Abstract
The exon-junction complex (EJC) performs essential RNA processing tasks1-5. Here, we describe the first human disorder, Thrombocytopenia with Absent Radii6 (TAR), caused by deficiency in one of the four EJC subunits. A compound inheritance mechanism of a rare null allele and one of two low-frequency SNPs in the regulatory regions of RBM8A, encoding the Y14 subunit of EJC, causes TAR. We found that this mechanism explained 53 of 55 cases (P<5×10−228) with the rare congenital malformation syndrome. Fifty-one of those 53 carried a previously associated7 submicroscopic deletion of 1q21.1; two carried a truncation or frameshift null mutation in RBM8A. We show that the two regulatory SNPs result in reduction of RBM8A transcription in vitro and that Y14 expression is reduced in platelets from TAR cases. Our data implicate Y14 insufficiency, and presumably EJC defect, as the cause of TAR syndrome.
The Thrombocytopenia with Absent Radii (TAR) syndrome is characterized by a reduction in the number of platelets (the cells that make the blood clot) (generally below 50×109/L, normal range 150-350×109/L) and the absence of one of the bones in the forearm (the radius) but with preservation of the thumb, which distinguishes TAR from other syndromes that combine blood abnormalities with absence of the radius, such as Fanconi anemia6,8. TAR cases have low numbers of megakaryocytes, the platelet precursor cells that reside in the bone-marrow, and cases frequently present with bleeding episodes in the first year of life, which diminish with age. The severity of skeletal abnormalities varies from absence of radii to virtual absence of upper limbs with or without lower limb defects, such as malformations of the hip and knee9. An inherited or de novo deletion on 1q21.1 is present in the majority of cases7, but the apparent autosomal recessive nature of the syndrome requires the existence of an additional causative allele. This additional allele has remained elusive in spite of the sequencing of the protein-coding exons of 10 genes (including RBM8A) in the minimally deleted region (chr1:145399075-145594214, Fig. 1a, Supplementary Fig. 1, Supplementary Note) in a previous study7.
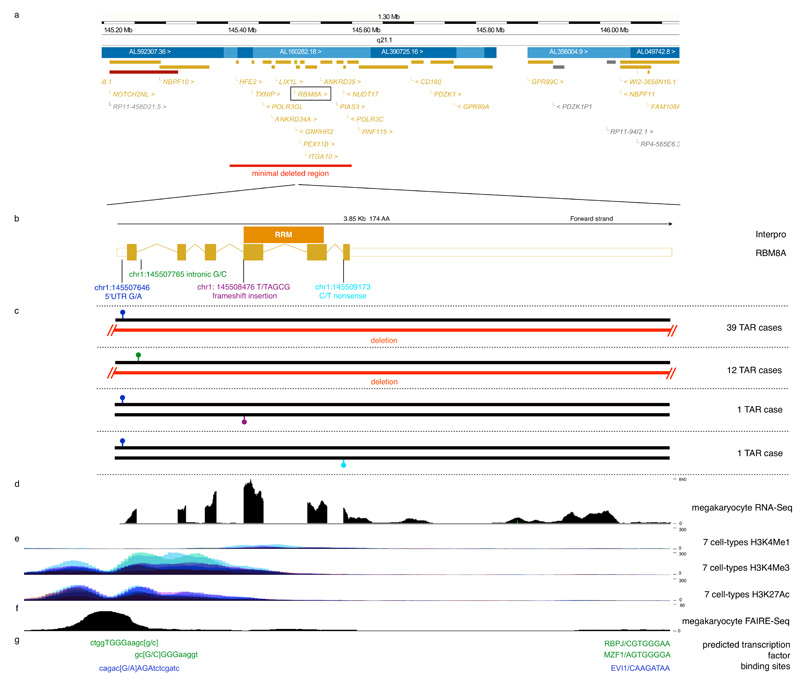
Figure 1. Most TAR syndrome cases have a low-frequency regulatory variant and a rare null allele in the RBM8A locus.
a) Fifty-three out of 55 TAR cases are heterozygous carriers of a rare 1q21.1 deletion of varying size. The red bar indicates the region that is deleted in all 53 cases with a deletion.
b) The RBM8A transcript is shown in genomic coordinates with the RNA binding domain (RRM) indicated by the orange bar above the transcript.
c) We identified two low-frequency regulatory SNPs in 53 of a total of 55 TAR cases studied. The first, 145507646 G/A, is located in the 5′UTR of RBM8A and has a population minor allele frequency of 3.05% (shown in blue). The second, 145507765 G/C is located in the first intron of RBM8A and has a population minor allele frequency of 0.41% (shown in green). Thirty-nine TAR cases carried the minor allele of the 5′UTR SNP on one chromosome and the 1q21.1 deletion on the other; 12 TAR cases carried the minor allele of the intronic SNP on one chromosome and the 1q21.1 deletion on the other. The compound inheritance of the 1q21.1 deletion and one of the two regulatory SNPs is strongly associated with TAR with a p-value <5 × 10×228. Two further TAR cases were found to have the 5′UTR minor allele in combination with either a frameshift insertion (shown in purple) and a nonsense mutation in RBM8A (shown in light blue) instead of the 1q21.1 deletion, implicating RBM8A as the causative gene for TAR syndrome.
