Abstract
The application of paired-end next generation sequencing approaches has made it possible to systematically characterize rearrangements of the cancer genome to base-pair level. Utilizing this approach, we report the first detailed analysis of ovarian cancer rearrangements, comparing high-grade serous and clear cell cancers, and these histotypes with other solid cancers. Somatic rearrangements were systematically characterized in eight high-grade serous and five clear cell ovarian cancer genomes and we report here the identification of > 600 somatic rearrangements. Recurrent rearrangements of the transcriptional regulator gene, TSHZ3, were found in three of eight serous cases. Comparison to breast, pancreatic and prostate cancer genomes revealed that a subset of ovarian cancers share a marked tandem duplication phenotype with triple-negative breast cancers. The tandem duplication phenotype was not linked to BRCA1/2 mutation, suggesting that other common mechanisms or carcinogenic exposures are operative. High-grade serous cancers arising in women with germline BRCA1 or BRCA2 mutation showed a high frequency of small chromosomal deletions. These findings indicate that BRCA1/2 germline mutation may contribute to widespread structural change and that other undefined mechanism(s), which are potentially shared with triple-negative breast cancer, promote tandem chromosomal duplications that sculpt the ovarian cancer genome. Copyright © 2012 Pathological Society of Great Britain and Ireland.
Keywords: ovarian cancer, structural rearrangements, TSHZ3
Introduction
Somatically acquired structural genomic rearrangements are common in most solid cancers and are especially so in ovarian cancer 1. Ovarian cancer is a collective term for a number of distinctly different histotypes and tumours of varying degrees of malignancy 2. The most common epithelial ovarian cancer histotypes include serous, clear cell, mucinous and endometrioid. Amongst these, high-grade serous cancers (HGSCs) are the most common, accounting for approximately 70% of all cases of invasive epithelial ovarian cancer and the majority of deaths.
Increasingly comprehensive genomic analyses of HGSCs and clear cell cancers are providing insights into pathways of transformation and the molecular determinants of response to therapy. Our previous gene expression profiling of HGSCs identified four molecular subtypes 3, which are associated with different clinical outcomes 3, 4. A signalling pathway involving MYCN, LIN28B and LET7 is associated with one of the four subtypes 4, suggesting that specific pathway activation may drive the development and behaviour of one or more HGSCs molecular subtypes. DNA copy number analyses have shown that gains and losses are particularly frequent in HGSCs 5, 6, with a high level of genomic disorganization apparent in most cancer samples. A recent detailed genomic analysis of 500 HGSCs by the TCGA consortium identified frequently amplified and deleted regions of the HGSC genome 1. Approximately 50% of HGSCs had defects in the BRCA1/2 pathway, either through germline or somatic mutation or methylation of pathway members.
Clear cell cancers are associated with endometriosis 7, and have been recently found to harbour somatic mutations in the ARID1A gene in approximately 50% of tumours 8, 9. The patterns of gene expression and copy number change seen in clear cell ovarian cancer are distinct from HGSCs, and frequently involve amplification and over expression of cytokines including IL6, receptor tyrosine kinases and other downstream signalling components 10, 11. The gene expression profiles of ovarian clear cell cancers are similar to renal and uterine clear cell tumours 12. Favourable responses have been observed in a small number of ovarian clear cell patients to sunitinib 10, a drug with considerable activity in renal clear cell cancer.
Next-generation DNA sequence analysis is providing an unprecedented level of information about the cancer genome, identifying new mutations 9, 13, 14, the impact of mutagens 15, 16 and novel processes that sculpt the cancer genome 17. Here we used paired-end DNA sequencing to seek novel gene fusions and characterize structural changes in ovarian cancer samples, comparing and contrasting HGSCs and clear cell genomes.
Materials and methods
Patient samples and ethics
Tumour samples and clinical data were obtained from women enrolled in the Australian Ovarian Cancer Study (http://www.aocstudy.org). All participants provided written informed consent and Human Research Ethics Committee approval was obtained at the Peter MacCallum Cancer Centre (Queensland Institute of Medical Research, University of Melbourne, Australia) and all participating hospitals for the study. Further clinical data, information on biospecimens and microarray analysis are described in the Supplementary methods (see Supporting information).
Next-generation sequencing
Next-generation sequencing and structural variant analysis was carried out as described previously 18. Briefly, 37 bp paired-end reads generated on the Illumina GA2 were aligned to the human reference genome (hg19) using BWA. Rearrangement breakpoints were called when two or more discordantly mapped read pairs supported the same underlying event. Breakpoints were classified according to the relative orientations and insert sizes of the read pairs into those suggesting deletion, translocation, inversion or tandem duplication (insertion). Candidate breakpoints were confirmed as somatic by PCR on both tumour and matched normal DNA and mapped to base pair resolution by capillary sequencing. Breakpoints were classified according to the relative orientations and insert sizes of the read pairs into those suggesting deletion, translocation, inversion or tandem duplication (insertion). Rearrangements were further classified based on integration with copy number data to include amplicon junctions, fold-back inversions and genomic shards 19.
Validation of gene rearrangements
cDNA from total RNA for sequenced samples and validation samples was synthesized using M-MLV reverse transcriptase (Promega), as described previously 20. Endpoint RT–PCR was performed according to standard protocols using Thermo-Start DNA polymerase (ThermoScientific). Products were resolved by agarose gel electrophoresis and samples with visible products (for CCNY/CREM and ATP9B rearrangements only) were subjected to a second independent PCR reaction with alternate primer sets. No products were confirmed by a second PCR reaction except for control samples, where rearrangements were initially identified. Gene expression of TSHZ3 was measured by quantitative reverse-transcription PCR (qRT–PCR) normalized to ACTB and HPRT1 control genes and median CT (threshold cycle) values obtained across eight serous samples, using a SYBR Green qPCR assay on the 7900HT Fast Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). RNA was treated with DNase I (Promega) prior to cDNA synthesis to eliminate background amplification of genomic DNA. Primer details are given in Table S4 (see Supporting information).
