ml) and Annexin V-FITC (Biolegend). Living cells were characterised as being negative for both markers. To determine metabolic activity intracellular ATP content was quantified using the CellTiter-Glo Luminenscent Cell Viability Assay (Promega) according to the manufacturer’s protocol (3×104 cells were seeded per 96 well in triplicates). Release of lactate dehydrogenase (LDH) was monitored using the CytoTox 96 Non-Radioactive Cytotoxicity Assay (Promega) according to the company’s protocol. (2×104 thymocytes per 96-well were seeded in triplicates).
Lactate Assay
Measurement of intracellular L-lactate was performed by the usage of the Lacate Colorimetric Assay Kit from Abcam. 1×107 thymocytes per treatment were cultured for 24h in a 24well plate, harvested sedimented by centrifugation and washed twice in PBS. After repeated centrifugation, the pellet was lysed in enzyme containing reaction buffer. To correct for non-selective enzymatic background activity, cells were mock-treated and lysed in reaction buffer without enzyme. Absolute lactate concentration in cell lysates was calculated based on OD-values obtained using the provided lactate standards.
Preparation of histological sections
Organs were fixed in 4% PFA (paraformaldehyde) in PBS, processed according to standard procedures, and stained in H&E (hematoxylin and eosin).
Statistical analysis
Estimation of statistical differences between groups was carried out using the unpaired Student t-test or ANOVA analysis, where appropriate. Comparison of tumor onset was performed using a log-rank test, the χ2-test and the Fisher’s exact test was used for comparison of frequency distributions. P-values of <0.05 were considered to indicate statistically significant differences.
Supplementary Material
Suppl. Fig. 1: Organ weight analysis of mice lacking Bad and/or Bmf
Organ weight of 8 to 10 week old wt, bmf−/−, bad−/−, and bmf−/−bad−/− mice was compared (a), or split according to gender prior to comparison; female (b) and male (c). * p<0.05; ** p<0.0001.
Suppl. Fig. 2: Apoptosis induction in primary thymocytes
Primary thymocytes of the indicated genotypes were cultured for 12h in DMEM medium in the presence or absence of 10% serum and graded concentrations of glucose. To discriminate apoptosis from necroptosis the caspase inhibitor Q-VD-OPh (QVD; 20μM) or the necroptosis inhibitor Nec-1 (50μM) were added to the cultures. Addition of DMSO (0.5%) served as solvent control. Survival was monitored by AnnexinV/PI staining and flow cytometric analysis. Bars represent means ± the SEM from three independent experiments using n ≥ 3 animals per genotype are shown. * p<0.05; QVD treated vs. solvent control.
Suppl. Fig. 3: Assessment of necrosis in primary thymocytes
Cells were cultured in different media as described in suppl. Fig. 2, and the percentage of PI-positive dead cells was performed by measuring PI-uptake over time in a flow cytometer. Means ± the SEM from four independent experiments using n ≥ 4 animals per genotype are shown. No significant differences were observed between genotypes or between controls, solvent or Nec-1 treated cells.
Suppl. Fig. 4: Evaluation of cell death by LDH-release
Primary thymocytes of the indicated genotypes were cultured for 24h in DMEM medium in the presence or absence of 10% serum and graded glucose concentrations. Loss of viability was assessed by the release of lactate-dehydrogenase (LDH) in the cell culture supernatant after 24h and 48h by colorimetric analysis in microtiter plates. Means ± the SEM from three independent experiments using n ≥ 3 animals per genotype are shown. No significant differences were observed across genotypes. * p<0.05 compared to wt.
Suppl. Fig. 5: Quantification of ATP and lactate levels in primary thymocytes
(a) Primary thymocytes of the indicated genotypes were cultured for 24h or 48h in the presence of serum and graded glucose concentrations. To determine the metabolic capacity of these cells intracellular ATP levels were analyzed after cell harvest or after 24h or 48h in culture. Symbols represent means ± the SEM of n ≥ 3 animals per genotype. * p<0.05; ** p<0.01 (wt compared to other genotypes at 24h) (b) Intracellular L-lactate levels were measured by colorimetric analysis to monitor differences between the genotypes. Cells were cultured as in (a) for 24h. Control refers to lactate levels present in thymocytes immediately after isolation. No significant differences were observed across genotypes.
