Abstract
Structural variation is widespread in mammalian genomes1,2 and is an important cause of disease3, but just how abundant and important structural variants (SVs) are in shaping phenotypic variation remains unclear4,5. Without knowing how many SVs there are, and how they arise, it is difficult to discover what they do. Combining experimental with automated analyses, we identified 0.71M SVs at 0.28M sites in the genomes of thirteen classical and four wild-derived inbred mouse strains. The majority of SVs are less than 1 kilobase in size and 98% are deletions or insertions. The breakpoints of 0.16M SVs were mapped to base pair resolution allowing us to infer that insertion of retrotransposons causes more than half of SVs. Yet, despite their prevalence, SVs are less likely than other sequence variants to cause gene-expression or quantitative phenotypic variation. We identified 24 SVs that disrupt coding exons, acting as rare variants of large effect on gene function. One third of the genes so affected have immunological functions.
The preeminent organism for modeling the relationship between phenotype and genotype, including SVs, is the mouse, but our catalogue of SVs in this animal is incomplete6 and most of what we know about the impact of SVs on phenotypes comes from analyses of gene expression7,8. Up to 28% of the between-strain variation in gene expression in hematopoietic stem and progenitor cells has been attributed to SVs7; SVs may account for between 66% to 74% of between-strain expression variation in kidney, liver, lung and testis8. Since gene expression variation is believed to contribute to variation in phenotypes in the whole organism9, SVs may turn out to have a major role in the genetic determination of many aspects of mouse biology.
Combining short-read paired-end mapping with experimental analyses (Supplementary Methods), we found SVs greater than 100 bp at 0.28M sites in the mouse genome, amounting to 0.71M SVs in thirteen classical and four wild-derived inbred strains of mice (Supplementary Table 1a), affecting 1.2% (33.0 Mb) and 3.7% (98.6 Mb) of the genome respectively (Supplementary Table 1b). Deletions, a category we can measure accurately, have a median size of 349 bp with modes at 100 bp and 6,400 bp (Supplementary Fig. 1a).
Our catalogue contains far more SVs than previously recognized: 99.4% of SVs are simple and 0.6% are complex (Supplementary Table 1a), where simple SVs include insertions, deletions, inversions and copy number gains, and complex SVs consist of a mixture of events that abut each other. From experimental analyses of simple deletion SVs, we estimated an average false negative rate of 17% in the classical inbred strains (Supplementary Table 2a, 2b and 3a) and 24% in the wild-derived strains (Supplementary Table 2b); false positive rates were below 5% for all strains (Supplementary Table 2c). False negative rates for non-deletion simple SVs as well as complex SVs were higher than for simple deletions, ranging from 24% to 31% and 35% to 54% per strain, respectively (Supplementary Table 3b).
It proved difficult to obtain robust estimates of SVs smaller than 100 bp. Our best estimate of the rate of SVs between 30 and 100 bp is based on combining manual and automated methods over a region of 7.2 Mb (Supplementary Methods). Assuming that this region is typical, the rest of the genome (in classical laboratory strains) contains approximately 49,000 SVs in this size range.
Microhomology at SV breakpoints, as well as the sequence content within SVs and the SV’s ancestral state, were used to infer the likely mechanism of formation for simple SVs. To obtain breakpoint sequence, we performed de novo local assembly for 80.3% of deletions. Comparison of 1,314 predicted deletion breakpoints to the breakpoint delineated by PCR and sequencing (Supplementary Table 4) revealed that 57.7% of breakpoint predictions are exact and 86.5% are within 20 bp (Supplementary Table 5a). In cases where the local assembly strategy failed, we relied on the original breakpoint estimates obtained from the mapping of reads to the reference genome: 83.3% of these estimates are within 100 bp of the actual breakpoint (Supplementary Tables 5b). Breakpoint accuracy for insertions, inversions and copy number gains is presented in Supplementary Tables 5c, 5d and 5e, respectively.
