Abstract
The inhabitants of the mammalian gut are not always relatively benign commensal bacteria but may also include larger and more parasitic organisms, such as worms and protozoa. At some level, all these organisms are capable of interacting with each other. We found that successful establishment of the chronically infecting parasitic nematode Trichuris muris in the large intestine of mice is dependent on microflora and coincident with modulation of the host immune response. By reducing the number of bacteria in the host animal, we significantly reduced the number of hatched T. muris eggs. Critical interactions between bacteria (microflora) and parasites (macrofauna) introduced a new dynamic to the intestinal niche, which has fundamental implications for our current concepts of intestinal homeostasis and regulation of immunity.
The mammalian gut contains around 1013 bacteria (1), the majority of which belong to the phyla Bacteroidetes or Firmicutes (1, 2). Coevolution with these microbes has driven the functional morphology and immune function of the gastrointestinal tract (3-5). Without microbes, aberrant physiology develops together with problems in host defense. Both can be rectified upon reintroduction of bacteria (6). Additionally, childhood exposure to microbes can direct the maturing immune system to develop a tolerance to environmental antigens, the so-called “hygiene hypothesis” (7). More recently this concept has been extended to include “macrofauna” of the gut, such as helminth parasites (8). A helminth-driven TH2 and TH cell regulatory response has evolved to counter infection and repair the damage that these parasites cause (9). Dysregulation of these immune responses leads to prolonged infection and disease. Indeed, helminths have been found to be a major force underlying the evolution and selection of interleukin genes (10). Thus, gut commensal bacteria and gastrointestinal-dwelling helminths have lived in close association throughout evolution. Relationships between bacteria and metazoa have already been documented, such as filarial worms and the endosymbiont Wolbachia (11); however, a functional nonendosymbiotic relationship between prokaryotes and parasitic metazoa within the infected host remains to be defined. This complex intestinal ecology will have major implications for the immunoregulatory mechanisms of gut inflammation and autoimmune disease (12).
Trichuris is a genus comprising more than 50 species of whipworm, an extremely prevalent and successful group of intestinal-dwelling nematode parasites infecting many diverse mammalian hosts, with infection by T. trichiura estimated to infect almost a billion people (13). All Trichuris species inhabit the large intestine (cecum and colon). Infection proceeds upon ingestion of embryonated eggs from the external environment. Upon hatching, the larvae emerge from polar egg opercula and establish infection within the epithelium of the crypts of Lieberkühn of the cecum and colon. Following the characteristic four molts, dioecious adult parasites develop to patency (at a rate dependent on the host), mate, and release unembryonated eggs into the environment via the feces. Here, we investigated a bacterially driven mechanism of hatching of mouse whipworm, T. muris, that facilitates infection of the mammalian host and subsequent immune response.
T. muris eggs were induced to hatch in vitro when incubated for at least 30 min with explants of mouse cecum containing substantial numbers of bacteria at 37°C (Fig. 1A). To define the role of bacteria in this process, we incubated eggs in a culture of Escherichia coli, a common gut commensal. In the presence of E. coli, hatching was observed at a similar level to that seen within gut explants; 0.4-μm filtration to remove E. coli from cultures prevented hatching and showed that a structural component of the bacteria, not a secreted molecule, was responsible for the hatching (Fig. 1A). Further analysis confirmed that a variety of microorganisms (five strains of bacteria and one of yeast) could induce efficient hatching over 2 hours to levels comparable with that found with gut explants over 18 hours (Fig. 1, B and C). By using transwells of different sizes, it was possible to confirm that direct contact between the bacteria and the eggs was required for hatching (Fig. 1, C and D). Bacterially promoted hatching only occurred at 37°C, suggesting that temperature is also a hatching cue (Fig. 1E), presumably to prevent hatching in the external environment where T. muris eggs embryonate. To locate the site of interaction between the bacteria and parasite eggs, we incubated the green fluorescent protein (GFP)–expressing E. coli strain PK1162 (14) with embryonated eggs. Bacteria clearly cluster around the opercula at the poles of the eggs (Fig. 1, F to H) where the worms emerge (Movie S1).
