Binding of Smad3 to the foxp3 enhancer is not required for thymic T reg cell development, but thymic involution with aging reveals the contribution of TGF-β–Smad2 signaling in gut T reg cell development.
Abstract
Regulatory T cells (T reg cells) are essential for the prevention of autoimmunity throughout life. T reg cell development occurs intrathymically but a subset of T reg cells can also differentiate from naive T cells in the periphery. In vitro, Smad signaling facilitates conversion of naive T cells into T reg cells but results in unstable Foxp3 expression. The TGF-β–Smad response element in the foxp3 locus is located in the CNS1 region in close proximity to binding sites for transcription factors implicated in TCR and retinoic acid signaling. From in vitro experiments it was previously postulated that foxp3 transcription represents a hierarchical process of transcription factor binding in which Smad3 would play a central role in transcription initiation. However, in vitro conditions generate T reg cells that differ from T reg cells encountered in vivo. To address the relevance of Smad3 binding to the CNS1 enhancer in vivo, we generated mice that exclusively lack the Smad binding site (foxp3CNS1mut). We show that binding of Smad3 to the foxp3 enhancer is dispensable for T reg cell development in newborn and adult mice with the exception of the gut.
The TGF-β–Smad pathway plays a central role in development, maintenance, survival, and function of T cells (Li and Flavell, 2008). Numerous studies have reported severe inflammation and death of mice with complete or T cell–specific lack of individual components of this pathway attributable to increased T cell activation. Foxp3-expressing regulatory T cells (T reg cells) suppress uncontrolled activation and expansion of T cells. The role of TGF-β in the generation of T reg cells is therefore of special interest. There is, however, no consensus on T reg cell dysfunction in various mutant mice. Intrathymic T reg cell development in mice mutant in the TGF-β pathway was described to be reduced by some (Li et al., 2006; Liu et al., 2008) but not others (Marie et al., 2005, 2006; Takimoto et al., 2010). These studies are problematic because it is not clear whether the effects of a deficiency in TGF-β signaling were cell intrinsic or not.
Experiments addressing extrathymic T reg cell generation suffer from the same uncertainty, even though there are convincing data on the conversion of naive T cells into T reg cells in the gut where TGF-β produced by antigen-presenting DCs appears to enhance the conversion process (Annacker et al., 2005; Coombes et al., 2007; Sun et al., 2007). DCs in the gut also produce retinoic acid (RA), and RA has been implicated to provide access for transcription factors like Smad3 to the foxp3 locus (Xu et al., 2010b).
In vitro, TGF-β helps the efficient conversion of naive T cells into T reg cells but Foxp3 expression in these T reg cells is unstable (Polansky et al., 2008). In addition, in vitro experiments can be misleading and are particularly misleading in this case as binding of phosphorylated Smad3 to the CNS1 enhancer is dispensable for T reg cell development (see subsequent text).
TGF-β signaling includes the phosphorylation and subsequent nuclear translocalization of Smad2 and Smad3 transcription factors (Moustakas et al., 2001). Curiously, although Smad2 lacks DNA binding activity (Shi et al., 1998), it has been shown that both Smad2 and Smad3 contribute to TGF-β–mediated foxp3 induction and maintenance (Nolting et al., 2009; Takimoto et al., 2010; Xu et al., 2010b).
Initiation of foxp3 expression appears to require some delicate regulation, the mechanism of which is not fully understood. Foxp3 gene expression is controlled by a core promoter and at least three enhancers (conserved noncoding sequences, CNS1-3; Mantel et al., 2006; Tone et al., 2008; Ruan et al., 2009; Xu et al., 2010b; Zheng et al., 2010). Each regulatory element contains a variety of transcription factor binding sites. CNS1 is located between exons −2b and −1 and regulates foxp3 promoter activity. Besides a consensus sequence for Smad3, CNS1 harbors transcription factor binding sites for Rel-NFAT, CREB, RAR/RXR, and AP-1 (Tone et al., 2008; Ruan et al., 2009; Xu et al., 2010b). It has been suggested that numerous transcription factors function in a chronological and functional hierarchy (Ruan et al., 2009; Xu et al., 2010b) but these experiments rely on in vitro conditions that are not met in vivo.