d) Sequencing of RNA from cord-blood-derived megakaryocytes shows that RBM8A is expressed in megakaryocytes.
e) Histone modifications from the ENCODE Project13 in 7 cell lines (GM12878, H1-hESC, HSMM, HUVEC, K562, NHEK, and NHLF) indicate the presence of regulatory elements at the promoter and first intron of RBM8A. H3K4Me1 is often found near regulatory elements, H3K4Me3 is often found near promoters and H3K27Ac is often found near active regulatory elements (UCSC Genome Browser).
f) FAIRE marking regions of open chromatin shows that the 5′UTR SNP and first intron SNP are accessible in megakaryocytes.
g) Computational modeling predicts that the 5′UTR SNP minor allele creates a binding site for the transcription factor Evi1, whilst the intronic SNP minor allele is predicted to disrupt binding of Mzf1 and Rbpj.
To identify the additional causative allele, we selected five cases of European ancestry with the deletion and sequenced their exomes (Online Methods). We failed to find associated coding mutations in a gene. However, four of the cases carried the minor allele of a low-frequency single nucleotide polymorphism (SNP) in the 5′ UnTranslated Region (5′UTR) of the RBM8A gene, whilst the remaining one carried a novel SNP in the first intron of the same gene (Fig. 1b). Genotyping by Sanger sequencing of another 48 cases of European ancestry with the 1q21.1 deletion identified the two SNPs in 35 and 11 samples, respectively (Fig. 1c, Supplementary Table 1, 2 and Supplementary Note). A mother of non-European ancestry with TAR and her fetus aborted on grounds of prenatal diagnosis of TAR both did not carry the 5′UTR or the intronic SNP (Supplementary Note), and we suggest that in this case there is a different additional causative allele that we have failed to identify. In the 25 trios where the deletion in the child was not a de novo event, we confirmed that the deletion and the newly identified SNPs were inherited from different parents (Supplementary Table 1). The minor allele frequencies (MAFs) of the 5′UTR and intronic SNP were 3.05% and 0.42% respectively in 7504 healthy individuals of the Cambridge BioResource10 (Supplementary Note) and the deletion was absent from 5919 shared healthy controls of the Wellcome Trust Case Control Consortium10. Thus, the concurrent presence of one of the two non-coding SNPs on one allele, combined with a 1q21.1 deletion on the other, is unequivocally associated with TAR syndrome with an estimated p-value of <5×10−228 (Supplementary Note). Next, we sequenced all exons of RBM8A in two additional TAR cases that do not carry the 1q21.1 deletion but were found to carry the 5′UTR SNP. We identified a 4-b(ase)p(air) frameshift insertion at the start of the fourth exon in the first case and established that the non-coding SNP and insertion were on different chromosomes; in the second case we identified a nonsense mutation in the last exon of RBM8A (Fig. 1b,c). Both mutations were absent from 458 exome samples of the 1000 Genomes Project11 and 416 samples from the CoLaus cohort12. We conclude that, in the vast majority of cases, compound inheritance of a rare null allele (a deletion, frameshift mutation or premature stop codon), and one of two low-frequency non-coding SNPs in RBM8A causes TAR syndrome.
Based on the genetic results we postulated a hypomorphic mechanism, whereby one copy of the RBM8A gene is not functional due to a null allele and the expression of the other copy is reduced as a result of the non-coding SNPs in the 5′UTR or in the first intron. Histone modifications in seven human cell lines from the Encode project indicate that both SNPs are localized in potential active regulatory elements (Fig. 1d,e ref. 13). Annotation of open chromatin structure using the FAIRE (formaldehyde-assisted isolation of regulatory elements) technique provided further evidence in megakaryocytes (Fig. 1f, ref. 14) to support this. Computational predictions suggest that the 5′UTR SNP introduces a binding site for the transcriptional repressor EVI1 and that the intronic SNP disrupts a binding site for the transcription factors MZF1 and RBPJ (Fig. 1g). The EVI1 binding prediction was confirmed by electrophoretic mobility shift assays (EMSA) assays in the megakaryocytic cell line CHRF-288-11 (or CHRF in short), showing Evi1 protein binding to the minor but only weak binding to the major allele (Fig. 2a). EMSA studies for the intronic SNP showed specific decreased binding of nuclear proteins to the minor allele, although we could not confirm the presence of either MZF1 or RBPJ in the supershift experiment (Supplementary Fig. 2). The results of luciferase reporter assays in cell lines representative of megakaryocytes and osteoblasts showed that the differential binding detected by EMSA was functionally relevant and that both the 5′UTR and intronic SNP significantly reduced RMB8A promoter activity. The minor alleles were associated with significantly lower luciferase activity in human megakaryocytic CHRF and DAMI cell lines and the murine osteoblast cell line MC3T3. No effect of the 5′UTR SNP was observed in human endothelial EAHY926 and HEK293 cells; the minor allele of the intronic SNP did exert an effect in the latter but not the former (Fig. 2b). We next performed immunoblot staining in platelet lysates from three TAR cases (UCN 10, 13 and 16, all with the deletion 1.q21.1 and 5′UTR SNP combination) and their parents and a further 4 cases where parental samples were not available: three with the deletion 1.q21.1 and either 5′UTR SNP (UCN 83 and 113) or the intronic SNP (UCN 64), and one with the 4 bp insertion in RBM8A in combination with the 5′UTR (UCN 33) (Supplementary Fig. 3). Densitometry analysis showed a significant reduction of the level of Y14, the protein encoded by RBM8A, when compared to the parents and healthy controls (Fig. 2c). Taken together the genetic and biological data strongly support our hypothesis that TAR results from an insufficiency of the Y14 protein. The results from the luciferase assay suggest that the minor allele of the 5′ UTR SNP may encode for lower transcription than the major allele. In vivo expression assays in platelet RNA samples from 12 healthy volunteers heterozygous for the 5′ UTR SNP however showed no significant differences between the allelic transcript levels (P=0.91, paired T-test on allelic ratios, Supplementary Fig. 4). It therefore still leaves open the question of the exact mechanism by which the non-coding SNPs lead to the decreased protein expression observed in TAR cases.