Fig. 1.
T. muris eggs are induced to hatch in vitro by contact with bacteria. (A) Hatching of T. muris eggs for 2 hours at 37°C incubated with 5-cm sections of mouse cecum, E. coli bacterial suspension, or 0.22-μm filtered (overnight) bacterial suspension. Luria-Bertani (LB) broth was used as negative control. (B) T. muris eggs were cultured with four strains of bacteria (E. coli, S. aureus, S. typhimurium, or P. aeruginosa) for 2 hours at 37°C. (C) T. muris eggs incubated with Saccharomyces cerevisiae, with or without 0.4-μm transwell or 0.22-μm filtered S. cerevisiae cultures for 2 hours at 37°C. (D) Hatching of T. muris eggs in E. coli bacterial suspension or with 0.4-μm or 3-μm transwells. (E) T. muris eggs incubated for 2 hours with E. coli at 4°C, room temperature (RT), or 37°C. Eggs that did not hatch at 4°C or room temperature were then incubated for a further 2 hours at 37°C (4/37 and RT/37, respectively). (F to H) T. muris eggs were cultured at 37°C for 1 hour with GFP-expressing bacteria, then washed in phosphate-buffered saline before further incubation for 1 hour at 37°C. (F) Light-field image; (G) fluorescence image; (H) combined image. Scale bar, 10 μm. All figures show means ± SEM, *P < 0.01, using analysis of variance (ANOVA).
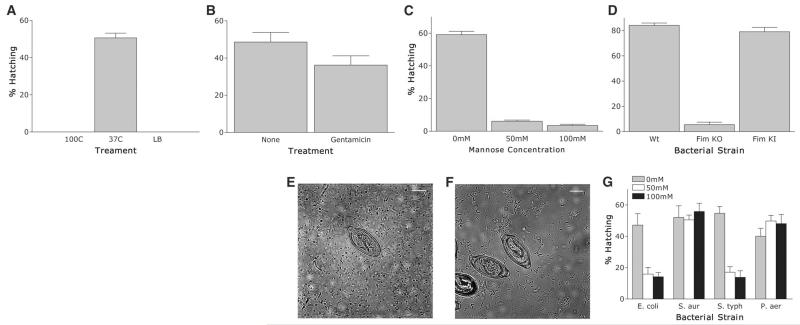
Structural disruption of E. coli (boiling, 10 min) prevented hatching (Fig. 2A), although after bacteriostatic antibiotic treatment (gentamicin), E. coli were still able to induce hatching (Fig. 2B) but were not viable. Taken together, the data support a role for the intact bacterial surface as a critical component of the hatching process.
Fig. 2.
T. muris eggs are induced to hatch in vitro after contact with type 1 fimbriae. (A) T. muris eggs were incubated for 2 hours at 37°C with E. coli preincubated for 10 min at 37° or 100°C. LB broth alone was used as a negative control. (B) T. muris eggs were incubated for 2 hours at 37°C with E. coli pretreated for 30 min with gentamicin (600 μg/ml) or untreated as a control. (C) T. muris eggs were incubated for 2 hours at 37°C with E. coli treated with 0, 50, or 100 mM mannose 30 min before and during hatching. (D) T. muris eggs were incubated for 2 hours at 37°C with wild-type E. coli (strain PK1162), FimKO E. coli, or FimKI E. coli. (E and F) T. muris eggs were incubated with FimKO (E) or FimKI (F) E. coli for 1 hour at 37°C. Scale bar, 10 μm. (G) T. muris eggs after 2 hours of incubation at 37°C with E. coli, S. aureus, S. typhimurium, or P. aeruginosa plus 0, 50, or 100 mM mannose before and during hatching. All figures show means ± SEM, *P < 0.001 using ANOVA.