In TGF-β–dependent T reg cell–promoting cultures Smad3 binding was first observed at CNS1, whereas the promoter was devoid of Smad3 (Ruan et al., 2009). Later, Smad3 dissociated from the enhancer and temporarily emerged at the promoter as part of a c-Rel–dependent enhanceosome. A model in which all components of the enhanceosome have to be present to turn on foxp3 expression has been suggested; lack of one (e.g., Smad3) would leave the promoter dormant. A conserved Smad consensus sequence in the promoter has been found; however, its role in TGF-β–mediated conversion remains to be determined (Samon et al., 2008). In the CNS1, an inverted-repeat Smad3-binding site has been described (Fig. 1; Tone et al., 2008). In vitro, Tone et al. (2008) showed that mutation (replacement with random sequence) of this highly conserved Smad3-binding site results in loss of Smad3 binding and strong but incomplete reduction of promoter activity. Although these studies provide a mechanistic scenario for how TGF-β signaling can be incorporated in signaling events under TGF-β–dependent in vitro conditions, they may be partially or entirely irrelevant for the generation of T reg cells in vivo.
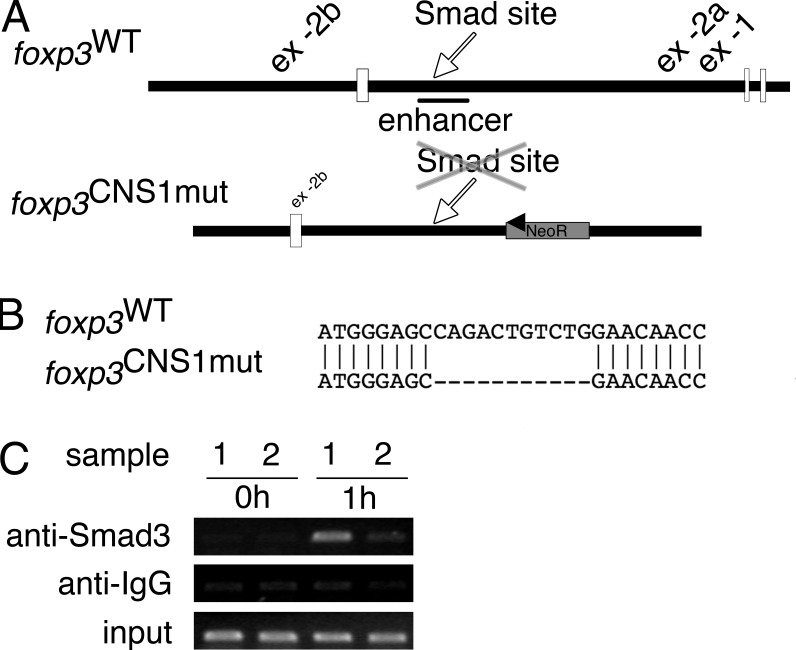
Figure 1.
Deletion of Smad-binding site in foxp3 enhancer/CNS1. (A) Schematic depiction of upstream region of wild-type foxp3 locus and the gene targeting vector. The CNS1 and the position of the Smad-binding site in the wild type or deleted binding site in the foxp3CNS1mut allele are indicated. (B) Alignment of the wild type and the foxp3CNS1mut allele at the position of the Smad-binding site, showing the deleted sequence. (C) ChIP analysis of Smad3 binding to the foxp3 CNS1 region. foxp3WT (sample 1) or foxp3CNS1mut (sample 2) CD4+CD25− splenocytes were not stimulated (0 h) or stimulated for 1 h with TGF-β, anti-CD3, and anti-CD28. Immunoprecipitated DNA was analyzed by PCR. Data are representative of two independent experiments.
In vivo, deletion of the complete CNS1 region (foxp3CNS1ko) was only correlated with reduced T reg cell numbers in gut-associated lymphoid tissue (GALT) in aged mice, leading to a late onset of inflammation in the gut, as well as a defect in maternal-fetal tolerance resulting in increased abortion of allogenic embryos (Zheng et al., 2010; Samstein et al., 2012). The authors suggested that lack of Smad3 signaling via the CNS1 could be a major cause for the reduction in T reg cell numbers in these experiments, and in fact animals without a placenta do not have the CNS1 enhancer.
We generated foxp3CNS1mut mice that dissect TGF-β–Smad signaling from other signaling pathways that affect the CNS1 region. In these mice, only Smad binding to the enhancer is abolished. We report here that in vitro efficient conversion of naive T cells into T reg cells requires binding of Smad3 to the enhancer binding site. In vivo, however, TGF-β signaling through binding of Smad3 to the CNS1 is dispensable for T reg cell development with the exception of the gut.