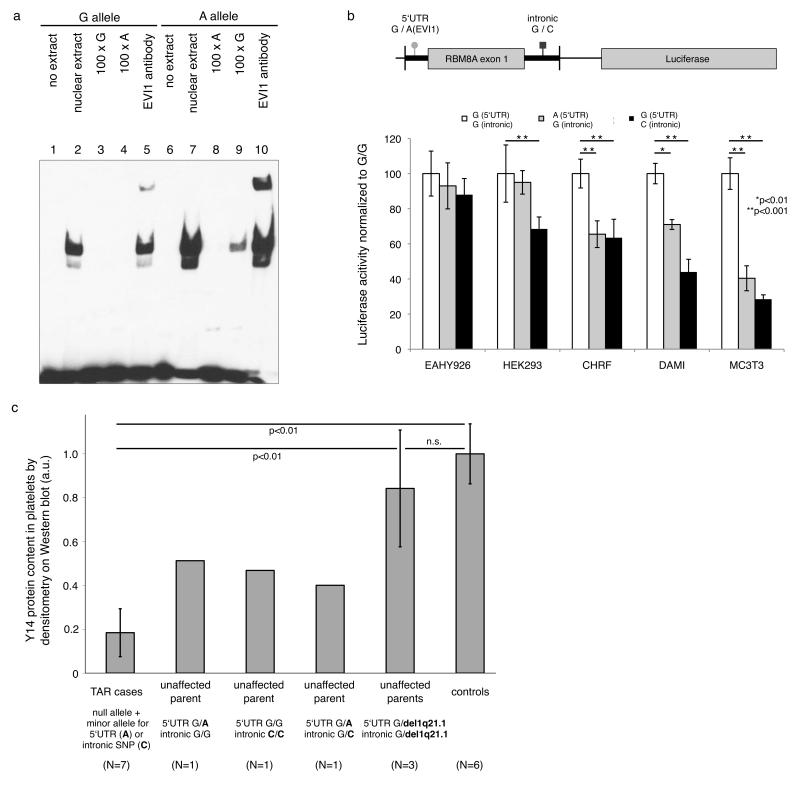
Figure 2. Effect of the regulatory SNPs on transcription factor binding, RBM8A promoter activity and protein expression in platelets.
a) Electrophoretic mobility shift assays (EMSA) in nuclear protein extracts from the megakaryocytic cell line CHRF-288-11 showed higher protein affinity of the probe containing the A-allele (lane 7) than the G-allele (lane 2) of the 5′UTR non-coding SNP. Protein binding of A-allele-probes was competed by specific unlabeled probes (lane 8), but not by unspecific G-allele-probes (lane 9). We observed a supershift with Evi1 antibodies for DNA-protein complexes containing the A-allele-probe (lane 10), indicating that the 5′UTR SNP minor allele increases binding affinity for the transcription factor EVI1 in vitro.
b) Luciferase reporter assays in cell lines representative of megakaryocytes (CHRF, DAMI) and osteoblasts (mouse MC3T3) showed significantly decreased RBM8A promoter activity for the minor alleles of both the 5′UTR and intronic non-coding SNPs (indicated by asterisks,). No effect of the 5′UTR SNP was observed in human endothelial EAHY926 and HEK293 cells. Error bars represent s.d. Statistical analysis by Tukey–Kramer multiple comparisons test.
c) Densitometry analysis of immunoblot staining for Y14, the protein encoded by the RBM8A gene, in platelet lysates from seven TAR cases, six parents (three with the 1q21.1 deletion, one heterozygote for the 5′UTR SNP, one homozygote for the 5′UTR SNP and one compound heterozygote for the 5′UTR and intronic SNPs) and six controls. Results show a significantly reduced Y14 protein level in TAR cases when compared to parental and control samples. Immunoblots are presented in Supplementary Fig. 3. Error bars represent s.d. Statistical analysis by heteroscedastic t-test. Only genotype configurations indicated by lines were compared. n.s., not significant.