Acknowledgments
We thank A. Strasser and the late S. Korsemeyer for mice and reagents. I. Gaggl for mouse genotyping, C. Soratroi for technical assistance, K. Rossi for animal care, G. Böck for cell sorting. This work was supported by grants from the “Tiroler Krebshilfe” to FB and CW, the “Tiroler Wissenschaftsfond” (TWF) to VL, as well as the Austrian Science Fund, FWF (Y212-B12; P23510-B19) to AV and the Integrated Center for Research and Therapy (IFTZ) of the Innsbruck Medical University to AV and FB. CW is recipient of a DOC-fFORTE doctoral fellowship sponsored by the Austrian Academy of Science (ÖAW).
Footnotes
Conflict of interest: The authors declare no conflict of interest.
The authors have no competing financial interests.
Reference List
- (1).Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008 Jan;9(1):47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- (2).Chipuk JE, Green DR. How do BCL-2 proteins induce mitochondrial outer membrane permeabilization? Trends Cell Biol. 2008 Apr;18(4):157–64. doi: 10.1016/j.tcb.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002 Sep;2(3):183–92. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- (4).Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR, et al. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol Cell. 2005 Feb 18;17(4):525–35. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- (5).Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005 Feb 4;17(3):393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- (6).Maiuri MC, Le TG, Criollo A, Rain JC, Gautier F, Juin P, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007 May 16;26(10):2527–39. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Danial NN. BAD: undertaker by night, candyman by day. Oncogene. 2008 Dec;27(Suppl 1):S53–S70. doi: 10.1038/onc.2009.44. [DOI] [PubMed] [Google Scholar]
- (8).Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, et al. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 2008 Dec 26;135(7):1311–23. doi: 10.1016/j.cell.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Ranger AM, Zha J, Harada H, Datta SR, Danial NN, Gilmore AP, et al. Bad-deficient mice develop diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2003 Aug 5;100(16):9324–9. doi: 10.1073/pnas.1533446100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Kelly PN, White MJ, Goschnick MW, Fairfax KA, Tarlinton DM, Kinkel SA, et al. Individual and overlapping roles of BH3-only proteins Bim and Bad in apoptosis of lymphocytes and platelets and in suppression of thymic lymphoma development. Cell Death Differ. 2010 Oct;17(10):1655–64. doi: 10.1038/cdd.2010.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Frenzel A, Labi V, Chmelewskij W, Ploner C, Geley S, Fiegl H, et al. Suppression of B-cell lymphomagenesis by the BH3-only proteins Bmf and Bad. Blood. 2010 Feb 4;115(5):995–1005. doi: 10.1182/blood-2009-03-212670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, Jang Y, et al. Regulation of PTEN transcription by p53. Mol Cell. 2001 Aug;8(2):317–25. doi: 10.1016/s1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- (13).Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002 Jul;2(7):489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- (14).Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009 Aug;9(8):550–62. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- (15).Datta SR, Katsov A, Hu L, Petros A, Fesik SW, Yaffe MB, et al. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol Cell. 2000 Jul;6(1):41–51. [PubMed] [Google Scholar]
- (16).Lee JW, Soung YH, Kim SY, Nam SW, Kim CJ, Cho YG, et al. Inactivating mutations of proapoptotic Bad gene in human colon cancers. Carcinogenesis. 2004 Aug;25(8):1371–6. doi: 10.1093/carcin/bgh145. [DOI] [PubMed] [Google Scholar]
- (17).Teo K, Gemmell L, Mukherjee R, Traynor P, Edwards J. Bad expression influences time to androgen escape in prostate cancer. BJU Int. 2007 Sep;100(3):691–6. doi: 10.1111/j.1464-410X.2007.07001.x. [DOI] [PubMed] [Google Scholar]