Type 1 fimbriae facilitate mannose-sensitive adherence of E. coli to cells and mucosal surfaces (15). They are encoded by the fim gene cluster and consist of a major structural subunit (FimA) and several minor components including the adhesin FimH located at the fimbrial tip that recognizes terminally located D-mannose moieties on cell-bound and secreted glycoproteins (16, 17). To elucidate whether type 1 fimbriae played a role in hatching, we investigated the effect of exogenous mannose on hatching. The addition of mannose significantly inhibited hatching without affecting the numbers or viability of bacteria (Fig. 2C and fig. S1). Likewise, strain AAEC072A (FimKO), which lacks the fim gene cluster and does not express type 1 fimbriae (18) (fig. S2), did not promote hatching (Fig. 2D), although viability of these bacteria was comparable to that of wild-type Fim+ E. coli strain PK1162 (fig. S3). In contrast, mannose-sensitive hatching was seen with strain AAEC072A harboring plasmid pLB254 (FimKI) encoding the cloned fim gene cluster (Fig. 2D and fig. S2). In contrast to the strains expressing type 1 fimbriae that associate with the opercula at the poles (Fig. 1, F to H, and Fig. 2, E and F), strain AAEC072A failed to adhere to the eggs (Fig. 2, E and F), confirming a role for type 1 fimbriae in mediating interactions between the parasite eggs and the bacterium. The observation that Salmonella typhimurium also exhibited mannose-sensitive hatching similar to that seen with E. coli (Fig. 2G) indicates that bacterially induced hatching is partly mediated by type 1 fimbriae. However, the Gram-negative bacterium Pseudomonas aeruginosa induced hatching in the presence of mannose but does not express type 1 fimbriae. Similarly, the ability of the Gram-positive bacterium Staphylococcus aureus to promote hatching indicates that other mechanisms also exist.
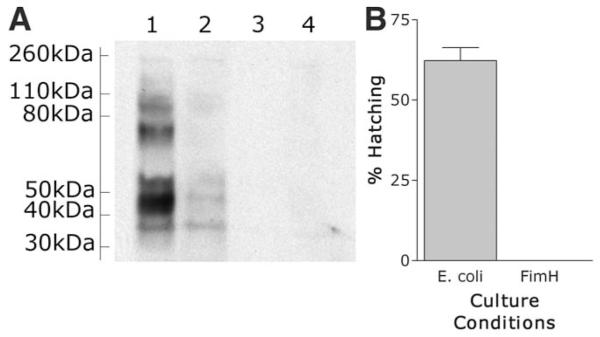
His-tagged FimH was purified (fig. S4) and used in a far Western blot analysis of solubilized surface egg proteins. Five to seven surface proteins were detected that bound to purified FimH, with the most intense signal generated from a protein of 45 kD (Fig. 3A). Mass spectrophotometry of these different bands and data mining of an expressed sequence tag database (NEMBASE) have identified putative proteins of T. muris and other parasitic nematodes, although further categorization will depend on sequencing of the T. muris genome. Purified FimH itself did not cause hatching of T. muris eggs (Fig. 3B), suggesting that some structural conformation, perhaps cross-linking by multiple FimH adhesins, is required. In addition, it is known that type 1 fimbriae mediate shear-dependent adhesion typified by weak binding under conditions of low flow that strengthens as the shear forces increase (17). Thus, it is perhaps not surprising that FimH on the bacterial surface behaves differently from the purified molecule. It is known that interaction between FimH and uroplakin molecules on the cell surface of uroepithelial umbrella cells or β1 integrins on fibroblast cells is known to induce a signal transduction cascade (19). It is possible that the interaction between FimH and a mannosylated receptor on the surface of the T. muris eggs stimulates a signal transduction cascade that leads to the emergence of the worm out of the egg.
Fig. 3.
Western blot analysis of purified FimH binding to surface egg proteins. T. muris eggs are induced to hatch in vitro after contact with FimH adhesion on type 1 fimbriae. Egg proteins were subjected to SDS–polyacrylamide gel electrophoresis under reducing conditions and electrophoretically transferred onto nitrocellulose. The migration positions of protein standards are indicated on the left. Lane 1, purified FimH and secondary antibody; lane 2, purified FimH preincubated with mannose to specifically block binding; lane 3, purified FimH only (no secondary); lane 4, secondary only.