RESULTS AND DISCUSSION
Generation of foxp3CNS1mut mice
To study the role of Smad3 binding to the CNS1 in vivo, we generated a mutant mouse that selectively lacks the inverted-repeat binding site (foxp3CNS1mut; Fig. 1). The binding site (CAGACtGTCTG) was removed by site-directed mutagenesis (Fig. 1 B) and this mutation of the CNS1 has been introduced into the foxp3 allele by gene targeting. Correct integration of the deletion was verified by sequencing (Fig. 1 B). The neomycin resistance gene was removed in vivo by crossing chimeras to Cre recombinase germ line deleter mice, leaving a single loxP site downstream of CNS1. Consequently, Smad3 was no longer able to bind the CNS1 as shown by ChIP analysis (Fig. 1 C). foxp3CNS1mut mice did not display any apparent abnormalities compared with foxp3WT mice in terms of fertility, growth, life span, or health.
Smad3 binding contributes to TGF-β–dependent in vitro conversion
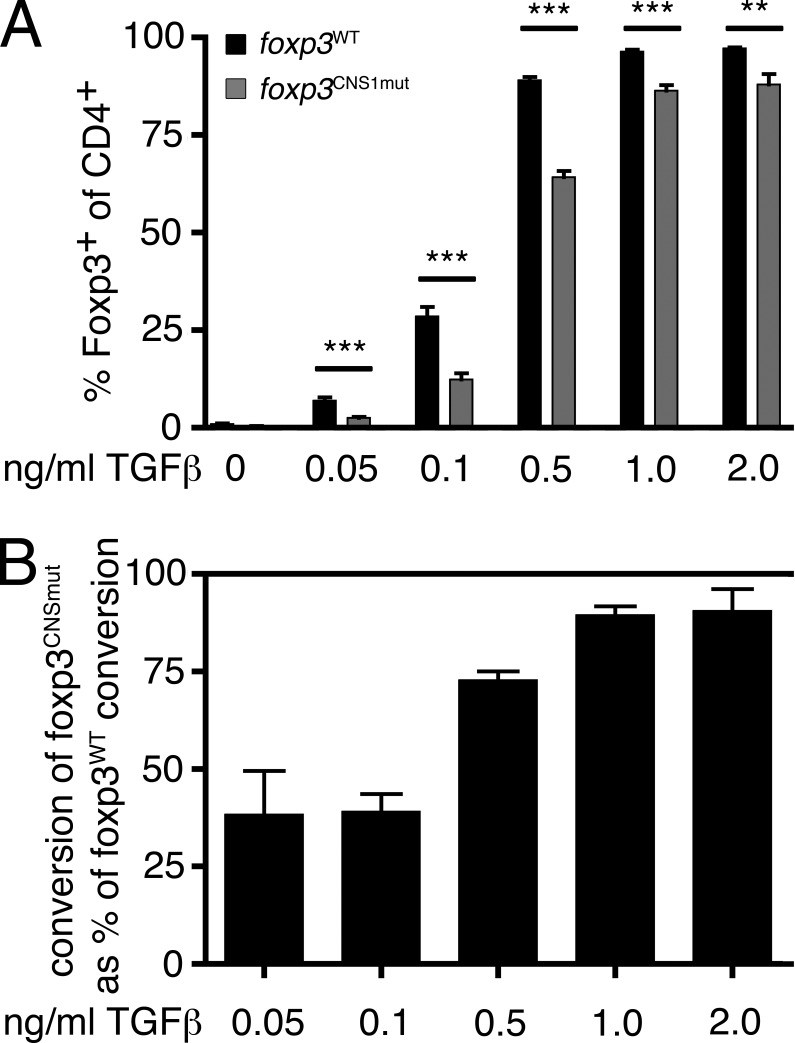
Initially, conversion of naive T cells into T reg cells in vitro in the presence of TGF-β and TCR stimulation was used to assess the requirement of Smad binding to CNS1 in TGF-β–dependent T reg cell development. We compared the ability of foxp3CNS1mut and foxp3WT naive T cells to convert into T reg cells at different concentrations of TGF-β (0–2 ng/ml; Fig. 2). In the absence of TGF-β only marginal spontaneous conversion into T reg cells took place (foxp3WT vs. foxp3CNS1mut, 0 ng/ml TGF-β, 1 ± 0.5% vs. 0.4 ± 0.2%; ±SD). foxp3CNS1mut naive T cells were disadvantaged under conversion conditions that require TGF-β signaling (Fig. 2 A). Particularly at low concentrations of TGF-β, a strong impairment in conversion of foxp3CNS1mut naive T cells was observed (Fig. 2, A and B). Conversion of foxp3CNS1mut naive T cells was reduced by 61% at 0.05 and 0.1 ng/ml TGF-β, and 27% at 0.5 ng/ml TGF-β, respectively (foxp3WT vs. foxp3CNS1mut, 0.05 ng/ml TGF-β, 6.9 ± 2.1% vs. 2.5 ± 0.7%; 0.1 ng/ml TGF-β, 28.6 ± 7.3% vs. 12.4 ± 4.6%; 0.5 ng/ml TGF-β, 89 ± 2.5% vs. 64.2 ± 4.5%; ±SD; Fig. 2 B). At a TGF-β concentration of 1 ng/ml, conversion rates of foxp3CNS1mut as well