Germline and somatic analysis of BRCA pathway dysfunction
Complete germline sequencing of BRCA1 and BRCA2 and multiplex ligation-dependent probe amplification (MLPA) for detection of large chromosomal aberrations were undertaken in the National Association of Testing Authorities (NATA) accredited Molecular Pathology Laboratory at the Peter MacCallum Cancer Centre. Detailed methods are provided elsewhere (Alsop et al, in press). Somatic mutations in tumour samples were screened by high-resolution melt (HRM) analysis of all coding exons of BRCA1 and BRCA2, as described by others 21. Gene promoter methylation of tumour DNA was assessed for BRCA1, FANCF and PALB2 using methylation-sensitive HRM, as described previously 22. Reported percentage of methylated allele present in tumour DNA was estimated by comparison to a panel of control samples. No promoter methylation of FANCF or PALB2 was identified in the 13 samples screened, and only one sample showed promoter methylation of BRCA1 (see Supporting information, Table S1).
Results
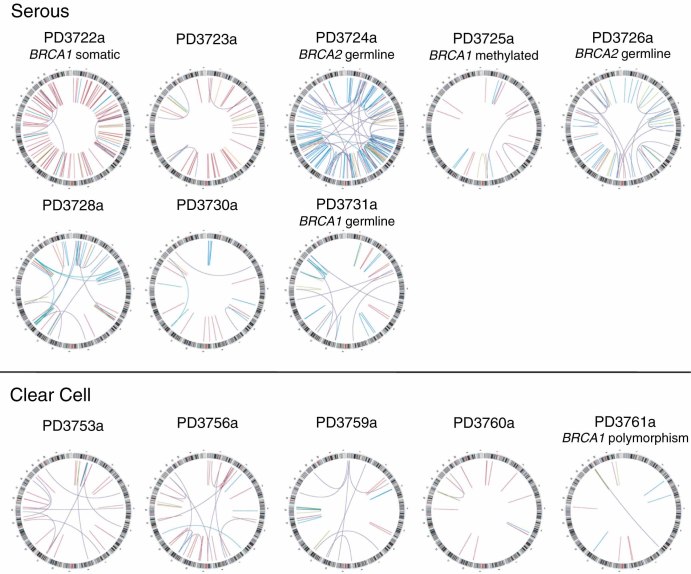
To identify DNA rearrangements in ovarian cancer genomes, we carried out paired end sequencing of tumour DNA from 13 ovarian tumour samples (eight HGSCs and five clear cell; see Supporting information, Table S1). A total of 1.1 × 109 37 bp read pairs generated 7 × 1010 base pairs that could be assigned uniquely to the reference genome equating to 4.3–16.5-fold physical coverage per sample. Putative structural rearrangements were identified from incorrectly mapping read pairs 18, 23 and 634 confirmed somatically acquired DNA rearrangements were identified across 13 ovarian cancers (range 13–150/sample), with base pair sequence level resolution of 598 individual breakpoints (94%; see Supporting information, Table S2). Each genome displayed a different spectrum of unique somatically acquired rearrangements (Figure 1), with varying proportions of rearrangements in each class (Table 1).
Figure 1.
Genomic rearrangements of 13 ovarian cancer genomes. The chromosomes of the reference genome are drawn around the circumference of each circos plot 42. Rearrangements are represented by lines linking somatically acquired breakpoints: orange, fold back inversions; light blue, one or both end map to an amplicon; purple, translocations; green, inversions; red, tandem duplications; dark blue, deletions. BRCA1/2 mutation status or mechanism of inactivation indicated
Table 1.
Breakpoint frequencies in 13 ovarian cancer cases illustrating correlations with BRCA1/2 status and histotype
| Sample | Histotype | BRCA status | Totalbreakpoints | Translocations (%) | Deletions (%) | Tandem duplications (%) | Amplicons (%) | Inversions (%) | Fold-back | Shards (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| PD3722a | Serous | Somatic (BRCA1) | 114 | 4 | 7 | 71 | 7 | 8 | 2 | 2 |
| PD3723a | Serous | Wt | 37 | 8 | 3 | 68 | 3 | 11 | 8 | 0 |
| PD3724a | Serous | Germline (BRCA2) | 150 | 24 | 51 | 13 | 3 | 6 | 1 | 3 |
| PD3725a | Serous | Methylated (BRCA1) | 19 | 11 | 16 | 68 | 0 | 5 | 0 | 0 |
| PD3726a | Serous | Germline (BRCA2) | 60 | 22 | 42 | 17 | 0 | 18 | 0 | 2 |
| PD3728a | Serous | Wt | 75 | 9 | 19 | 16 | 36 | 5 | 11 | 4 |
| PD3730a | Serous | Wt | 21 | 10 | 29 | 48 | 10 | 0 | 5 | 0 |
| PD3731a | Serous | Germline (BRCA1) | 30 | 23 | 27 | 20 | 17 | 7 | 3 | 3 |
| PD3753a | Clear cell | Wt | 31 | 32 | 3 | 58 | 0 | 6 | 0 | 0 |
| PD3756a | Clear cell | Wt | 39 | 23 | 10 | 62 | 3 | 3 | 0 | 0 |
| PD3759a | Clear cell | Wt | 30 | 27 | 17 | 30 | 0 | 20 | 0 | 7 |
| PD3760a | Clear cell | Wt | 15 | 7 | 7 | 73 | 0 | 7 | 0 | 7 |
| PD3761a | Clear cell | Polymorphism of likely low clinical significance | 13 | 8 | 23 | 38 | 0 | 23 | 8 | 0 |
Categories with the highest proportion of breaks for each tumour are in bold italics.