Although the data clearly show that hatching in vitro requires an interaction between bacteria and the egg via type 1 fimbriae, it is unclear whether this is also important in vivo. Thus, we treated C57BL/6 mice with enrofloxacin from 2 days before infection throughout the course of infection to ascertain whether there was any effect on worm establishment. At day 21 post-infection (p.i.), the worm burden was significantly (P < 0.001) decreased in animals that had been treated with antibiotics (Fig. 4A). Interestingly, these animals made a stronger TH2 response to infection, with increased levels of interleukin-4 (IL-4) (Fig. 4B) and IL-13 (Fig. 4C). Interferon-γ (IFN-γ) secretion was unchanged (Fig. 4D). Parasite-specific immunoglobulin G1 (IgG1) antibody production was increased in antibiotic-treated animals, reflecting the increased TH2 response, whereas IFN-γ–controlled IgG2a levels were unaffected, reflecting the IFN-γ response (fig. S5). IL-17 has been found to be related to bacterial presence in the small intestine (20) and is significantly reduced in animals treated with antibiotics. Similarly, in our experiments, IL-17 expression was reduced in antibiotic-treated animals (Fig. 4F), as was IL-6, which drives IL-17 production (Fig. 4E).
Fig. 4.
T. muris eggs are induced to hatch in vivo by the presence of bacteria. (A) Worm burden at day 18 p.i.. in C57BL/6 mice treated with enrofloxacin (Baytril) from day −2 p.i. to day 18 p.i. Untreated C57BL/6 mice were used as controls. (B) IL-4 secretion by antigen restimulated MLN cells from enrofloxacin-treated and untreated control C57BL/6 mice at day 18 p.i. assessed by CBA assay. (C) IL-13 secretion. (D) IFN-γ secretion. (E) IL-6 secretion. (F) IL-17 secretion. (G) Worm burden at day 18 p.i. in SCID mice treated with enrofloxacin from day −2 p.i. to day 18 p.i. Untreated SCID mice were used as controls. (H) Worm burden at day 14 p.i. in AKR mice treated with enrofloxacin at the beginning only (day −5 to day 5) or end only (day 5 to day 13) of infection. Untreated AKR mice used as controls. All figures show means ± SEM, P < 0.01 by ANOVA.
To rule out a major role for acquired immunity in worm reduction, we infected severe combined immunodeficient (SCID) mice and treated them with enrofloxacin throughout. At day 18 p.i., worm burdens were reduced in antibiotic-treated mice (Fig. 4G), demonstrating that it is not the increased TH2 response that is solely responsible for the worm expulsion observed. A comparison of antibiotic treatment regimes in susceptible mice (treatment from day −5 to day +5 p.i. or day +7 to day 13 p.i.) clearly showed that depleting the microflora affected the establishment and not the subsequent survival of the worms within the intestine at day 14 p.i. (Fig. 4H). All animals treated with antibiotics had decreased aerobic and anaerobic flora as measured by growth on LB agar (fig. S6).
The paradigm that the adaptive immune system has evolved to control microbes has been modified to include the concept that the immune system is in fact controlled by microorganisms. Our results constitute evidence to suggest that we must extend this paradigm again to include “macrofauna,” the metazoan parasites such as Trichuris. We can clearly see a critical relationship between bacteria and T. muris that has a marked effect on the host immune response to this parasite. We propose that temperature, together with an increase in bacterial load at these sites, provides optimum conditions for efficient hatching of Trichuris species eggs. Additionally, controlled damage of the intestinal epithelium provides an enhanced potential route of access for microflora to the adaptive immune system. This has subsequent beneficial consequences for the host, in that antibacterial TH17 responses and TH1 responses are generated that are known to control pathogenic bacterial pathogens (20, 21). The induction of TH17 and TH1 responses in the gut is closely associated with the induction of regulatory T cell subsets (22). These are known to have beneficial effects for survival of longlived chronic helminth infections in the absence of overt pathology and indeed are central to the tenet of the modified hygiene hypothesis (8). It is clear that in nature, coevolution of microflora of the gut and host should not be considered in the absence of the influence of macrofauna such as gastrointestinal-dwelling nematode parasites.