as foxp3WT cells plateaued. High concentrations of TGF-β compensated for some of the observed impairment; however, foxp3CNS1mut naive T cells did not reach foxp3WT conversion rates (foxp3WT vs. foxp3CNS1mut, 1 ng/ml TGF-β, 96.4 ± 1.4% vs. 86.3 ± 4.4%; 2 ng/ml TGF-β, 97.2 ± 0.6% vs. 88 ± 6.6%; ±SD). These data are consistent with data by Tone et al. (2008) showing that mutation of this Smad binding site does not result in complete loss of foxp3 promoter activity. ChIP experiments (Fig. 1 C) excluded the possibility that phosphorylated Smad3 in foxp3CNS1mut T cells was able to promote T reg cell conversion as a component of the enhanceosome at CNS1 without binding to DNA. It is possible, however, that other Smad-binding sites compensate for its inability to bind to CNS1 in foxp3CNS1mut mice. Besides a possible binding site in the promoter, such sites have not been identified to date (Samon et al., 2008). We conclude that Smad3 binding to the foxp3 enhancer is required for efficient TGF-β–dependent T reg cell conversion in vitro at low, i.e., at physiologically relevant TGF-β levels.
Figure 2.
TGF-β–dependent in vitro conversion of naive T cells into T reg cells. (A) Frequencies of Foxp3-expressing cells among CD4+ T cells in 2.5-d cultures containing the indicated concentrations of TGF-β have been measured. (B) Conversion rate of foxp3CNS1mut T cells normalized on conversion rate of foxp3WT T cells at the indicated TGF-β concentrations. Data are representative of two (0.05 and 2 ng/ml TGF-β) or three (0, 0.1, 0.5, and 1 ng/ml TGF-β) independent experiments. Error bars indicate means ± SEM. ***, P < 0.001; **, P < 0.01.
Normal thymic T reg cell development in foxp3CNS1mut neonates
Next, we analyzed T reg cell development in vivo at two time points after birth but before weaning. It has been described that thymic T reg cell development is diminished in mice that lack a signaling-competent TGF-βR on thymocytes and, therefore, have defective TGF-β–Smad signaling (Li et al., 2006; Xu et al., 2010a). In these studies, increased proliferation appeared to compensate for the reduction of thymic T reg cells.