By classifying tumours according to the dominant class of rearrangement, three distinct mutation profiles were observed in HGSCs. In four of the HGSCs, half or more of the chromosomal rearrangements involved tandem duplications (median 410 kb), including one case with 81 tandem duplication events detected (PD3722a). For three HGSCs, deletions were the dominant rearrangement class, with the remaining serous cancer sample having a genome dominated by junctions within amplicons (Table 1). The three cases showing a high frequency of deletions had either a germline BRCA1 or BRCA2 mutation (PD3724a, PD3726a and PD3731a) and the deletions in these cases were small (median 3.2 kb) when compared to deletions in cases without germline BRCA1/2 mutations (median 288 kb). Over-representation of deletions in BRCA1/2 mutant compared to wild-type cases as a proportion of all rearrangements was statistically significant (p < 3.91 × 10−11 by a Poisson regression test). Two HGSCs cases with somatic alteration of BRCA1, either through point mutation or promoter hypermethylation (PD3722a and PD3725a, respectively), did not show the deletion/translocation phenotype. Fewer rearrangements were observed in ovarian clear cell cancers compared with HGSCs (Table 1), consistent with our previous copy number analysis 10. The types of rearrangement present within each genome were more evenly distributed between the classes in the clear cell histotype. Tandem duplications were the dominant class in all five clear cell cases, but relatively high proportions of translocations, deletions and inversions were also seen (Table 1). Statistical analyses utilizing a generalized linear model (GLM) approach indicated that the occurrence of deletions (p = 0.0053) and amplicons (p = 0.010) was significantly different between clear cell and HGSCs cases. Further, the overall p value for the probability that the distributions differ (from the log likelihood, when compared against the null model of no interaction between cancer type and rearrangement class distribution) was 3.13 × 10−6.
We have previously observed high proportions of tandem duplications in a subset of breast cancer genomes 23. Given the apparent similarity in pattern and distribution of this class of rearrangement, we undertook a comparative analysis of ovarian and other solid cancers for which data was available. Although HGSCs and clear cell cancers are considered to have different aetiologies and patterns of somatic mutation, we considered them as a single group for the purposes of the analysis, as tandem duplications were found in both histotypes. We compared 634 ovarian cancer rearrangements identified in this study, with 994 rearrangements identified in primary breast cancers 23, 352 rearrangements in pancreatic cancers 19 and 464 rearrangements recently reported in an analysis of seven prostate cancer genomes 24. Comparing the proportions of each class of rearrangement in each cancer type revealed significant differences (χ2p value for effects of interaction between cancer type and rearrangement type = 1.88 × 10−68). There were significantly more tandem duplications in breast and ovarian cancers compared to pancreatic and prostate cancers (p = 1.6 × 10−12). Amplified fold back inversions were much more common in pancreatic than breast or ovarian cancers (p = 4.94 × 10−04).
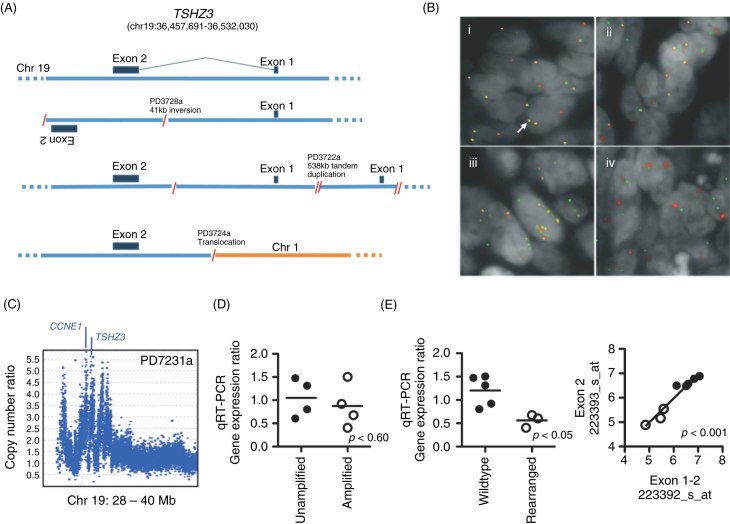
In total, 371 (59%) of the somatic rearrangement breakpoints identified in this study fell in or within 10 kb of gene ‘footprints’ (ie within Ensembl gene coordinate boundaries), with 16 gene footprints disrupted in more than one sample (see Supporting information, Table S3). While these mostly occurred in genes with large genomic footprints, suggesting that a proportion are likely passenger events, several warrant further consideration. TSHZ3, a homeobox transcription factor that has been implicated in ureter formation 25 and development of respiratory neurons 26, was the most commonly disrupted gene, occurring in 3/8 HGSC samples. The breakpoints (caused by translocation, inversion or tandem duplication) all lie in the only intron and map within 29 kb of each other (Figure 2A). The gene is approximately 74 kb in length and we have not found it deleted in other cancer types previously (unpublished data).
Figure 2.