We investigated if there are any variants in strong linkage disequilibrium (LD) with either the 5′UTR or the intronic SNP (Supplementary Fig. 5). We could identify no such candidates for the 5′UTR SNP, and haplotype analysis using the four exome-sequenced TAR cases carrying the 5′UTR SNP minor allele showed that this allele was present on at least two distinct haplotype backgrounds. This provides an additional line of evidence that the 5′UTR SNP minor allele is causative. We did identify a rare non-coding SNP (chr1:145483747 C/T) 25 kb upstream of RBM8A in high LD with the intronic SNP; Sanger sequencing confirmed that this variant was present in all 11 genotyped TAR cases carrying the minor allele of the intronic SNP. The data from the ENCODE Project and our own FAIRE-Seq open-chromatin data in megakaryocytes indicate that this additional SNP is not located in a regulatory region, whereas the intronic SNP is. Increased protein binding to the minor allele further corroborates the assumption that the intronic SNP is causative. We cannot exclude the possibility that the 5′UTR SNP or the intronic SNP are not the causative variants; however, in light of the biological and genetic evidence we believe this is unlikely.
Y14 is one of the four components of the exon junction splicing complex (EJC), which is involved in basic cellular functions such as nuclear export and subcellular localization of specific transcripts2,4, translational enhancement5 and nonsense-mediated RNA decay (NMD)1,3,4. The RBM8A transcript is widely expressed15, is present in all hemopoietic lineages (Supplementary Fig. 6) and the protein sequence is highly conserved between species (Supplementary Fig. 7). Considering the important function of the EJC, it is likely that a complete lack of Y14 in humans is not viable. Indeed, in Drosophila, knockdown of its ortholog tsu leads to major defects in abdomen formation16 and we found that knock-down of the orthologous rbm8a transcript in Danio rerio by antisense morpholinos resulted in extreme malformations and death two days post fertilization (Supplementary Fig. 8). This is comparable with studies of a knock-down model of the companion EJC protein Eif4a3 showing that the EJC plays a central role in vertebrate embryogenesis17. In this context, our results are compatible with both a dose-effect phenomenon and a lineage dependent deficiency of Y14. The dose-effect phenomenon is supported by the observation that simple haploinsufficiency is not sufficient to create an aberrant phenotype as evidenced by the apparently healthy carriers of the 1q21.1 deletion. We also did not observe an effect on platelet count of either the 5′UTR SNP or the intronic SNP in the 403 and 59 individuals from the Cambridge BioResource who carried the minor allele for each SNP respectively (Supplementary Table 3). This suggests that compound inheritance of a null allele together with the minor allele of one of the two regulatory SNPs brings Y14 levels below a critical threshold in certain tissues. The cell-line dependent effect shown in the luciferase assay is likely to be the result of differences in RBM8A gene expression regulation by combinatorial binding of transcription factors (including Evi1) in the context of the regulatory SNP. A further mechanism by which a deficiency in Y14 (and therefore of the EJC) may not be ubiquitous is suggested by studies showing that NMD not only targets nonsense mRNAs but also regulates physiological mRNA abundance in a gene-specific manner (reviewed in ref. 18). Hemopoietic-specific knock-down of the core NMD protein Upf2 in the mouse for example resulted in complete disappearance of the hemopoietic stem cell compartment, whilst more differentiated cells were only mildly affected19. Finally, in addition to a tissue-dependent effect, it is possible that the regulatory SNPs have a developmental stage-dependent effect: in Mus musculus, Evi1 is expressed in a transient manner in emerging limb buds20. This may provide an explanation for the skeletal abnormalities observed in TAR.
In conclusion, we have used DNA sequencing to uncover the genetic basis of TAR syndrome and identified a genetic mechanism of compound inheritance involving a null allele together with a low-frequency regulatory variant. In the case of TAR syndrome the combination of a rare deletion and a low frequency regulatory SNP reduces Y14 abundance, probably in a cell-type- and developmental stage-dependent manner. Whether the same mechanism underlies other Mendelian disorders, in particular in other microdeletion syndromes showing variable penetrance and expression, remains to be established but it highlights the importance of analyzing regulatory regions for causative mutations. While we have demonstrated altered protein binding affinity for the minor alleles of the regulatory SNPs, the mechanisms by which these SNPs lead to reduced Y14 expression in platelets are not clear, and may be different for the 5′UTR SNP and the intronic SNP. Although genetic defects in the minor spliceosome21,22 and nonsense-mediated decay23 have been linked to human disease, to the best of our knowledge, TAR syndrome is the first human disorder caused by a defect affecting one of the four EJC subunits.
Methods
Exome sequencing
We applied the Agilent SureSelect protocol (Agilent, South Queensferry, UK, catalogue no. G3362A) to enrich for 39.3 Mb of exonic sequence24. The enriched DNA was sequenced on the Illumina GAII platform (Illumina, Little Chesterford, UK). We generated 13.1-13.5 Gb of sequence per individual, resulting in a mean coverage of 123-127-fold and 89.9-90.5% of the targets were covered at least 10-fold.