- (18).Kuroda J, Puthalakath H, Cragg MS, Kelly PN, Bouillet P, Huang DC, et al. Bim and Bad mediate imatinib-induced killing of Bcr/Abl+ leukemic cells, and resistance due to their loss is overcome by a BH3 mimetic. Proc Natl Acad Sci U S A. 2006 Oct 3;103(40):14907–12. doi: 10.1073/pnas.0606176103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Gilmore AP, Valentijn AJ, Wang P, Ranger AM, Bundred N, O’Hare MJ, et al. Activation of BAD by therapeutic inhibition of epidermal growth factor receptor and transactivation by insulin-like growth factor receptor. J Biol Chem. 2002 Aug 2;277(31):27643–50. doi: 10.1074/jbc.M108863200. [DOI] [PubMed] [Google Scholar]
- (20).Danial NN, Gramm CF, Scorrano L, Zhang CY, Krauss S, Ranger AM, et al. BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature. 2003 Aug 21;424(6951):952–6. doi: 10.1038/nature01825. [DOI] [PubMed] [Google Scholar]
- (21).Puthalakath H, Villunger A, O’Reilly LA, Beaumont JG, Coultas L, Cheney RE, et al. Bmf: a proapoptotic BH3-only protein regulated by interaction with the myosin V actin motor complex, activated by anoikis. Science. 2001 Sep 7;293(5536):1829–32. doi: 10.1126/science.1062257. [DOI] [PubMed] [Google Scholar]
- (22).Pinon JD, Labi V, Egle A, Villunger A. Bim and Bmf in tissue homeostasis and malignant disease. Oncogene. 2008 Dec;27(Suppl 1):S41–S52. doi: 10.1038/onc.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Labi V, Erlacher M, Kiessling S, Manzl C, Frenzel A, O’Reilly L, et al. Loss of the BH3-only protein Bmf impairs B cell homeostasis and accelerates gamma irradiation-induced thymic lymphoma development. J Exp Med. 2008 Mar 17;205(3):641–55. doi: 10.1084/jem.20071658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Ploner C, Rainer J, Niederegger H, Eduardoff M, Villunger A, Geley S, et al. The BCL2 rheostat in glucocorticoid-induced apoptosis of acute lymphoblastic leukemia. Leukemia. 2008 Feb;22(2):370–7. doi: 10.1038/sj.leu.2405039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, et al. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 1999 Nov 26;286(5445):1735–8. doi: 10.1126/science.286.5445.1735. [DOI] [PubMed] [Google Scholar]
- (26).Erlacher M, Michalak EM, Kelly PN, Labi V, Niederegger H, Coultas L, et al. BH3-only proteins Puma and Bim are rate-limiting for gamma-radiation- and glucocorticoid-induced apoptosis of lymphoid cells in vivo. Blood. 2005 Dec 15;106(13):4131–8. doi: 10.1182/blood-2005-04-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Erlacher M, Labi V, Manzl C, Bock G, Tzankov A, Hacker G, et al. Puma cooperates with Bim, the rate-limiting BH3-only protein in cell death during lymphocyte development, in apoptosis induction. J Exp Med. 2006 Dec 25;203(13):2939–51. doi: 10.1084/jem.20061552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Coultas L, Bouillet P, Stanley EG, Brodnicki TC, Adams JM, Strasser A. Proapoptotic BH3-only Bcl-2 family member Bik/Blk/Nbk is expressed in hemopoietic and endothelial cells but is redundant for their programmed death. Mol Cell Biol. 2004 Feb;24(4):1570–81. doi: 10.1128/MCB.24.4.1570-1581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005 Feb 4;17(3):393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- (30).Hubner A, Cavanagh-Kyros J, Rincon M, Flavell RA, Davis RJ. Functional cooperation of the proapoptotic Bcl2 family proteins Bmf and Bim in vivo. Mol Cell Biol. 2010 Jan;30(1):98–105. doi: 10.1128/MCB.01155-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Zhao Y, Coloff JL, Ferguson EC, Jacobs SR, Cui K, Rathmell JC. Glucose metabolism attenuates p53 and Puma-dependent cell death upon growth factor deprivation. J Biol Chem. 2008 Dec 26;283(52):36344–53. doi: 10.1074/jbc.M803580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Wang P, Kang D, Cao W, Wang Y, Liu Z. Diabetes mellitus and risk of hepatocellular carcinoma: a systematic review and meta-analysis. Diabetes Metab Res Rev. 2011 Sep 5; doi: 10.1002/dmrr.1291. [DOI] [PubMed] [Google Scholar]