(A) Wild-type TSHZ3 locus (top) and schematics of the three rearrangements identified by next-generation sequencing. (B) Tissue microarray images of four cases with varying TSHZ3 rearrangements: (1) loss of the 5′ end of TSHZ3 with example of locus without rearrangement indicated by a white arrow; (ii) balanced break; (iii) amplification; and (iv) breakage with amplification of the 3′ end of TSHZ3. Green, BAC probes RP11-280H11 and RP11-241C16 (5′ TSHZ3); red, RP11-161K19 and RP11-164O11 (3′ TSHZ3). (C) Affymetrix SNP 6.0 copy number data for tumour sample PD7231a, showing multiple focal amplifications incorporating CCNE1 and TSHZ3. (D) Scatter plot of TSHZ3 expression by gene copy number status, where amplification is > three copies. (E) Scatter plot showing TSHZ3 gene expression by qRT–PCR (left) and gene expression microarray (right) in rearranged tumours (open circles) compared to samples without gene rearrangement (closed circles)
Rearrangement of TSHZ3 was further explored by two-colour FISH analysis of 90 HGSCs samples on tissue microarrays (TMA), which included three cases (PD3722a, PD3724a, PD2728a) identified in the rearrangement screen. In total, 11/90 HGSC cases were found to have rearrangements involving the TSHZ3 locus. The small tandem duplication in PD3722a was not detectable on TMA FISH, whilst the translocation in PD3724a was readily confirmed. The inversion in PD3728a was not discernable on FISH; however, the locus was amplified. In the remaining cases, there were two main patterns of rearrangement, split roughly equally between amplification and rearrangement breaks in the gene, including two cases with apparently balanced breaks, confirming TSHZ3 as recurrently rearranged (Figure 2B).
Assessing the importance of TSHZ3 in ovarian cancer is confounded by its proximity to CCNE1, mapping 1.45 Mb telomeric on 19q12 (Figure 2C). CCNE1 is known to be amplified and operative as a cancer gene in serous ovarian cancer 27, and the breakpoints in TSHZ3 could be collateral to amplification involving CCNE1. To further evaluate this, those TMA cases with TSHZ3 breakpoints and available DNA (n = 10) were evaluated using Affymetrix SNP 6.0 gene arrays for evidence of independence from CCNE1 amplification. Five of 10 cases had high-level amplification of CCNE1 with associated gain and breaks in TSHZ3, a frequency substantially above the 15–20% seen in unselected cases 20. Sample PD7231a shows a complex pattern of copy number change, including multiple, focal, high-level amplifications incorporating both CCNE1 and TSHZ3 (Figure 2C). In this case, TSHZ3 and CCNE1 are on separate segments, although they are presumably co-amplified. Of the five cases without CCNE1 amplification, four had evidence for low-level copy-number gain (copy numbers 3 or 4) of both TSHZ3 and CCNE1, with TSHZ3 breakpoints detectable on SNP array in two of the cases; PD7229a and PD7730a both had balanced rearrangements on FISH and were not detectable on SNP array, as expected. In the remaining case, PD3722a, there was no copy number gain for TSHZ3 other than the small tandem duplication of the locus, whereas CCNE1 had a single copy gain. These data suggest that TSHZ3 rearrangement can occur at least in some instances independently of CCNE1 amplification. Additionally, Affymetrix SNP 6.0 data obtained from the TCGA ovarian cancer study (see Methods) was also evaluated for independent breakpoints affecting TSHZ3 in the absence of CCNE1 amplification. This analysis revealed five cases with evidence for TSHZ3 breakpoints apparently unrelated to CCNE1 gain or amplification.
Quantitative RT–PCR analysis showed TSHZ3 expression was not associated with gene amplification (Figure 2D); however, there was evidence of reduced expression of TSHZ3 in rearranged cases compared with HGSCs without disruption of the locus (Figure 2E). We also compared gene expression by microarray, using probe sets detecting transcripts across exon 1 and 2 boundaries (223392_s_at) and within exon 2 only (223393_s_at). No differences between probe sets were detected in rearranged versus wild-type samples, suggesting that there is no aberrant transcription of short transcripts (Figure 2E). To further identify potential fusion transcripts we employed 5′ RACE across all eight sequenced samples. We were, however, only able to detect wild-type products (data not shown). If novel transcripts were generated in the rearranged samples, it is possible that low expression may have limited their detection.
These findings suggest that if TSHZ3 is contributing to ovarian cancer, it may be through down-regulation or loss of function. However, arguing against this is the lack of truncating mutations reported in the recently released data from whole-exome sequencing of more than 250 serous ovarian cancers, where only two missense and two silent somatic mutations were identified 1.
We examined TSHZ3 gene expression further in two large independent datasets of HGSCs (see Supporting information, Figure S1). Although rearrangement of TSHZ3 is associated with reduced gene expression, we found that a high level of TSHZ3 was associated with shorter progression-free survival only in the AOCS dataset (p < 0.04) 3. In both AOCS and TCGA 1 cohorts, high TSHZ3 expression was associated with the C1 molecular subtype, which is characterized by intense tumour stromal and epithelial–mesenchymal transition gene expression signatures and poor clinical outcome.
Non-identical tandem duplications of approximately 1 and 0.5 Mb were identified in 2/8 HGSCs samples that disrupted the gene encoding Rho GTPase activating protein 6 (ARHGAP6) on the X chromosome (see Supporting information, Table S3). These two cases were also rearranged for TSHZ3. ARHGAP6 functions as a GAP for RhoA GTPase, which has been implicated in cell motility, and angiogenesis and has been reported to be up-regulated in multiple cancers, including serous ovarian carcinomas 28. Normally, ARHGAP6 would serve to limit the activity of RhoA by activating the intrinsic GTPase activity resulting in conversion to the inactive GDP-bound form of the enzyme 29. The locus encodes multiple splice forms, but no significant expression differences were discernable in the rearranged versus non-rearranged cases.