Genome-wide estimates of the contribution of each mechanism to SV formation were derived from analysis of breakpoint sequence of deletions relative to C57BL/6J. We have highly accurate breakpoint sequence for this SV category, which should be unbiased with respect to ancestry. Using rat as an outgroup, we classified 19% of relative deletion SVs as ancestral deletions, 57% as ancestral insertions and the remainder (24%) were indeterminate (Supplementary Fig. 2).
SVs are most often due to retrotransposons (LINEs (25%), LTRs (14%) and SINEs (15%)), followed by variable number tandem repeats (VNTRs) (15%) and pseudogenes (2%). Other mechanisms, not involving retrotransposons, account for 29% of SVs. Outgroup analysis showed that the transposon-associated SVs arose almost exclusively from ancestral insertions events (98.8%). Target site duplications (12-16 bp) surround the breakpoints of LINE and SINE derived SVs; shorter (6-8 bp) sequences are associated with LTR SVs (Supplementary Fig. 1b). Non-repeat mediated SVs are mainly a result of ancestral deletion events (79%), and are associated with microhomologies up to 7 bp in length (Supplementary Fig. 1b), consistent with either microhomology-mediated break-induced replication (MMBIR)10 or microhomology-mediated end joining (MMEJ)11.
Given their potential role in human disease12, we were interested to document the occurrence of SVs that arise at the same genomic locus independently in unrelated strains (recurrent SVs). Non-allelic homologous recombination (NAHR) is the major mechanism for recurrent SVs13, while fork stalling and template switching (FoSTeS) and/or microhomology-mediated break-induced replication (MMBIR) mechanisms may be important for non-recurrent SVs14.
Using the SV breakpoints obtained from PCR sequencing (249 SV sites in eight strains, accounting for over 4,000 breakpoints, Supplementary Table 4), we identified SVs occurring at the same locus in different strains, but with different breakpoints, indicating independent origins. In the classical strains, only 2.5% of deletions at the same locus had different breakpoint sequences. However within all 17 strains we found multiple alleles at 12% of SVs, due almost entirely to the presence of different alleles originating from the wild-derived inbred strains. Consistent with the low frequency of recurrent SVs, breakpoint features associated with NAHR are rare. We estimated that 0.13% of deletions are due to NAHR, when we required a signature of >=200 bp of >=90% sequence identity.
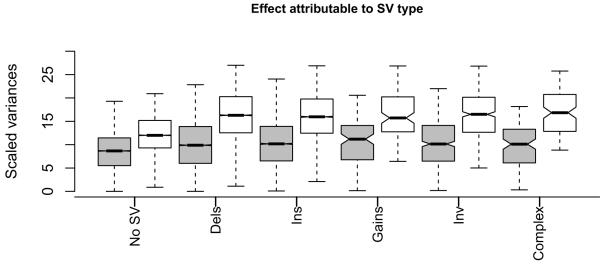
We assessed the impact of SVs on phenotypes by first estimating the proportion of heritability attributable to SVs8 from brain RNAseq and found that no category accounts for more than 10% (Fig. 1). To determine if these results were specific to brain tissue, we analysed gene expression data for the eight founder strains of the heterogeneous stock (HS) population (n = 5 for each) from liver, measured on Illumina gene-expression arrays15. Mean heritability attributable to an SV, for transcripts overlapping one or more SVs, was 9.5%. Since many transcripts overlap multiple small SVs (median of 3, maximum of 216), we hypothesized that SV heritability might be related to the amount of gene overlapped. For each transcript we summed the amount of DNA overlapping a gene and expressed this as a proportion of the total length of the gene. SVs that overlap 50% or more of a gene make a large contribution to heritability: in brain tissue, such SVs contribute to 25% of the variance, compared to 7.8% for transcripts where SVs overlap less than 50% of the gene. However, large overlaps (50% or more) are rare, affecting less than 3% of transcripts. Thus while SVs make a modest contribution to the overall heritability of expression variance, at individual transcripts they may be the main cause of between-strain differences in expression.
Figure 1. Impact of SVs on gene expression.