Sequence analysis
Sequence analysis was performed as described previously25, with the main difference that here we considered sequence variants with allele frequency up to 5% as inferred from variation data from dbSNP131, the 1000 Genomes Project11, and 354 exomes from the CoLaus cohort12.
Genotyping of the 5′UTR and intronic SNPs in Cambridge BioResource samples
The RBM8A 5′UTR and intronic SNPs were genotyped in 7504 individuals from the Cambridge BioResource with custom TaqMan SNP Genotyping Assays (Applied Biosystems, Warrington, UK) according to the manufacturers’ protocols. All genotyping data were scored twice by different operators. Supplementary Table 2 shows the genotype counts and the corresponding estimated minor allele frequencies (MAF) for both variants. There was no evidence for deviation from Hardy-Weinberg equilibrium (Supplementary Table 2).
Preparation of primary human megakaryocytes
Megakaryocytes (MKs) were obtained from cord blood-derived CD34+ hematopoietic stem cells (HSCs) by cultures for 7 days in a medium supplemented with human recombinant thrombopoietin (THPO) and interleukin-1ß (IL1B)26.
Sequencing of megakaryocyte RNA
Megakaryocyte RNA was sequenced as described previously25. Then reads were aligned to the February 2009 Homo sapiens high coverage assembly (hg19) using GSNAP27 version 2011-03-28. Read trimming was disabled and we allowed for up to 5 mismatches and novel splicing sites at most 100,000 bp apart. Visualization on the IGV browser28 showed that the RBM8A gene is transcribed in MKs, as confirmed by qPCR (see S.I.).
Formaldehyde-assisted isolation of regulatory elements (FAIRE)
Primary megakaryocytes from three unrelated individuals were obtained as described above. For each sample, we cross-linked approx. 15 million primary megakaryocytes with 1% formaldehyde for 12 min at room temperature, and subsequently performed FAIRE experiments as previously described14,29. FAIRE DNA was processed following the Illumina paired-end library generation protocol. Libaries were sequenced with 54 bp paired-end reads on Illumina GA II and aligned as described previously25. In order to reduce experimental noise of individual preparations, we pooled the read fragments from the three individuals. The coverage profile on the combined data was created using the R packages ShortRead30 and rtracklayer31.
Predicted transcription factor binding site analysis
Transcription factor binding sites were annotated using the software MatInspector32 with the following parameters: library version: 8.3 (October 2010); matrix group: general core promoter elements and vertebrates; core=1.00; matrix=optimized+0.02.
Electrophoretic mobility shift assay (EMSA)
CHRF-288-11 cells were cultured as previously described14. Nuclear protein extract was prepared with the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific, Waltham, USA) following manufacturer’s instructions. Oligonucleotides for gel shift assays were as follows: for the 5′UTR G/A SNP at position 1:145,507,646, probe sequence 5′-biotin-AGT GTC TGA GCG GCA CAG AC[G/A] AGA TCT CGA TCG AAG G; for the intronic G/C SNP at position 1:145,507,765, probe sequence 5′-biotin-AGA CGG CTG GTG GGA AGC [G/C]GG GAA GGT GCG AGA GAA GG. Competitor probes were prepared without biotin labels. The labeled strands were annealed with the unlabeled complementary strands using a standard protocol. All oligonucleotides were provided by Sigma-Aldrich (St. Louis, USA). We performed gel shift assays as previously decribed14. For competition assays, we used 100-fold molar excess of the unlabeled probes. For the 5′UTR SNP, supershift experiments were performed with EVI1 antibodies (sc-8707 X, Santa Cruz Biotechnology, Santa Cruz, USA). Reactions were incubated for 45 min at room temperature. The reaction products were separated via electrophoresis for 75 min on 6% DNA Retardation Gels (Invitrogen, Paisley, UK) in 1x Novex TBE Running Buffer (Invitrogen), followed by transfer onto nylon membranes (Biodyne B, Thermo Fisher Scientific) and detection using the Chemiluminescent Nucleic Acid Detection Module (Thermo Fisher Scientific). For the intronic SNP, supershift experiments were performed using MZF1 (sc-46179 X and sc-66991 X) and RBPJ (sc-28713 X, all Santa Cruz Biotechnology) antibodies with 0.1 mM EDTA in the binding reaction and 120 min incubation at room temperature.
RBM8A promoter activity by luciferase reporter assay
Co-transfection experiments of different cell lines (EAHY926, HEK296, MC3T3, CHRF-288-11 and DAMI) were performed with pEGFP (Clontech, Mountain View, CA) and reporter plasmid RBM8A (wild type, with 5′UTR or intronic SNP)-pGL3-luciferase (Promega, Madison, WI). The RBM8A promoter region starting from nucleotide position -303, exon 1 and the first 142 nucleotides of intron 1 were cloned 5′ of the luciferase gene. For each co-transfection assay, cells were transfected using lipofectamin (Life Technologies, Carlsbad, CA) with 2 μg pEGFP and 4 μg RBM8A-pGL3 plasmid for HEK293, EAHY926 and MC3T3 cells. DAMI and CHRF cells were transfected using the Amaxa electroporation system (method X-01; Lonza AG, Cologne, Germany). Luciferase activity was determined as described33. Each plasmid was assayed in six separate transfection experiments and Firefly luciferase activity was standardized with GFP expression. Statistical analysis was performed using InStat 3.01 software (GraphPad Software Inc., San Diego, CA).