Amongst the other genes broken, ARID1A, a member of the SWI/SNF chromatin remodelling complex, was found to be presumptively inactivated by a ∼130 kb inversion on chromosome 1 in the clear cell case, PD3753a. Frequent inactivating point mutations and a single rearrangement were recently reported in clear cell ovarian cancer 8, 9, consistent with the inactivating rearrangement identified here. Indeed, four of the five cases reported here were also included in Wiegand et al 9, with one case, PD3761a, having frameshift mutation in ARID1A. Also, ARID1A has been previously implicated in other human cancers 30, 31 and recently found to be mutated recurrently in clear cell renal cell carcinoma 32. Additionally, breakpoints in PPP2R2B, a component of the regulatory subunit B of serine/threonine phosphatase 2 (PP2A) involved in cell growth and division, were found in two samples (one serous and one clear cell tumour). Mutations in the PP2A regulatory subunit A component, PPP2R1A, have previously been shown in ovarian clear cell cancers 8.
Although all 13 HGSCs and clear cell cases had rearrangements, no recurrent gene fusions were detected amongst the discovery set of samples. Nine single instances of in-frame fusion genes and five in-frame internally rearranged genes were identified (Table 2), all occurring in HGSCs. To investigate whether any of these potential gene fusions and internally rearranged genes were transcribed, PCR assays were designed to the exons surrounding the genomic breakpoints and RNA from the relevant cases was assayed by reverse transcription (RT)–PCR. Products were successfully amplified for 7/9 gene fusions but none of the internally rearranged genes, indicating that a substantial proportion of in-frame fusions were expressed. RT–PCR product sequence was verified by conventional sequencing.
Table 2.
In-frame gene fusions and internal gene rearrangements
| Sample | 5′ Gene | 5′ Gene Accession No. | Exons in predicted fusion gene | 3′ Gene | 3′ Gene Accession No. | Exons in predicted fusion gene | Expressed? | Class of rearrangement |
|---|---|---|---|---|---|---|---|---|
| PD3722a | HGS* | NM_004712.3 | 1–8 | SLC16A5* | NM_004695.2 | 6–7 | Yes | Tandem duplication |
| PD3722a | CCNY | NM_145012.4 | 1–11 | CREM | NM_181571.1 | 4–14b | Yes | Other intrachromosomal |
| PD3723a | C2orf67 | NM_152519.2 | 1–3 | MARCH4 | NM_020814.2 | 3–4 | Yes | Other intrachromosomal |
| PD3723a | MRPL43 | NM_176792.1 | 1–5 | SH3PXD2A | NM_014631.2 | 5–14 | No | Tandem duplication |
| PD3723a | C11orf41 | NM_012194.1 | 1–18 | DNAJC24 | NM_181706.4 | 4–5 | Yes | Other intrachromosomal |
| PD3724a | AGGF1 | NM_018046.3 | 1 | SCAMP1 | NM_004866.4 | 3–9 | Yes | Deletion |
| PD3726a | NDUFA11 | NM_175614.2 | 1–2 | STX17 | NM_017919.2 | 5–8 | Yes | Translocation |
| PD3731a | MAP3K1 | NM_005921.1 | 1–13 | SIRPA | NM_001040022.1 | 9–10 | Yes | Translocation |
| PD3760a | SV2B | NM_014848.3 | 1–8 | CRTC3 | NM_022769.3 | 11–15 | No | Shard (TD) |
| PD3722a | PHACTR1 | NM_030948.1 | Duplication of exons 3 & 4 | No | Tandem duplication | |||
| PD3722a | SGCZ | NM_139167.2 | Duplication of exons 2 & 3 | No | Tandem duplication | |||
| PD3726a | CLSTN2 | NM_022131.2 | Deletion of exon 2 | No | Other intrachromosomal | |||
| PD3728a | ATP9B | NM_198531.3 | Duplication of exons 12–15 | ND | Tandem duplication | |||
| PD3760a | THSD4 | NM_024817.2 | Duplication of exons 6 & 7 | No | Tandem duplication |
Alternative splicing of the fusion transcript yields both in-frame and out-of-frame products. ND = not determined.
In order to detect recurrence of novel rearranged transcripts in ovarian cancer, we screened cDNA from an independent validation set of 109 HGSCs for the same exon joining events for six of the in-frame fusion genes found in HGSCs cases (CCNY/CREM, NDUFA11/STX17, AGGF1/SCAMP1, MAP3K1/SIRPA, C2orf67/MARCH4, C11orf41/DNAJC24) and one of the in-frame internally rearranged transcripts (ATP9B). We were unable to detect expression of any of the fusion transcripts in the validation cohort. We next looked for co-expression of CCNY/CREM in our previous analysis of ovarian tumours 3. We reasoned that oncogenic fusion transcripts would likely show gene over-expression and preselection of cases may increase the chance of detecting recurrent low-frequency fusions. We identified 25 samples from 215 HGSCs (independent of the 109-sample validation set) that showed high expression of both CREM and CCNY (expression above median + 0.5× median absolute deviation [MAD]), representing a statistically significantly higher proportion than would be expected by chance (p < 0.05). From these, RNA was available for 15 samples and analysed for expression of the CCNY/CREM fusion. No fusion products were detected in the selected samples, suggesting that if co-expression was the result of rearrangement/fusion, breakpoints varied from the originally defined boundaries.