Within-strain (grey boxes) and between-strain (white boxes) gene expression variances for transcripts which are not overlapped by any structural variant (No SV) and for those which are overlapped. Within-strain variance is due to environmental effects; between strain to environmental and genetic effects. The difference between the two variances is a measure of heritability. Six categories are shown: deletions (Dels), insertions (Ins), copy number gains (Gains), inversions (Inv), and complex rearrangements (Complex).
As another method to assess the impact of SVs on phenotype, we applied a test of functionality16 to 281,246 SVs in association with 100 phenotypes measured in over 2,000 HS mice17. We identified 290 QTLs where SVs were among the variants most likely to be functional, but in all these cases the SVs were only a subset of the total number of functional variants. We found a small but significant deficit in SVs among the functional variants (0.36% compared to 0.54% among the non-functional, P < 1E-16, χ 2 = 72.1).
While SVs make a relatively small contribution to the total amount of quantitative phenotypic variation, at a small number of QTLs they are the cause of variation. As shown in our companion paper18, larger effect QTLs are more likely to arise from SVs. We identified 12 QTLs where the SV overlapped a gene or flanking region (2 Kb up and downstream), and where the QTL effect size is in the top 5% of the distribution. Table 1 lists these SVs, the genes they affect and the putative phenotype with which they are associated. Two associations have been directly tested: complementation of the deletion of the H2-Ea promoter has confirmed the effect of this SV on the T-cell phenotype19; analysis of a knock out of Eps15 showed the predicted lower locomotor activity (Fig. 2a).
Table 1.
QTLs associated with SVs
| Phenotype | Chr | SV start | SV stop | Ancestral event | Gene | SV overlap |
|---|---|---|---|---|---|---|
| Mean platelet volume | 1 | 175158884* | 175158885* | ins (large) | Fcer1a | upstream |
| OFT Total activity | 2 | 144402760 | 144402971 | SINE ins | Sec23b | intron |
| Hippocampus cellular proliferation marker | 4 | 49690362 | 49690363 | del (137 bp) | Grin3a | intron |
| Home cage activity | 4 | 108951263 | 108951264 | IAP ins (~6400 bp) | Eps15 | upstream |
| T-cells: %CD3 | 4 | 130038388 | 130038389 | SINE ins (202 bp) | Snrnp40 | intron |
| Wound healing | 7 | 90731819 | 90731820 | IAP ins (~6400 bp) | Tmc3 | upstream |
| Red cells: mean cellular haemoglobin | 7 | 111397607 | 111479433 | ins | Trim5 | exon |
| Red cells: mean cellular haemoglobin | 7 | 111504989 | 111505193 | del | Trim30b | UTR |
| Red cells: mean cellular volume | 8 | 87957244 | 87957245 | LINE ins (~500 bp) | 4921524J17Rik | upstream |
| Serum urea concentration | 11 | 115106127 | 115106250 | del | Tmem104 | UTR |
| Hippocampus cellular proliferation marker | 13 | 113783196 | 113783359 | del | Gm6320 | upstream |
| T-cells: CD4/CD8 ratio | 17 | 34483681 | 34483682 | del (629 bp) | H2-Ea | upstream |
Ins: insertion; del: deletion; SINE: Short INterspersed repeat Elements; LINE: Long INterspersed repeat Elements; IAP: Intracisternal A Particle. Start and stop coordinates are given for Build37 of the mouse genome. Unless there is a plus sign (“+”), coordinates refer to the exact coordinates as delineated by Sanger sequencing.
Figure 2. Experimental analysis of SVs.
a) Locomotor activity in Eps15 -/- mice. Activity was recorded during a period of 10 minutes in an open field arena. N = 7 for Eps15 -/- male mice and n= 16 for matched control wildtype.
b) Schematic representation of the Trim6-Trim30 genes cluster on chromosome 7. Boxes represent the sequential positions of the Trim6, Trim34, Trim5/12 and Trim30 genes. Trim5 and Trim12a genes are only present in the C57BL/6J genome occurred by segmental duplication of the Trim12c gene present in all 17 strains. The flanking Trim34 and Trim30 genes do not vary between strains. Coordinates are given for Build37 of the mouse genome.