Y14 protein expression analysis in platelet extracts
Blood (20 ml) anticoagulated with 3.8% (wt/vol) trisodium citrate was centrifuged at 200 g and the platelet rich plasma (PRP) centrifuged at 700 g with 0.1 vol of ACD (2.5% trisodium citrate, 1.5% citric acid, 2% D-glucose) pH4.5 to obtain a platelet pellet subsequently lysed in ice-cold PBS containing 1% igepal CA-630 (Sigma Chemical, St Louis, MO), 1 mmol/L EDTA, 2 mmol/L DTE and 1 protease inhibitor cocktail tablet (Roche)/50 mL and cleared of insoluble debris by centrifugation at 16,100 g for 10 minutes at 4°C. Protein fractions were mixed with 5% SDS reducing sample buffer, separated by SDS/PAGE acrylamide gels and transferred to Hybond ECL-nitro-cellulose membrane (GE Heathcare, Buckinghamshire, UK). After blocking with Tris-buffered saline with Tween-20 supplemented with 5% non-fat dry milk, the blots were incubated with primary antibodies against Y14, Gsα or β-actin, followed with HRP-conjugated secondary antibody and detection with ECL reagent (Thermo Scientific Pierce, Rockford, IL). The following primary antibodies were used: rabbit polyclonal Y14 (Q-24), mouse monoclonal anti-Y14 (4C4) (both from Santa Cruz Biotechnology, Heidelberg, Germany), mouse monoclonal anti-Gsα(1) / mouse monoclonal anti-β-actin (Sigma Chemical, St Louis, Mo). Both Y14 antibodies were tested for their specificity using recombinant Y14-GST purified by sepharose beads as described34. Densitometry analysis was carried out using ImageJ64 software.
Allele-specific expression
Leucodepleted platelet pellets were generated from EDTA anticoagulated blood taken from Cambridge Bioresource donors heterozygotes for the 5′UTR SNP by means of serial spins and leukodepletion with anti-CD45 magnetic beads (Dynabeads CD45, 111.53D; Invitrogen, Paisley) as described previously35. The pellets were resuspended in 2mL Trizol (Invitrogen) and RNA was prepared essentially according to Trizol manufacturer’s instructions. After treatment with Turbo-DNA Free reagent (Applied Biosystems, Warrington, UK), cDNA was generated using the Superscript III method, with random hexamers (Invitrogen). Genomic DNA (gDNA) was prepared from whole blood using the Guanidine Hydrochloride - chloroform method. A PCR to amplify exon 1 of RBM8A from both gDNA and cDNA was then performed using AmpliTaq GOLD (Applied Biosystems, Warrington, UK), dNTPs (800nM; GE. Little Chalfont, UK) and the primers described in Supplementary Table 3 with the following cycling conditions; 95°C 10min; then cycling sequence of 95°C 15sec, 66°C 30sec for 5 cycles with the latter step decreasing by 1 degree per cycle; then standard cycling (30 cycles) of 95°C 15sec, 60°C 30sec; followed by incubations of 72°C 7 min, then 4°C. The PCR products were purified by spin column (D4014; Zymo Research, Irvine, CA) and ligated into TOPO vector (Invitrogen) at 20-25°C for 2 hours. Four μl of this was used to transform chemically competent TOP10 cells (Invitrogen) before plating onto FastMedia Amp XGal Agar (InvivoGen, San Diego, CA). After overnight growth, white colonies were picked into separate wells of 96-well PCR plates and colony PCR performed with AmpliTaq GOLD and the primers described in Supplementary Table 3 and the following cycling conditions: 95°C 10min; then cycling sequence of 95°C 15sec, 54°C 45sec, 72°C 15sec, for 5 cycles with the annealing step decreasing by 1 degree per cycle; then standard cycling (30 cycles) of 95°C 15sec, 48°C 45sec, 72°C 15sec; followed by incubations of 72°C 7 min, then 4°C. The PCR products were genotyped with custom TaqMan SNP Genotyping Assays (Applied Biosystems, Warrington, UK) according to the manufacturers’ protocols.
qPCR hemopoietic lineages
cDNA was prepared from leucodepleted pellets as described above. cDNA preparation from the other haemopoietic lineage has been described36. TaqMan gene expression analysis was performed on cDNA using proprietary reagents, according to the manufacturer’s instructions (Applied Biosystems, Warrington, UK.). The assay numbers were GAPDH (Hs99999905_m1) and RBM8A (Hs4234933_g1). Assays were conducted in 384-well format on a 7900HT Sequence Detection System (Applied Biosystems) and the threshold cycle number (Ct) for GAPDH was subtracted from that of the other genes assayed on that sample (ΔCt), to normalize for reaction loading.