- (33).Gramantieri L, Fornari F, Ferracin M, Veronese A, Sabbioni S, Calin GA, et al. MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin Cancer Res. 2009 Aug 15;15(16):5073–81. doi: 10.1158/1078-0432.CCR-09-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Kelly PN, White MJ, Goschnick MW, Fairfax KA, Tarlinton DM, Kinkel SA, et al. Individual and overlapping roles of BH3-only proteins Bim and Bad in apoptosis of lymphocytes and platelets and in suppression of thymic lymphoma development. Cell Death Differ. 2010 Oct;17(10):1655–64. doi: 10.1038/cdd.2010.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Villunger A, Huang DC, Holler N, Tschopp J, Strasser A. Fas ligand-induced c-Jun kinase activation in lymphoid cells requires extensive receptor aggregation but is independent of DAXX, and Fas-mediated cell death does not involve DAXX, RIP, or RAIDD. J Immunol. 2000 Aug 1;165(3):1337–43. doi: 10.4049/jimmunol.165.3.1337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Fig. 1: Organ weight analysis of mice lacking Bad and/or Bmf
Organ weight of 8 to 10 week old wt, bmf−/−, bad−/−, and bmf−/−bad−/− mice was compared (a), or split according to gender prior to comparison; female (b) and male (c). * p<0.05; ** p<0.0001.
Suppl. Fig. 2: Apoptosis induction in primary thymocytes
Primary thymocytes of the indicated genotypes were cultured for 12h in DMEM medium in the presence or absence of 10% serum and graded concentrations of glucose. To discriminate apoptosis from necroptosis the caspase inhibitor Q-VD-OPh (QVD; 20μM) or the necroptosis inhibitor Nec-1 (50μM) were added to the cultures. Addition of DMSO (0.5%) served as solvent control. Survival was monitored by AnnexinV/PI staining and flow cytometric analysis. Bars represent means ± the SEM from three independent experiments using n ≥ 3 animals per genotype are shown. * p<0.05; QVD treated vs. solvent control.
Suppl. Fig. 3: Assessment of necrosis in primary thymocytes
Cells were cultured in different media as described in suppl. Fig. 2, and the percentage of PI-positive dead cells was performed by measuring PI-uptake over time in a flow cytometer. Means ± the SEM from four independent experiments using n ≥ 4 animals per genotype are shown. No significant differences were observed between genotypes or between controls, solvent or Nec-1 treated cells.
Suppl. Fig. 4: Evaluation of cell death by LDH-release
Primary thymocytes of the indicated genotypes were cultured for 24h in DMEM medium in the presence or absence of 10% serum and graded glucose concentrations. Loss of viability was assessed by the release of lactate-dehydrogenase (LDH) in the cell culture supernatant after 24h and 48h by colorimetric analysis in microtiter plates. Means ± the SEM from three independent experiments using n ≥ 3 animals per genotype are shown. No significant differences were observed across genotypes. * p<0.05 compared to wt.
Suppl. Fig. 5: Quantification of ATP and lactate levels in primary thymocytes
(a) Primary thymocytes of the indicated genotypes were cultured for 24h or 48h in the presence of serum and graded glucose concentrations. To determine the metabolic capacity of these cells intracellular ATP levels were analyzed after cell harvest or after 24h or 48h in culture. Symbols represent means ± the SEM of n ≥ 3 animals per genotype. * p<0.05; ** p<0.01 (wt compared to other genotypes at 24h) (b) Intracellular L-lactate levels were measured by colorimetric analysis to monitor differences between the genotypes. Cells were cultured as in (a) for 24h. Control refers to lactate levels present in thymocytes immediately after isolation. No significant differences were observed across genotypes.