There are relatively few examples where an SV can be said unequivocally to delete one, or more, coding exons. Without nucleotide resolution accuracy we cannot be certain whether the breakpoint of an SV lies within an exon. Therefore to find SVs overlapping exons we used our most accurate and complete category of SV calls: deletions relative to C57BL/6J. We identified 210 that overlap exons (Ensembl Build 58); after removing pseudogenes, and genes not annotated as ‘protein coding’, we were left with 24 SVs that affect coding exons, including six that encompass a gene in its entirety (Table 2).
Table 2.
SVs affecting coding regions
| MGI gene | Chr | SV start | SV stop | Ancestral Event | Known function |
|---|---|---|---|---|---|
| Soat1 | 1 | 158394620 | 158401436 | del | Hair interior defects |
| Olfr1055 | 2 | 86179898 | 86186982 | IAP ins | Olfactory |
| Fcrl5 | 3 | 87245084 | 87245947 | del | Infection and immunity |
| Nes | 3 | 87780530 | 87780662 | VNTR | Brain development |
| Pglyrp3 | 3 | 91831862 | 91835385 | del | Infection and immunity |
| Skint4,3,9* | 4 | 111731004 | 112272814 | ins | Infection and immunity |
| Fv1 | 4 | 147244398 | 147245739 | del | Infection and immunity |
| Ugt2b38 | 5 | 87850554 | 87854999 | del | Metabolism |
| Klrb1a | 6 | 128559593 | 128559740 | del | Infection and immunity |
| Klri2 | 6 | 129689526 | 129691211 | del | Infection and immunity |
| Tas2r120* | 6 | 132580541 | 132613777 | del+linked ins (326 bp) | Taste |
| Tas2r103 | 6 | 132985563 | 132986696 | del | Taste |
| Zfp607* | 7 | 28646761 | 28671650 | del | DNA-binding |
| Krtap5-5 | 7 | 149415121 | 149415210 | VNTR | Hair formation |
| Trim5,12a* | 7 | 111397607 | 111479433 | ins | Infection and immunity |
| Defb8 | 8 | 19447465 | 19450575 | ins+del (54 bp) | Infection and immunity |
| Zfp872 | 9 | 22004856 | 22005023 | VNTR | DNA-binding |
| Olfr913 | 9 | 38402589 | 38403498 | del | Olfactory |
| Rtp3 | 9 | 110889280 | 110889465 | VNTR | Bone density |
| Nlrp1c* | 11 | 71046193 | 71101410 | ins | Embryonic development |
| Fam110c | 12 | 31759321 | 31759461 | VNTR | Cell migration |
| Olfr234 | 15 | 98328544 | 98328861 | del | Olfactory |
| Krtap16-1 | 16 | 88874294 | 88874392 | VNTR | Hair formation |
| Amd2* | 18 | 64607747 | 64609669 | ins | Biosynthesis of polyamines |
MGI: Mouse Genome Informatics; VNTR: Variable Number Tandem Repeat.
indicates that the structural variant overlaps the entire gene. Unless there is a plus sign (“+”), coordinates refer to the exact coordinates as delineated by Sanger sequencing.
Five of the 24 SVs are already known20,21,22,23,24; the remaining 19 are novel. A third of the genes affected are involved in immunity and infection. Our data expands current knowledge of the molecular architecture of these SVs. Fig. 2b shows antiviral genes Trim5 and Trim12a are unique to C57BL/6J, due to segmental duplication25. All the other strains contain only the Trim12c gene. Therefore the mouse contains a unique homologue of the human TRIM5 gene. A similar analysis revealed that documented exonic changes in the beta defensin 8 gene (Defb8)26 are linked to a previously undetected 3,192 bp deletion that includes the first exon of the gene.