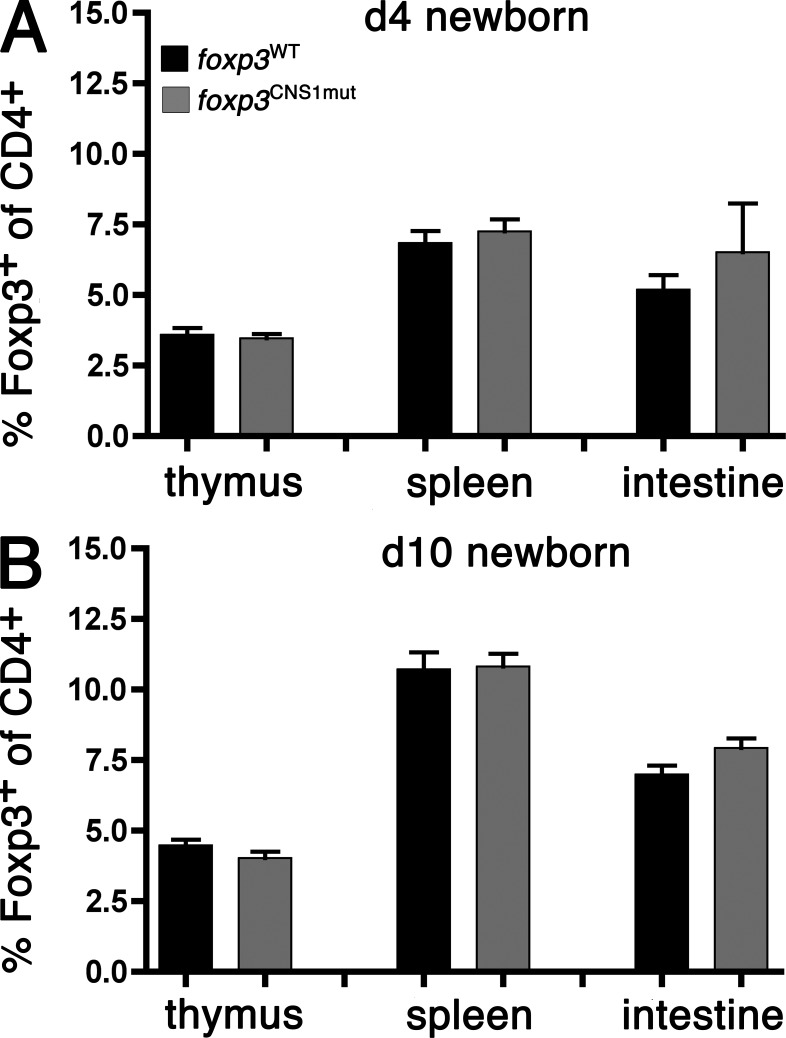
To investigate the role of TGF-β signaling through Smad3 binding to the foxp3 enhancer, we analyzed frequencies and proliferation status of T reg cells in foxp3CNS1mut males and wild-type littermates at days 4 and 10 after birth (Fig. 3). Around day 3 of age, T reg cells first appear in the thymus (Kim and Rudensky, 2006). We analyzed the appearance of thymic as well as splenic and intestinal T reg cells at day 4. Comparable frequencies of T reg cells were found in foxp3WT versus foxp3CNS1mut mice in all three organs (foxp3WT vs. foxp3CNS1mut thymi, 3.6 ± 0.8% vs. 3.5 ± 0.4%; spleens, 6.8 ± 1.4% vs. 7.3 ± 1.1%; intestines, 5.2 ± 1.6% vs. 6.5 ± 4.2%; ±SD; Fig. 3 A).
Figure 3.
T reg cell development in days 4 and 10 neonates. T reg cell frequencies among CD4+ cells have been measured in thymi, spleens, and intestines of day 4 (A) and day 10 (B) foxp3CNS1mut and foxp3WT mice. Data on day 4 T reg cell frequencies are representative of 6 foxp3CNS1mut and 10 foxp3WT mice and two independent experiments. Data on day 10 T reg cell frequencies in thymus and spleen are representative of 10 foxp3CNS1mut and 13 foxp3WT males and four independent experiments. Data for day 10 T reg cell frequencies in intestine are representative of six foxp3CNS1mut and eight foxp3WT males and two independent experiments. Error bars indicate means ± SEM.
As shown in Fig. 3 B, T reg cell frequencies increased in all three organs between 4- and 10-d-old mice with no apparent difference between foxp3CNS1mut mice and foxp3WT mice at day 10 (foxp3WT vs. foxp3CNS1mut thymi, 4.5 ± 0.7% vs. 4.0 ± 0.7%; spleens, 10.7 ± 2.2% vs. 10.8 ± 1.4%; intestines, 7 ± 0.9% vs. 7.9 ± 0.8%; ±SD). To analyze the proliferation of T reg cells, day 10 newborns were injected i.p. with EdU 16 h before analysis. There was no evidence that elevated proliferation contributed to similar T reg cell levels in different mice (foxp3WT vs. foxp3CNS1mut thymi, 23.3 ± 1.8% vs. 24 ± 3%; spleens, 21.4 ± 2.4% vs. 21.8 ± 1.6%; ±SD; unpublished data).