Discussion
The landscape of rearrangements in HGSCs and clear cell ovarian cancers is characterized by a substantial proportion of cases, showing a predominance of tandem duplications, overlapping a phenotype previously shown in breast cancer 23. In addition, there is evidence for a deletion/translocation phenotype potentially linked to germline BRCA1/2 mutations, with a high prevalence of small deletions being particularly marked in BRCA2 null tumours. Somatic missense mutation and hypermethylation of BRCA1 was not, however, associated with the same mutation phenotype. Therefore, somatic events are unlikely to be functionally equivalent to germline truncating mutations if abrogation of BRCA1/2 function is directly linked to the phenotype. Overall comparison between four different tumour types suggests that multiple different mechanisms are operative in the reconstruction of cancer genomes, likely involving as-yet unidentified exposures and/or genome maintenance defects.
The observed high frequency of tandem duplications in the ovarian cancer samples, especially HGSCs cases, adds to previous data suggesting molecular similarities between triple-negative and HGSCs 33, 34. Of note, the tandem duplication phenotype was present in both histological subtypes of ovarian cancer. This is of interest, as these subtypes show marked differences in cancer gene mutations found 35, expression clustering 10 and response to therapy. Given the apparently different histogenesis 36, our findings suggest convergent acquisition of the tandem duplication phenotype during oncogenesis. Further, tandem duplication was not associated with germline BRCA1 or BRCA2, suggesting that the phenotype is attributable to a yet to be identified common pathogenetic mechanism.
A recent study by Ng et al reported on the presence of a tandem duplicator phenotype in HGSCs 37. Genome paired-end sequencing of PEO1/PEO4 cell lines derived from a BRCA2 mutation carrier showed interchromosomal breakpoints and small deletions consistent with homologous recombination (HR) deficiency. In contrast, the PEO14/PEO23, BRCA wild-type cell lines had intact HR and a tandem duplicator phenotype. Using SNP copy number data, a ‘tandem duplicator-like’ (TD-like) pattern was inferred and used to estimate the phenotype frequency in the TCGA dataset. TD-like copy number aberrations were reported to occur in 12.8% of HGSCs and, consistent with our findings, these were mutually exclusive to BRCA1/2 carrier mutations.
We found no evidence of recurrent fusion genes in our study. While less common than in haematological malignancies and sarcomas, recurrent gene fusions have been identified in other solid tumours, such as TMPRSS2–ERG in prostate 38 and EML4–ALK in non-small cell lung cancers 39. Recently, Salzman et al reported on the identification of the novel ESRRA–C11orf20 gene fusion in serous ovarian cancer, using an ultra-high-throughput RNA sequencing approach 40. Recurrence was reported in approximately 15% of cases (n = 67); however, we saw no evidence of this rearrangement in our screen. Identification of ESRAA–C11orf20 fusions may have been limited by a low frequency of recurrence and the small number of serous tumours analysed in our study.
TSHZ3 was identified as a target of recurrent breakage in ovarian cancer. Gene expression analysis suggests that if TSHZ3 is contributing to pathogenesis, it may be through loss of function or simply down-regulation. Inactivation of TSHZ3 in vivo shows clear gene dosage effects, leading to haploinsufficiency, neonatal lethality and reduced heterozygote pup size compared to wild-type siblings 26. Additionally, TSHZ3 has recently been shown to be among the most down-regulated genes in breast and prostate cancer, suggesting likely relevance in various tumour types 41. Additional work is warranted to clarify mechanisms of deregulation and the role of TSHZ3 in ovarian cancer.
Data access
Next-generation read datasets are available under managed access from the European Genome-Phenome Archive (EGA; http://www.ebi.ac.uk/ega/). Microarray data GEO submission in progress.
Acknowledgments
PAF and MRS would like to acknowledge the Wellcome Trust for support (Grant No. 077012/Z/05/Z). DDLB acknowledges the National Health and Medical Research Council (NHMRC) for funding support (Project Grant No. 628779). JDB would like to acknowledge Cancer Research UK for funding, and support from the University of Cambridge, Hutchison Whampoa Ltd, the Cambridge Experimental Cancer Medicine Centre and the NIHR Biomedical Research Centre. The Australian Ovarian Cancer Study was supported by the US Army Medical Research and Materiel Command (Grant No. DAMD17-01-1-0729), the Cancer Council Tasmania, the Cancer Foundation of Western Australia and the National Health and Medical Research Council of Australia. We gratefully acknowledge the cooperation of the following institutions associated with the Australian Ovarian Cancer Study: New South Wales—John Hunter Hospital, North Shore Private Hospital, Royal Hospital for Women, Royal North Shore Hospital, Royal Prince Alfred Hospital and Westmead Hospital; Queensland—Mater Misericordiae Hospital, Royal Brisbane and Women's Hospital, Townsville Hospital and Wesley Hospital; South Australia—Flinders Medical Centre, Queen Elizabeth II, Royal Adelaide Hospital; Tasmania—Royal Hobart Hospital; Victoria—Freemasons Hospital, Mercy Hospital for Women, Monash Medical Centre and Royal Women's Hospital; Western Australia—King Edward Memorial Hospital, St John of God Hospitals Subiaco, Sir Charles Gairdner Hospital and Western Australia Research Tissue Network (WARTN) and the Westmead Gynaecological Oncology Tissue Bank, a member of the Australasian Biospecimens Network-Oncology group. We also acknowledge the contribution of the AOCS Management Group: D Bowtell, G Chenevix-Trench, A Green, P Webb, A deFazio, D Gertig, the study nurses and research assistants, and express our gratitude to all women who participated in the study. We are grateful for assistance provided by Alex Dobrovic and Thomas Mikeska for the MS-HRM studies.