Our results are important in three respects: first, we find an unexpectedly large number of SVs with diverse molecular architecture, thus providing a catalogue of the most dynamic and variable regions of the mouse genome. Second, we were able to map almost 60% of deletions to base pair resolution, allowing us to classify SVs by the mechanism that created them. In contrast to human SV studies, the great majority of SVs we have discovered are non-recurrent rearrangements, based on two observations: among the classical strains, only 2.5% of deletions at the same locus had different breakpoint sequences and less than 1% of deletions are due to NAHR12. Third, SVs have relatively little impact on gene function, a conclusion based on the following observations. We found that SVs overlapping a gene account for less than 10% of variation in gene expression, three to four times less than that found by studies using expression arrays7,8. SVs overlapping exons are rare: since the frequency of insertions is equal to that of deletions, and since these two categories make up 98% of all SVs, extrapolating from the 24 SVs that delete exons, we predict that there are only about 50 SVs that directly overlap exons, or about 0.2% of the total burden of SVs in the genome. Finally, our analysis of the phenotypic consequences of SVs on QTLs for multiple phenotypes points to a relative deficit of SVs as the molecular basis of complex phenotypes. For the classical laboratory strains, SNPs and indels affect 0.5% of the genome, while on average 33 Mb (2.5%) of each classical laboratory strain falls into structurally variant regions of the genome. This implies that SVs are about five fold more likely to have phenotypic consequences than the combined effect of SNPs and indels. Yet we find that SVs contribute only 10% to the heritability of gene expression, not the 80% implied by the genomic size argument.
It is important to note that conclusions based on our analysis of the HS outbred population may not apply to other outbred populations. The mouse population we tested is derived from inbred progenitors whose homozygosity will have purged their genomes of variants that could otherwise be maintained in heterozygous freely mating populations. Nevertheless, despite their relative rarity in the mouse genome, SVs that cause phenotype change are likely to provide biological insights out of proportion to their relative small contribution to phenotypic variance. We expect that the alleles we have described will provide a starting point for investigating the relationship between phenotype and genotype in mice.
Methods Summary
SV discovery
We used a combination of four computational methods: split-read mapping27, mate-pair analysis28, single-end cluster analysis (SECluster and RetroSeq, unpublished), and read-depth29. These methods identify deletions, insertions, inversions and copy number gains. We also derived methods to recognize other types of rearrangements, such as inversion plus insertion or inversion plus deletion, newly revealed from our experimental analysis.
Experimental analysis
We visually inspected short-read sequencing data using LookSeq30 and manually detected SVs across mouse chromosome 19 in its entirety and a random set of other chromosomal regions. We analysed molecular structures of these SVs at nucleotide-level resolution using PCR and Sanger-based sequencing.
Outgroup analysis
The rat was used as an outgroup species to classify each mouse SV as either an ancestral deletion or an ancestral insertion. We predicted the ancestral state in the rat by estimating the size of the region in the rat genome that was homologous to the region that encompassed the mouse SV.
SV classification
We developed a machine learning method to classify SVs. The method used a random forest classifier, trained using sequence features within the SVs. Microhomology between breakpoints was determined by recording the longest sequence of bases that was identical between each breakpoint of each SV.
Functional impact of SVs
We tested whether an SV is likely to be functional using merge analysis16. The variances of expression data were calculated using ANOVA in the statistical software R using formulae described in8 and also by comparing a model where the expression value is explained by the strain, to a model in which the expression is explained by strain and whether or not the animal has an SV.
Supplementary Material
Acknowledgements
We thank Adam Whitley, Giles Durrant, Andrew Marc Hammond, Danica Joy Fabrigar, Lucia Chen, Martina Johannesson, Enzhao Cong and Glòria Blázquez for helping B.Y. with various laboratory-based work. We also thank Chris P. Ponting for comments on the manuscript. This project was supported by The Medical Research Council, UK and the Wellcome Trust. DJA is supported by Cancer Research UK.