In agreement with previous studies by Thornton et al. (2010), virtually all T reg cells in newborn mice expressed the transcription factor Helios as assessed by intracellular staining in spleen and intestine of day 10 mice (unpublished data). These data show that the development of the first wave of thymic T reg cells does not require Smad3 binding to the CNS1 and refute implications of the mentioned studies on a role of TGF-β–Smad signaling in early intrathymic T reg cell development. It appears that T reg cells with an intrinsic TGF-β signaling defect develop normally in newborns.
Normal T reg cell frequencies in young adult foxp3CNS1mut mice
We then investigated mice at 4 (Fig. 4 A) and 8 (Fig. 4 B) wk of age. Thymus, spleen, axial/inguinal LN, mesenteric LN (MLN), Peyer’s patches (PPs), lamina propria lymphocytes (LPLs), and intraepithelial lymphocytes (IELs) were analyzed with regard to their frequency of T reg cells among CD4+ T cells (Fig. 4, A and B, top) as well as Helios expression among T reg cells isolated from these sites (Fig. 4, A and B, bottom). Foxp3CNS1mut and foxp3WT mice were furthermore injected with EdU 16 h before analysis, and the proliferation status of T reg cells was assessed (Fig. 4, A and B, middle).
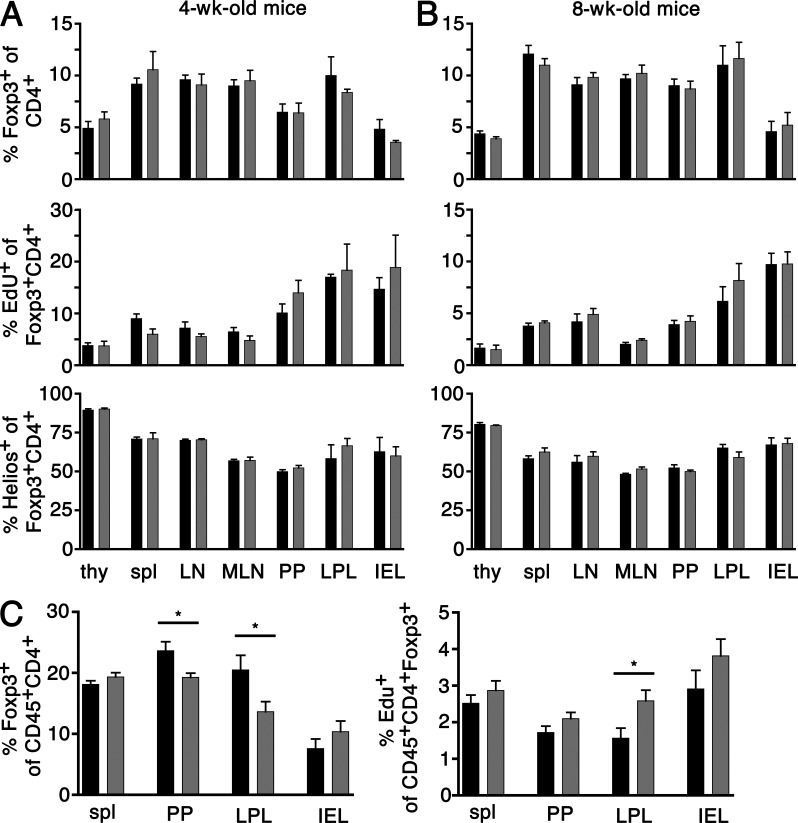
Figure 4.
Analysis of T reg cell frequencies, proliferation rate, and proportion of Helios-expressing T reg cells in adult males. 4-wk-old (A), 8-wk-old (B), and 6-mo-old (C) males have been injected with 1 mg EdU i.p. 16 h before analysis. (A and B) Thymi (thy), spleen (spl), axial/inguinal LN, MLN, PP, LPL, and IEL have been analyzed for frequency of Foxp3+ T reg cells among CD4+ T cells (top), EdU+ (middle), and Helios+ (bottom) cells among T reg cells. Data are representative of two to six males and two to three independent experiments per group. (C) Frequencies of T reg cells (left) and EdU+ T reg cells (right) were assessed in spleen, PP, LPL, and IEL. Data are representative of 7–10 males and five independent experiments per group. Data for foxp3WT mice are depicted in black and data for foxp3CNS1mut mice are depicted in gray. Error bars indicate means ± SEM. *, P < 0.03.
foxp3CNS1mut T reg cells appeared at normal frequencies and the proportion of Helios-expressing T reg cells among foxp3CNS1mut T reg cells appeared comparable to foxp3WT littermates at both time points. From 4 to 8 wk there is a mild increase in the Helios−/Helios+ ratio in several organs (thymus, spleen, LN, and MLN), whereas others remain unchanged (PP, LPL, and IEL). Helios expression has been suggested to distinguish peripherally converted from intrathymically generated T reg cells (Thornton et al., 2010). However, the mechanisms of Helios activation and regulation of expression are unclear, and Helios has also been associated with T cell activation and T helper cell differentiation (Akimova et al., 2011; Serre et al., 2011). Particularly interesting is the fact that gut-associated T reg cells appeared normal in terms of T reg cell numbers and T reg cells did not show any compensatory proliferation.