Author contributions
MRS, PAF, DDLB, DJM, DE and SLC conceived and designed the study; DJM, DE, SLC, KA and KDH, performed experiments; AOCS and XC provided reagents, biospecimens and clinical data; DJM, DE, SLC, JG, AB, JC, DG, CG, KDH, KWL, CKN, KR, JT and DCW analysed data; MRS, JDB, PJC, PAF, DDLB, DJM, DE and SLC interpreted the data; and DJM, DE, SLC, PAF and DDLB wrote the manuscript.
No conflicts of interest were declared.
Supporting Information on the Internet
The following supporting information may be found in the online version of this article:
Analysis of TSHZ3 gene expression in AOCS and TCGA datasets.
Sample details.
Summary of somatic rearrangements found in 13 ovarian cancers.
Breakpoints in known genes.
Primer sequences.
Tumour cell content predicted by ASCAT and PICNIC.
References
- 1.TCGA. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. DOI: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vaughan S, Coward JI, Bast RC, Jr, et al. Rethinking ovarian cancer: recommendations for improving outcomes. Nat Rev Cancer. 2011;11:719–725. doi: 10.1038/nrc3144. DOI: nrc3144 [pii] 10.1038/nrc3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tothill RW, Tinker AV, George J, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. DOI: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 4.Helland A, Anglesio MS, George J, et al. Deregulation of MYCN, LIN28B and LET7 in a molecular subtype of aggressive high-grade serous ovarian cancers. PloS One. 2011;6:e18064. doi: 10.1371/journal.pone.0018064. DOI: 10.1371/journal.pone.0018064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorringe KL, George J, Anglesio MS, et al. Copy number analysis identifies novel interactions between genomic loci in ovarian cancer. PloS One. 2010;5 doi: 10.1371/journal.pone.0011408. DOI: 10.1371/journal.pone.0011408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gorringe KL, Ramakrishna M, Williams LH, et al. Are there any more ovarian tumor suppressor genes? A new perspective using ultra high-resolution copy number and loss of heterozygosity analysis. Genes Chromosomes Cancer. 2009;48:931–942. doi: 10.1002/gcc.20694. DOI: 10.1002/gcc.20694. [DOI] [PubMed] [Google Scholar]
- 7.Crozier MA, Copeland LJ, Silva EG, et al. Clear cell carcinoma of the ovary: a study of 59 cases. Gynecol Oncol. 1989;35:199–203. doi: 10.1016/0090-8258(89)90043-7. [DOI] [PubMed] [Google Scholar]
- 8.Jones S, Wang TL, Ie M, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. DOI: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wiegand KC, Shah SP, Al-Agha OM, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. DOI: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anglesio MS, George J, Kulbe H, et al. IL6–STAT3–HIF signaling and therapeutic response to the angiogenesis inhibitor sunitinib in ovarian clear cell cancer. Clin Cancer Res. 2011;17:2538–2548. doi: 10.1158/1078-0432.CCR-10-3314. DOI: 10.1158/1078-0432.CCR-10-3314. [DOI] [PubMed] [Google Scholar]
- 11.Tan DS, Iravani M, McCluggage WG, et al. Genomic analysis reveals the molecular heterogeneity of ovarian clear cell carcinomas. Clin Cancer Res. 2011;17:1521–1534. doi: 10.1158/1078-0432.CCR-10-1688. DOI: 10.1158/1078-0432.CCR-10-1688. [DOI] [PubMed] [Google Scholar]
- 12.Zorn KK, Bonome T, Gangi L, et al. Gene expression profiles of serous, endometrioid, and clear cell subtypes of ovarian and endometrial cancer. Clin Cancer Res. 2005;11:6422–6430. doi: 10.1158/1078-0432.CCR-05-0508. DOI: 10.1158/1078-0432.CCR-05-0508. [DOI] [PubMed] [Google Scholar]
- 13.Dalgliesh GL, Furge K, Greenman C, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. DOI: nature08672 [pii] 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Haaften G, Dalgliesh GL, Davies H, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–523. doi: 10.1038/ng.349. DOI: ng.349 [pii] 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pleasance ED, Cheetham RK, Stephens PJ, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2009;463:191–196. doi: 10.1038/nature08658. DOI: nature08658 [pii] 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pleasance ED, Stephens PJ, O'Meara S, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2009;463:184–190. doi: 10.1038/nature08629. DOI: nature08629 [pii] 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stephens PJ, Greenman CD, Fu B, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. DOI: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell PJ, Stephens PJ, Pleasance ED, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. DOI: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell PJ, Yachida S, Mudie LJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. DOI: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Etemadmoghadam D, George J, Cowin PA, et al. Amplicon-dependent CCNE1 expression is critical for clonogenic survival after cisplatin treatment and is correlated with 20q11 gain in ovarian cancer. PloS One. 2010;5:e15498. doi: 10.1371/journal.pone.0015498. DOI: 10.1371/journal.pone.0015498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hondow HL, Fox SB, Mitchell G, et al. A high-throughput protocol for mutation scanning of the BRCA1 and BRCA2 genes. BMC Cancer. 2011;11:265. doi: 10.1186/1471-2407-11-265. DOI: 10.1186/1471-2407-11-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wojdacz TK, Dobrovic A, Hansen LL. Methylation-sensitive high-resolution melting. Nat Protoc. 2008;3:1903–1908. doi: 10.1038/nprot.2008.191. DOI: 10.1038/nprot.2008.191. [DOI] [PubMed] [Google Scholar]