Footnotes
Full methods are provided in Supplementary Information.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature. Supplementary Information contains Supplementary Figures and Tables, additional Methods, and Supplementary References.
Author information Data sets described here will be available under study accession number estd118 from the Database of Genomic Variants archive (DGVa) at http://www.ebi.ac.uk/dgva/page.php. Reprints and permissions information is available at www.nature.com/reprints. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- Mills RE, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, et al. Genome-wide mapping and assembly of structural variant breakpoints in the mouse genome. Genome Res. 2010;20:623–635. doi: 10.1101/gr.102970.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Ann. Rev. Genomics Hum. Gen. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agam A, et al. Elusive copy number variation in the mouse genome. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahan P, Li Y, Izumi M, Graubert TA. The impact of copy number variation on local gene expression in mouse hematopoietic stem and progenitor cells. Nat. Genet. 2009;41:430–437. doi: 10.1038/ng.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrichsen CN, et al. Segmental copy number variation shapes tissue transcriptomes. Nat. Genet. 2009;41:424–429. doi: 10.1038/ng.345. [DOI] [PubMed] [Google Scholar]
- Schadt EE, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat. Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, et al. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat. Genet. 2009;41:849–853. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JL, Kim EM, Haber JE, Lee SE. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 2003;23:8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Ann. Rev. Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GJ, et al. High resolution mapping of expression QTLs in heterogeneous stock mice in multiple tissues. Genome Res. 2009;19:1133–1140. doi: 10.1101/gr.088120.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin B, Flint J, Mott R. Using progenitor strain information to identify quantitative trait nucleotides in outbred mice. Genetics. 2005;171:673–681. doi: 10.1534/genetics.104.028902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W, et al. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat. Genet. 2006;38:879–887. doi: 10.1038/ng1840. [DOI] [PubMed] [Google Scholar]
- Keane T. Sequence variation amongst 17 laboratory and wild-derived mouse genomes and its affect on gene regulation and phenotypic variation. Nature. 2011 [Google Scholar]
- Yalcin B, et al. Commercially available outbred mice for genome-wide association studies. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best S, Le Tissier P, Towers G, Stoye JP. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- Boyden LM, et al. Skint1, the prototype of a newly identified immunoglobulin superfamily gene cluster, positively selects epidermal gammadelta T cells. Nat. Genet. 2008;40:656–662. doi: 10.1038/ng.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson TM, Munger SD, Boughter JD., Jr. Haplotypes at the Tas2r locus on distal chromosome 6 vary with quinine taste sensitivity in inbred mice. BMC Genet. 2005;6:32. doi: 10.1186/1471-2156-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson K, Heby O, Berger FG. The functional intronless S-adenosylmethionine decarboxylase gene of the mouse (Amd-2) is linked to the ornithine decarboxylase gene (Odc) on chromosome 12 and is present in distantly related species of the genus Mus. Mamm. Gen. 1999;10:784–788. doi: 10.1007/s003359901092. [DOI] [PubMed] [Google Scholar]
- Wu B, et al. Mutations in sterol O-acyltransferase 1 (Soat1) result in hair interior defects in AKR/J mice. J. Invest. Derm. 2010;130:2666–2668. doi: 10.1038/jid.2010.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tareen SU, Sawyer SL, Malik HS, Emerman M. An expanded clade of rodent Trim5 genes. Virology. 2009;385:473–483. doi: 10.1016/j.virol.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor K, et al. Defensin-related peptide 1 (Defr1) is allelic to Defb8 and chemoattracts immature DC and CD4+ T cells independently of CCR6. Eur. J. Immunol. 2009;39:1353–1360. doi: 10.1002/eji.200838566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JT, McIntyre RE, Adams DJ, Durbin R. Copy number variant detection in inbred strains from short read sequence data. Bioinformatics. 2010;26:565–567. doi: 10.1093/bioinformatics/btp693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manske HM, Kwiatkowski DP. LookSeq: a browser-based viewer for deep sequencing data. Genome Res. 2009;19:2125–2132. doi: 10.1101/gr.093443.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.