Abstract
Calculating absolute copy numbers in cancer genome sequences identifies disease-associated genes and provides insights into tumor evolution and heterogeneity.
Cancer genomes may contain a wide array of aberrations—point mutations, small insertions and deletions, genomic rearrangements and viral-genome insertions—all of which can be detected by deep sequencing of tumor samples. However, we are only just beginning to understand how to use such data to study the processes underlying oncogenesis. In a recent paper in Nature Biotechnology, Carter et al.1 show that quantifying copy number changes and point mutations on an absolute, rather than relative, scale facilitates the identification of oncogenes and tumor suppressors and provides insight into the subclonal architecture of tumors and into tumor evolution. Analytical approaches such as this should be useful for interpreting data from large cancer genome sequencing projects and from individual genomes sequenced as part of clinical care.
Copy number analysis represents arguably the simplest view on the cancer genome. Since the 1960s, it has been possible to count the number of copies of a chromosome or of large chromosomal regions in cancer cells using cytogenetic techniques such as karyotyping. The resolution of copy number detection has increased greatly with newer methods that compare tumor and normal genomes by comparative genomic hybridization to microarrays2 or by massively parallel sequencing. These methods have identified several recurrent copy number changes3,4, some of which clearly drive carcinogenesis and can serve as valid therapeutic targets (e.g., amplification of ERBB2 (HER2) in breast cancer).
But the higher resolution of comparative copy number analyses comes with a price: an array or sequencing experiment does not directly report the number of copies of a given genomic locus in a cancer cell but instead provides a relative number compared to a reference state, such as the average signal (e.g., array fluorescence intensity or sequencing read count) across the genome. This lack of absolute quantification arises because cancers are infiltrated with an unknown fraction of normal cells and because the amount of DNA per cancer cell can deviate substantially from the ~6.4 billion base pairs in a normal human diploid cell. The fraction of tumor versus normal cells—the purity of a sample—determines the variation in signal levels. The amount of DNA in a cell—the ploidy of a sample—determines the copy number state to which the average signal level corresponds. In addition, the cancerous cells in a tumor may themselves be heterogeneous, further complicating the picture. The genomic make-up of a cancer cell can, however, only be dissected in detail when the absolute copy numbers are known.
In principle, it should be possible to determine tumor purity and ploidy from relative copy number data and to use this information to calculate absolute copy numbers. Several methods to make such inferences from comparative genomic hybridization array data have recently been published5-7. These methods all use a mathematical framework that models the observed data as a mix of measurements from a heterogeneous population of cells: tumor cells may contain an unknown amount of DNA per cell, and the tumor sample may contain an unknown proportion of normal cells, which have a known amount of DNA per cell. Mathematically, the resulting system of equations is underdetermined. But the key insight exploited by the methods is that only a few combinations of purity and ploidy result in biologically meaningful solutions (Fig. 1, light blue boxes). For example, the number of copies of each chromosome cannot be negative and (barring subclonal copy number variation) must be a whole number. This line of reasoning can then be used to select the most likely combination of purity and ploidy.
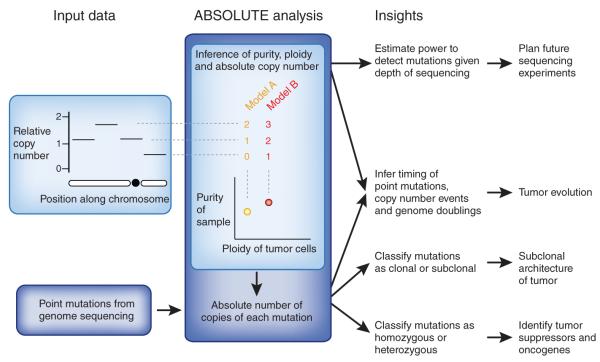
Figure 1.
Biological insights obtained from sophisticated methods for copy number inference. From SNP array or high-throughput sequencing data, methods such as ABSOLUTE can infer tumor purity and ploidy and genome-wide, absolute copy-number profiles. These methods leverage mathematical relationships between purity, ploidy and copy number at a locus and across the genome. Two alternative models are shown, corresponding to different combinations of purity and ploidy, for assignment of relative copy number to absolute copy number states. Purity and ploidy can be used to estimate the required depth of coverage to detect (clonal or subclonal) mutations with a given power. This is useful for designing subsequent sequencing experiments. Integrating copy number data with point mutation data makes it possible to calculate the absolute number of copies of each mutation in the genome. Tumor evolution can be studied by deducing the relative order in which point mutations, copy number events and genome doublings arose in the cancer genome. Separation of clonal from subclonal mutations provides insight into the subclonal architecture of tumors. Finally, homozygous mutations can be distinguished from heterozygous ones, facilitating the identification of tumor suppressors and oncogenes, respectively.
The work of Carter et al.1 takes these principles further by using purity and ploidy to estimate the absolute number of mutated copies (the ‘multiplicity’) of somatic point mutations called from massively parallel sequencing data. This step is based on the same mathematical principles as for inferring absolute copy numbers. Mutation multiplicity is a function of the purity of the sample, the ploidy of the tumor cells and the relative frequency of the mutation (called the ‘allelic fraction’, defined as the number of times a mutated base is observed divided by the total number of times any base is observed at the locus). Therefore, once purity and ploidy have been estimated from relative copy-number data, mutation multiplicity can be estimated as well. Carter et al.1 provide their software tools in a package called ABSOLUTE.