Supplementary Material
Acknowledgments
We thank Cordelia Langford and Peter Ellis for performing the enrichment for the exome sequencing, Senduran Balasubramaniam for assistance with data processing, and the Wellcome Sanger Institute sequencing core for sequencing. We thank Vincent Mooser from GlaxoSmithKline, Gerard Waeber and Peter Vollenweider from the CoLaus Cohort (Lausanne, Switzerland) and Jillian Durham, Carol Scott, and colleagues at the Sanger Institute for providing access to their collection of whole-exome sequencing data of the CoLaus cohort12. We thank Richard Durbin, Bertie Göttgens, and Isabel Palacios for comments on the manuscript. This study makes use of data generated by the UK10K Consortium, derived from samples from the TwinsUK cohort. A full list of the investigators who contributed to the generation of the data is available from www.UK10K.org. The study was supported by grants from the National Institute for Health Research (NIHR) (RP-PG-0310-1002, to CG, GK, PAS & WHO), from the British Heart Foundation (FS/09/039 to CG, RG/09/12/28096 to CAA), project grants from the Wellcome Trust (WT-082597/Z/07/Z to AC) and (WT-084183/2/07/2 to JS), grants by the Deutsche Forschungsgemeinschaft (SCHU1421/3-1) to H.S.) and the Sanitätsrat Dr. Emil Alexander Hübner-und-Gemahlin-Stiftung (T114/17644/2008/sm, to HS), by the ‘Excellentie financiering KULeuven’ (EF/05/013), by research grants G.0490.10N and G.0743.09 from the Fonds Wetenschappelijk Onderzoek-Vlaanderen (Belgium) and by GOA/2009/13 from the Research Council of the University of Leuven (Onderzoeksraad K.U.Leuven, Belgium) (to CT, KF, CVG). The project made use of NHS Blood and Transplant donors from the Cambridge BioResource (http://www.cambridgebioresource.org.uk/). This local resource for genotype-phenotype association studies is supported by a grant from the NIHR to the Cambridge Biomedical Research Centre to JS and JDJ. DSP and MK were supported by the Marie-Curie NetSim Initial Training Network grant EC-215820. The French cases were collected with support from Gis-Maladies Rares, DIATROC program, INSERM (ANR-08-GENO-028-03). Funding for UK10K was provided by the Wellcome Trust under award WT091310.
Footnotes
Author contributions CAA performed next-generation sequence analysis, Sanger sequence analysis, genetic analysis and statistical analysis. DSP performed EMSA experiments, FAIRE-Seq experiments and analysis, and in silico transcription factor binding analysis, under supervision of PD. HS, KF, JF, KS, CT, and RNE ascertained deletion status for TAR cases. KF and CT performed luciferase assays. HS, KF, CT, CG, and CMH performed Western blot experiments. JCS performed the Sanger sequencing and analyzed the data. PAS performed qPCR and allele-specific expression experiments. JDD performed allele-specific expression experiments. AC performed the zebrafish knockdown study with input from DLS. MK analyzed the megakaryocyte RNA-Seq data, under supervision of PB. GK supervised exome sequencing. JGS supervised the Cambridge BioResource study. NH and MH performed the CNV analyses. HS, MB, ND, RF, IK, PN, CALR, GS, CVG, RNE and CG clinically characterized TAR cases. CAA, KF, WHO and CG wrote the paper.
Accession IDs RBM8A, NCBI reference sequence NM_005105. The sequence data has been submitted to the European Genotype-Phenotype Archive under accession ID EGAD00001000018.
URLs Cambridge BioResource, http://www.cambridgebioresource.org.uk; The European Genome-phenome Archive (EGA), http://www.ebi.ac.uk/ega; UK10K, http://www.uk10k.org.