Supplementary Material
Acknowledgments
We thank L. Cliffe for numerous helpful discussions; J. Kott and R. Fernandez for help with microscopy; the staff in the Faculty of Life Sciences EM Facility for their assistance, particularly A. Mironov and the Wellcome Trust for equipment grant support to the EM Facility; the Faculty of Life Sciences Mass Spectrometry Unit and E. Keevill for help with protein identification; and P. Klemm for bacterial strains and plasmids. The Bioimaging Facility microscopes used in this study were purchased with grants from the UK Biotechnology and Biological Sciences Research Council, Wellcome, and the University of Manchester Strategic Fund. Accession numbers for proteins identified by mass spectrometry are Q9N5A6 CAEEL, TMP00496_1, TMP00827_1, TMP00007_1, and TMP00821_1. Supported by Wellcome Trust grant 083620Z and UK Medical Research Council grant G0601205.
Footnotes
Supporting Online Material
References
- 1.Ley RE, et al. Science. 2008;320:1647. doi: 10.1126/science.1155725. published online 22 May 2008 (10.1126/science.1155725) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turnbaugh PJ, et al. Nature. 2007;449:804. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hooper LV, Gordon JI. Science. 2001;292:1115. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 4.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Science. 2005;307:1915. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 5.Edelman SM, Kasper DL. Curr. Opin. Gastro. 2008;24:720. doi: 10.1097/MOG.0b013e32830c4355. [DOI] [PubMed] [Google Scholar]
- 6.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Nat. Immunol. 2003;4:269. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 7.Wold AE. Allergy. 1998;53(suppl.):20. doi: 10.1111/j.1398-9995.1998.tb04953.x. [DOI] [PubMed] [Google Scholar]
- 8.Yazdanbakhsh M, Matricardi PM. Clin. Rev. Allergy Immunol. 2004;26:15. doi: 10.1385/CRIAI:26:1:15. [DOI] [PubMed] [Google Scholar]
- 9.Maizels RM, Yazdanbakhsh M. Nat. Rev. Immunol. 2003;3:733. doi: 10.1038/nri1183. [DOI] [PubMed] [Google Scholar]
- 10.Fumagalli M, et al. J. Exp. Med. 2009;206:1395. doi: 10.1084/jem.20082779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor MJ, Bandi C, Hoerauf A. Adv. Parasitol. 2005;60:245. doi: 10.1016/S0065-308X(05)60004-8. [DOI] [PubMed] [Google Scholar]
- 12.Gaboriau-Routhiau V, et al. Immunity. 2009;31:677. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 13.Prevention and Control of Schistosomiasis and Soil-Transmitted Helminthiasis. 2002. p. 912. WHO Tech. Rep. [PubMed] [Google Scholar]
- 14.Reisner A, Haagensen JA, Schembri MA, Zechner EL. S. Molin, Mol. Microbiol. 2003;48:933. doi: 10.1046/j.1365-2958.2003.03490.x. [DOI] [PubMed] [Google Scholar]
- 15.Ofek IB, Beachey EH. Infect. Immun. 1978;22:247. doi: 10.1128/iai.22.1.247-254.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eto DS, Jones TA, Sundsbak JL, Mulvey MA. PLoS Pathog. 2007;3:e100. doi: 10.1371/journal.ppat.0030100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bouckaert J, et al. Mol. Microbiol. 2006;61:1556. doi: 10.1111/j.1365-2958.2006.05352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Breines DM, Burnham JC. J. Antimicrob. Chemother. 1994;34:205. doi: 10.1093/jac/34.2.205. [DOI] [PubMed] [Google Scholar]
- 19.Thumbikat P, et al. PLoS Pathog. 2009;5:e1000415. doi: 10.1371/journal.ppat.1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivanov II, et al. Cell Host Microbe. 2008;4:337. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivanov II, et al. Cell. 2009;139:485. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaudhry A, et al. Science. 2009;326:986. doi: 10.1126/science.1172702. published online 1 October 2009 (10.1126/science.1172702) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.