Generally, although proliferation of T reg cells strongly declines in all organs within 4–8 wk of age, and considering that thymic output does not further increase, it appears remarkable that T reg cell proportions among CD4+ T cells are held constant in all organs analyzed in foxp3CNS1mut and foxp3WT mice. Possible explanations for this observation could be that a plateau is reached by thymic T reg cells or the contribution of peripheral conversion increases during 4–8 wk of age.
Smad3 binding contributes to T reg cell conversion in GALT in older mice
We hypothesized that a possible positive impact of Smad3 binding on foxp3 promoter activation would become apparent at later ages. We analyzed 6-mo-old mice in which thymic output had declined. The frequency and proliferation of splenic T reg cells as well as T reg cells among IEL, LPL, and PP lymphocytes of foxp3CNS1mut mice were compared with their wild-type counterparts (Fig. 4 C). Although splenic T reg cell numbers appeared to be similar, an appreciable reduction in T reg cell frequency was found to be apparent in PP and LPL (foxp3WT vs. foxp3CNS1mut spleen, 18.2 ± 1.9% vs. 19.3 ± 2.2% [P = 0.206]; PP, 23.6 ± 4.7% vs. 19.3 ± 2.2% [P = 0.0157]; LPL, 20.5 ± 7.2% vs. 13.7 ± 5.2% [P = 0.0288]; IEL, 7.6 ± 4.5% vs. 10.4 ± 5.3% [P = 0.2654]; ±SD; Fig. 4 C, left).
A reduction of T reg cells in PP and LPL has also been reported in aged foxp3CNS1ko mice (Zheng et al., 2010). The reduction observed by Zheng et al. (2010) in foxp3CNS1ko mice (∼50%) was more profound than T reg cell reduction in foxp3CNS1mut mice (∼20% in PP and ∼35% in LPL), which might be explained by the contribution of other transcription factor binding sites present in the CNS1. Consistent with the possibility of compensation for reduced T reg cell numbers by increased proliferation we observed a tendency of T reg cells, particularly among LPL, for slightly elevated proliferative activity (foxp3WT vs. foxp3CNS1mut spleen, 2.5 ± 0.7% vs. 2.9 ± 0.8% [P = 0.3226]; PP, 1.7 ± 0.6% vs. 2.1 ± 0.5% [P = 0.1353]; LPL, 1.6 ± 0.8% vs. 2.6 ± 0.8% [P = 0.0241]; IEL, 2.9 ± 1.4% vs. 3.8 ± 1.3% [P = 0.2103]; ±SD; Fig. 4 C, right). This may mask a more profound defect in gut-associated T reg cell generation in foxp3CNS1mut mice as well as in foxp3CNS1ko mice.
We furthermore found the frequency of activated T cells (CD4+CD62L−/loCD44hi) in axial/brachial LN to be comparable in foxp3CNS1mut and foxp3WT mice. In addition, no histological evidence for increased lymphocyte migration into liver, lung, kidney, pancreas, stomach, or colon in foxp3CNS1mut mice could be found (unpublished data). In conclusion, TGF-β–Smad3 signaling via binding of Smad3 to the foxp3 enhancer partially helps the conversion of CD4+ T cells into T reg cells in the gut.