References
- 1.Kim VN, Kataoka N, Dreyfuss G. Role of the Nonsense-Mediated Decay Factor hUpf3 in the Splicing-Dependent Exon-Exon Junction Complex. Science. 2001;293:1832–1836. doi: 10.1126/science.1062829. [DOI] [PubMed] [Google Scholar]
- 2.Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lykke-Andersen J, Shu M-D, Steitz JA. Communication of the Position of Exon-Exon Junctions to the mRNA Surveillance Machinery by the Protein RNPS1. Science. 2001;293:1836–1839. doi: 10.1126/science.1062786. [DOI] [PubMed] [Google Scholar]
- 4.Palacios IM, Gatfield D, St Johnston D, Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427:753–757. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- 5.Wiegand HL, Lu S, Cullen BR. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proceedings of the National Academy of Sciences. 2003;100:11327–11332. doi: 10.1073/pnas.1934877100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall J, et al. Thrombocytopenia with absent radius (TAR) Medicine (Baltimore) 1969;48:411–439. doi: 10.1097/00005792-196948060-00001. [DOI] [PubMed] [Google Scholar]
- 7.Klopocki E, et al. Complex Inheritance Pattern Resembling Autosomal Recessive Inheritance Involving a Microdeletion in Thrombocytopenia-Absent Radius Syndrome. The American Journal of Human Genetics. 2007;80:232–240. doi: 10.1086/510919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geddis AE. Inherited Thrombocytopenia: Congenital Amegakaryocytic Thrombocytopenia and Thrombocytopenia With Absent Radii. Seminars in Hematology. 2006;43:196–203. doi: 10.1053/j.seminhematol.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Greenhalgh KL, et al. Thrombocytopenia-absent radius syndrome: a clinical genetic study. Journal of Medical Genetics. 2002;39:876–881. doi: 10.1136/jmg.39.12.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soranzo N, et al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat Genet. 2009;41:1182–1190. doi: 10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The 1000 Genomes Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Firmann M, et al. The CoLaus study: a population-based study to investigate the epidemiology and genetic determinants of cardiovascular risk factors and metabolic syndrome. BMC Cardiovasc Disord. 2008;8:6. doi: 10.1186/1471-2261-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The ENCODE Consortium Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paul DS, et al. Maps of Open Chromatin Guide the Functional Follow-Up of Genome-Wide Association Signals: Application to Hematological Traits. PLoS Genet. 2011;7:e1002139. doi: 10.1371/journal.pgen.1002139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salicioni AM, et al. Identification and Structural Analysis of Human RBM8A and RBM8B: Two Highly Conserved RNA-Binding Motif Proteins That Interact with OVCA1, a Candidate Tumor Suppressor. Genomics. 2000;69:54–62. doi: 10.1006/geno.2000.6315. [DOI] [PubMed] [Google Scholar]
- 16.Hachet O, Ephrussi A. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Current biology : CB. 2001;11:1666–1674. doi: 10.1016/s0960-9822(01)00508-5. [DOI] [PubMed] [Google Scholar]
- 17.Haremaki T, Sridharan J, Dvora S, Weinstein DC. Regulation of vertebrate embryogenesis by the exon junction complex core component Eif4a3. Developmental Dynamics. 2010;239:1977–1987. doi: 10.1002/dvdy.22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicholson P, et al. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cellular and Molecular Life Sciences. 2010;67:677–700. doi: 10.1007/s00018-009-0177-1. 700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weischenfeldt J, et al. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes & Development. 2008;22:1381–1396. doi: 10.1101/gad.468808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perkins AS, Mercer JA, Jenkins NA, Copeland NG. Patterns of Evi-1 expression in embryonic and adult tissues suggest that Evi-1 plays an important regulatory role in mouse development. Development. 1991;111:479–487. doi: 10.1242/dev.111.2.479. [DOI] [PubMed] [Google Scholar]
- 21.Edery P, et al. Association of TALS Developmental Disorder with Defect in Minor Splicing Component U4atac snRNA. Science. 2011;332:240–243. doi: 10.1126/science.1202205. [DOI] [PubMed] [Google Scholar]
- 22.He H, et al. Mutations in U4atac snRNA, a Component of the Minor Spliceosome, in the Developmental Disorder MOPD I. Science. 2011;332:238–240. doi: 10.1126/science.1200587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tarpey PS, et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet. 2007;39:1127–1133. doi: 10.1038/ng2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coffey AJ, et al. The GENCODE exome: sequencing the complete human exome. Eur J Hum Genet. 2011;19:827–831. doi: 10.1038/ejhg.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Albers CA, et al. Exome sequencing identifies NBEAL2 as the causative gene for gray platelet syndrome. Nat Genet. 2011;43:735–737. doi: 10.1038/ng.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tijssen MR, et al. Genome-wide Analysis of Simultaneous GATA1/2, RUNX1, FLI1, and SCL Binding in Megakaryocytes Identifies Hematopoietic Regulators. Developmental Cell. 2011;20:597–609. doi: 10.1016/j.devcel.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giresi PG, Lieb JD. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements) Methods. 2009;48:233–239. doi: 10.1016/j.ymeth.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan ShortRead: a Bioconductor package for input, quality assessment, and exploration of high throughput sequence data. Bioinformatics. 2011;25:2607–9. doi: 10.1093/bioinformatics/btp450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawrence M, Gentleman R, Carey V. rtracklayer: an R package for interfacing with genome browsers. Bioinformatics. 2009;25:1841–1842. doi: 10.1093/bioinformatics/btp328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cartharius K, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 33.Freson K, et al. 391 C to G substitution in the regulator of G-protein signalling-2 promoter increases susceptibility to the metabolic syndrome in white European men: consistency between molecular and epidemiological studies. J Hypertens. 2007;25:117–25. doi: 10.1097/HJH.0b013e3280109c6c. [DOI] [PubMed] [Google Scholar]
- 34.Freson K, et al. Platelet characteristics in patients with X-linked macrothrombocytopenia because of a novel GATA1mutation. Blood. 2001;98:85–92. doi: 10.1182/blood.v98.1.85. [DOI] [PubMed] [Google Scholar]
- 35.Gnatenko DV, et al. Transcript profiling of human platelets using microarray and serial analysis of gene expression. Blood. 2003;101:2285–2293. doi: 10.1182/blood-2002-09-2797. [DOI] [PubMed] [Google Scholar]
- 36.Watkins NA, et al. A HaemAtlas: characterizing gene expression in differentiated human blood cells. Blood. 2009;113:e1–9. doi: 10.1182/blood-2008-06-162958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.