To the Editor: A recent editorial of Nature Methods1 stated that proteomics raw “data can be reprocessed with new questions in mind, such as examining different post-translational modifications than the original study.” In our view this will be the main contribution to biology from the reprocessing of raw data. A significant percentage of fragmentation spectra generated by shotgun proteomics remains unassigned, which leaves much space for the creativity and knowledge of other scientists to extract additional information by developing and implementing sophisticated data analysis strategies. However, the lack of a suitable repository that allows easy deposition and access to this data has discouraged many scientists from sharing and reprocessing raw data. Here we show the utility of raw data by providing insight into adenosine diphosphate (ADP)-ribosylation via reanalysis of a phosphoproteomics dataset.
ADP-ribosylation is an evolutionarily ancient post-translational modification (PTM) that is generated by poly(ADP-ribose) polymerases and mono ADP-ribose transferases. All these enzymes control important cellular processes, and some of them have attracted great attention as potential targets in treatment of human disease. Despite the strong need to characterize the sites of this PTM mass spectrometric identification of ADP-ribosylated peptides has been restricted to studies on synthetic peptides or recombinant proteins2. Because phosphopeptide enrichment techniques can be applied to the isolation of ADP-ribosylated peptides3, we reasoned that some spectra generated in phosphoproteomics studies could match ADP-ribosylated peptides. We added this modification to the standard search engine Andromeda and interrogated the raw data from a large-scale mouse tissue phosphoproteomics study4. The partial inadequacy of traditional search algorithms to interpret the atypical fragmentation pattern of this technically challenging PTM2 was overcome by extensive manual analysis to identify ADP-ribosylation-specific fragment ions and to obtain good sequence coverage of the peptide backbone (Supplementary Methods). We confidently identified a total of 88 mono ADP-ribosylation sites from 79 proteins, with eight sites found also to be modified by ribose phosphate, a modification derived from ADP-ribose5 (Fig. 1; Supplementary Table 1; Supplementary Data). Arginine is the modified residue in all cases except for one glutamate ADP-ribosylation site. Interestingly, the vast majority of the identified sites were found on proteins expressed in the liver and no modified peptides were detected in 5 out of 9 tissues. The identified protein targets revealed that intracellular ADP-ribosylation on arginine residues is much more common than previously thought (all known arginine-specific ADP-ribose transferases in mammals modify extracellular proteins)5. Most notably, arginine-specific ADP-ribosylation was found prominently on tubulins and on translation initiation factors.
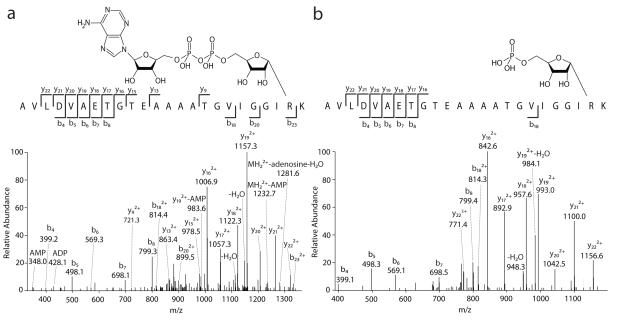
Figure 1.
Fragmentation spectra of apeptide modified by ADP-ribose (a; triply charged precursor at m/z 937.7702) and ribose-phosphate (b; triply charged precursor at m/z 828.0861).
This work shows that the re-interrogation of existing proteomics raw data files is a valid approach for the discovery of new, biologically important PTMs. We provide a dataset for the ADP-ribosylation research community, and expect that easier access to existing and future raw data will substantially increase the numbers of identified sites. We urge the proteomics community to find a solution to the pressing problem of raw file storage so that these valuable resources are not left untapped.
Supplementary Material
Acknowledgements
The authors thank M. H. Tatham and M. Trost for comments on the manuscript. This work was supported by a Sir Henry Wellcome Fellowship awarded by the Wellcome Trust (Sponsor Reference 088957/Z/09/Z, to I.M.)
Footnotes
Competing financial interest The authors declare no financial interests.
References
- 1.Nat. Methods. 2012;9:419–419. doi: 10.1038/nmeth.2011. [DOI] [PubMed] [Google Scholar]
- 2.Hengel SM, Goodlett DR. Int. J. Mass Spectrom. 2012;312:114–121. doi: 10.1016/j.ijms.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laing S, Koch-Nolte F, Haag F, Buck F. J. Proteomics. 2011;75:169–176. doi: 10.1016/j.jprot.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Huttlin EL, et al. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laing S, Unger M, Koch-Nolte F, Haag F. Amino Acids. 2011;41:257–269. doi: 10.1007/s00726-010-0676-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.