The most exciting advance in the paper involves applications of ABSOLUTE to derive novel insights into cancer biology (Fig. 1). The authors begin by analyzing exome sequencing data from 214 ovarian cancers. Copy-number profiles from array data on the same samples are used to estimate purity and ploidy, which are then applied to estimate multiplicity from the allelic fractions of 29,268 point mutations. These results allow prediction of which mutations are heterozygous (not all copies in the cancer cells are mutated) and which are homozygous (all copies in the cancer cells are mutated). Tumor suppressor genes should be inactivated by homozygous mutations, and this is indeed what is found: the known tumor suppressors TP53 and NF1 often contain homozygous mutations, as does CDK12, identifying it as a novel candidate tumor suppressor. The authors also show that it is possible to predict which mutations are clonal (multiplicity ≥ 1) and which are subclonal (multiplicity < 1) from relative copy number data, an analysis that reveals considerable subclonal diversity in ovarian cancer.
These results provide insights into tumor evolution. Clonal mutations that are present in all tumor cells must have occurred before subclonal mutations found in only a subset of tumor cells. Similarly, in segments of the genome with copy number gains, mutations with higher multiplicity values must have occurred before mutations with lower multiplicity values. Such reasoning allows one to deduce when mutations arose relative to one another and compared to copy number events. Under the right conditions, phylogenetic trees of subclonal diversification can be built8.
Next, Carter et al.1 apply ABSOLUTE to a data set of array comparative genome hybridization measurements of 3,155 samples from 25 different types of cancers. The results indicate that cancers with higher ploidy (a class that represents more than half of most epithelial cancers) have gained DNA by whole-genome duplication rather than by multiple independent gains of genomic material. Briefly, the authors reasoned that whole-genome duplication would cause over-representation of even-numbered, parent-specific copy number states. In contrast, a tumor genome shaped by successive smaller gains would contain equal proportions of odd- and even-numbered copy number states. Carter et al.1 use absolute copy number profiles to identify which tumors had undergone whole-genome duplication. Then, by comparing the frequency of gains and losses of entire chromosome arms between tumor samples with and without whole-genome duplication, they infer that many gains and losses occur before the genome doubling event. Such analyses show that genomic profiling of a cancer genome—even at only one time point—can tell us much about a tumor’s history.
Results derived from such approaches can have clinical implications. Clonal driver mutations would be preferred targets for therapy as targeting subclonal driver mutations would likely affect only a subset of cancer cells9. Yet subclonal evolution should clearly not be ignored because tumors with ongoing subclonal divergence are more likely to evolve mutations conferring resistance to therapy or to harbor rare subclones that already carry such mutations before treatment. The authors show different evolutionary trajectories of genome-doubled versus non-genome-doubled ovarian cancers. Genome-doubled cases have a higher number of focal copy number changes, a lower incidence of homozygous deletions (e.g., tumor suppressor NF1 is rarely hit in genome-doubled cases) and a higher cancer recurrence rate.
As ABSOLUTE and similar methods are refined in the future, it is worth considering why they cannot deduce tumor purity and ploidy in all cases. In the pan-cancer data set, ABSOLUTE judged 9.1% of samples to be non-aberrant. These cancers have genomes with perfectly normal copy number profiles (at least at the resolution of the assay). It is therefore impossible to determine the sample purity from copy-number data alone, although this could theoretically be resolved using point mutation data. Another reason for failure is insufficient purity (observed in 7.3% of cases) because an excess of normal cells contaminated the sample. This is inherently a sample problem, and will affect some tumor types more than others. Finally, 6.9% of samples failed because they were too complex to interpret. For instance, when every region in the genome is subject to subclonal copy-number changes, it may become mathematically impossible to calculate absolute purity and ploidy. However, these may be very interesting cases biologically because of their ongoing evolution and the competition between subclones. Improved methods, possibly combining copy number and point mutation data, may be able to handle this complexity.
Tens of thousands of cancer genomes will be sequenced over the next few years. Methods such as ABSOLUTE will undoubtedly contribute toward their analysis, generating new insights into oncogenesis, tumor evolution and subclonal diversification. Future methods will likely be run directly on sequencing data, eliminating the cost and extra sample requirements of SNP arrays, which are now almost routinely performed alongside massively parallel sequencing experiments. Methods that integrate across different ‘views’ of the data and across different classes of mutation will become increasingly important to understand the complexity of cancer genomes. The work of Carter et al.1 exemplifies such a strategy, which combines multiple sources of data to analyze cancer genomes, revealing considerable biological insights.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Carter SL, et al. Nat. Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kallioniemi A, et al. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 3.Bignell GR, et al. Nature. 2010;463:893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beroukhim R, et al. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Popova T, et al. Genome Biol. 2009;10:R128. doi: 10.1186/gb-2009-10-11-r128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Loo P, et al. Proc. Natl. Acad. Sci. USA. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yau C, et al. Genome Biol. 2010;11:R92. doi: 10.1186/gb-2010-11-9-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nik-Zainal S, et al. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerlinger M, et al. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]