- 23.Stephens PJ, McBride DJ, Lin ML, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. DOI: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berger MF, Lawrence MS, Demichelis F, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. DOI: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caubit X, Lye CM, Martin E, et al. Teashirt 3 is necessary for ureteral smooth muscle differentiation downstream of SHH and BMP4. Development. 2008;135:3301–3310. doi: 10.1242/dev.022442. DOI: 10.1242/dev.022442. [DOI] [PubMed] [Google Scholar]
- 26.Caubit X, Thoby-Brisson M, Voituron N, et al. Teashirt 3 regulates development of neurons involved in both respiratory rhythm and airflow control. J Neurosci. 2010;30:9465–9476. doi: 10.1523/JNEUROSCI.1765-10.2010. DOI: 10.1523/JNEUROSCI.1765-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Etemadmoghadam D, deFazio A, Beroukhim R, et al. Integrated genome-wide DNA copy number and expression analysis identifies distinct mechanisms of primary chemoresistance in ovarian carcinomas. Clin Cancer Res. 2009;15:1417–1427. doi: 10.1158/1078-0432.CCR-08-1564. DOI: 10.1158/1078-0432.CCR-08-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karlsson R, Pedersen ED, Wang Z, et al. Rho GTPase function in tumorigenesis. Biochim Biophys Acta. 2009;1796:91–98. doi: 10.1016/j.bbcan.2009.03.003. DOI: 10.1016/j.bbcan.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 29.Prakash SK, Paylor R, Jenna S, et al. Functional analysis of ARHGAP6, a novel GTPase-activating protein for RhoA. Hum Mol Genet. 2000;9:477–488. doi: 10.1093/hmg/9.4.477. [DOI] [PubMed] [Google Scholar]
- 30.Huang J, Zhao YL, Li Y, et al. Genomic and functional evidence for an ARID1A tumor suppressor role. Genes Chromosomes Cancer. 2007;46:745–750. doi: 10.1002/gcc.20459. DOI: 10.1002/gcc.20459. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Nagl NG, Jr, Flowers S, et al. Expression of p270 (ARID1A), a component of human SWI/SNF complexes, in human tumors. Int J Cancer. 2004;112:636. doi: 10.1002/ijc.20450. DOI: 10.1002/ijc.20450. [DOI] [PubMed] [Google Scholar]
- 32.Varela I, Tarpey P, Raine K, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. DOI: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahmed AA, Etemadmoghadam D, Temple J, et al. Driver mutations in TP53 are ubiquitous in high-grade serous carcinoma of the ovary. J Pathol. 2010;221:49–56. doi: 10.1002/path.2696. DOI: 10.1002/path.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bowtell DD. The genesis and evolution of high-grade serous ovarian cancer. Nat Rev Cancer. 2010;10:803–808. doi: 10.1038/nrc2946. DOI: 10.1038/nrc2946. [DOI] [PubMed] [Google Scholar]
- 35.Tan DS, Kaye S. Ovarian clear cell adenocarcinoma: a continuing enigma. J Clin Pathol. 2007;60:355–360. doi: 10.1136/jcp.2006.040030. DOI: 10.1136/jcp.2006.040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobel M, Kalloger SE, Boyd N, et al. Ovarian carcinoma subtypes are different diseases: implications for biomarker studies. PLoS Med. 2008;5:e232. doi: 10.1371/journal.pmed.0050232. DOI: 10.1371/journal.pmed.0050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng CK, Cooke SL, Howe K, et al. The role of tandem duplicator phenotype in tumour evolution in high-grade serous ovarian cancer. J Pathol. 2012;226:703–712. doi: 10.1002/path.3980. DOI: 10.1002/path.3980. [DOI] [PubMed] [Google Scholar]
- 38.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. DOI: 310/5748/644 [pii] 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 39.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. DOI: nature05945 [pii] 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 40.Salzman J, Marinelli RJ, Wang PL, et al. ESRRA–C11orf20 is a recurrent gene fusion in serous ovarian carcinoma. PLoS Biol. 2011;9:e1001156. doi: 10.1371/journal.pbio.1001156. DOI: 10.1371/journal.pbio.1001156 PBIOLOGY-D-11-02761 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamamoto M, Cid E, Bru S, et al. Rare and frequent promoter methylation, respectively, of TSHZ2 and − 3 genes that are both downregulated in expression in breast and prostate cancers. PLoS One. 2011;6:e17149. doi: 10.1371/journal.pone.0017149. DOI: 10.1371/journal.pone.0017149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krzywinski M, Schein J, Birol I, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. DOI: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.*Chin SF, Daigo Y, Huang HE, et al. A simple and reliable pretreatment protocol facilitates fluorescent in situ hybridisation on tissue microarrays of paraffin wax embedded tumour samples. Mol Pathol. 2003;56:275–279. doi: 10.1136/mp.56.5.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.*Van Loo P, Nordgard SH, Lingjaerde OC, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci USA. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. DOI: 1009843107 [pii] 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.*Greenman CD, Bignell G, Butler A, et al. PICNIC: an algorithm to predict absolute allelic copy number variation with microarray cancer data. Biostatistics. 2010;11:164–175. doi: 10.1093/biostatistics/kxp045. DOI: kxp045 [pii] 10.1093/biostatistics/kxp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of TSHZ3 gene expression in AOCS and TCGA datasets.
Sample details.
Summary of somatic rearrangements found in 13 ovarian cancers.
Breakpoints in known genes.
Primer sequences.
Tumour cell content predicted by ASCAT and PICNIC.