Partial reduction in GALT T reg cells does not increase susceptibility of foxp3CNS1mut mice to colitis
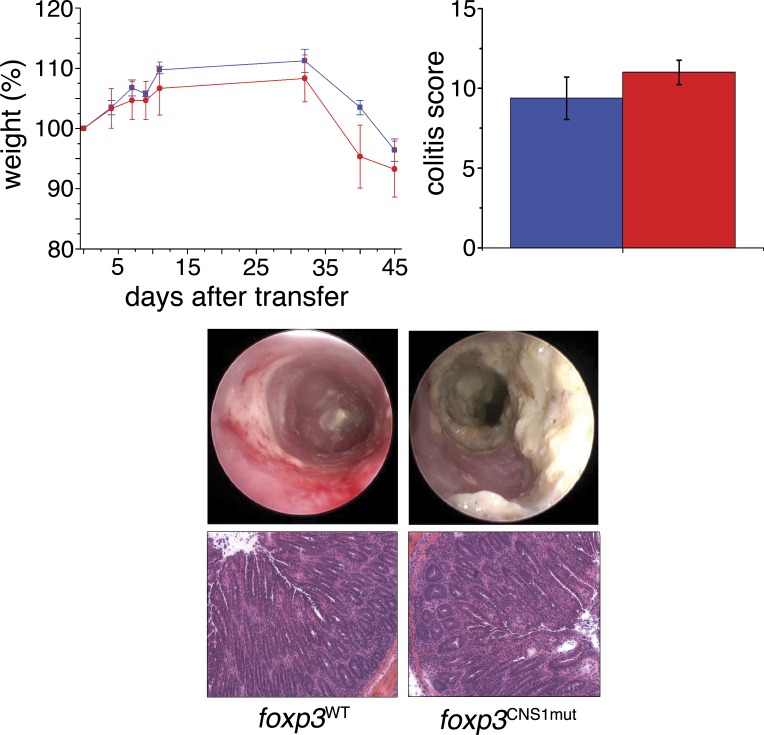
To investigate whether a mild defect in the generation or maintenance of gut T reg cells affects regulation of T cell activation under pathological conditions, we investigated two colitis models in age-matched foxp3CNS1mut and foxp3WT mice (Fig. 5). 3-mo-old mice were subjected to dextran sulfate sodium (DSS) in drinking water. DSS-induced colitis is a common colitis model in which DSS induces inflammation by its toxic effects on colonic epithelium leading to activation of innate immune cells, followed by activation of the adaptive immune system. T reg cells residing in the gut mucosa are able to interfere with the inflammation. Because DSS-induced colitis appears to be indirectly mediated by T cells, we also investigated the T cell–transfer colitis model transferring foxp3CNS1mut or foxp3WT naive T cells, respectively, into Rag-deficient hosts (Fig. 5). In this model, some of the transferred naive T cells undergo acute conversion into T reg cells, which can interfere with the developing colonic inflammation.
Figure 5.
Analysis of T cell transfer colitis. Naive T cells from foxp3WT mice (blue) or foxp3CNS1mut mice (red), respectively, have been transferred into Rag-deficient recipient mice. Body weight was measured (top left) and, at day 45, colitis activity was determined by mini-endoscopy and analyzed in colitis score (top right). Histological analysis of colons at day 45 after transfer showed similar signs of inflammation like in DSS-induced colitis. Data are representative of eight mice and two independent experiments per group. Error bars indicate means ± SEM.
In both conditions, similar weight losses were observed between both groups (Fig. 5, top left graph; and not depicted) and similar colitis severity scores were obtained (Fig. 5, top right graph). Furthermore, mini-endoscopy and HE staining of colon (day 7 in DSS-induced colitis, day 45 in T cell transfer colitis) revealed similar signs of inflammation such as thickening of the wall, loss of crypt architecture, ulcers, and accumulation of mononuclear cells. In conclusion, although the gut T reg cell frequency in mice older than 2 mo is partially negatively affected by mutations of the enhancer region, this defect does not increase the susceptibility of mutant mice to T cell–independent or T cell–dependent colitis. Similar results were previously obtained in CNS1-deficient mice (Zheng et al., 2010).
TGF-β influences all stages of a T cell’s life. Regarding the TGF-β–Smad signaling in T reg cells, previous studies suggested a dominant role of Smad3 binding to the foxp3 locus in T reg cell development. The Smad3 binding site in CNS1 is considered the crucial regulatory element by which TGF-β signals to the foxp3 locus. In vitro, CNS1-bound Smad3 contributes to foxp3 initiation (Tone et al., 2008; Ruan et al., 2009). Consistently, deletion of the entire CNS1 (removal of several transcription factor binding sites) resulted in decreased TGF-β–dependent in vitro conversion of naive T cells into T reg cells (Zheng et al., 2010). Contrary to previous in vivo studies, despite abrogated TGF-β signaling in T reg cells, CNS1-deficient mice were healthy. By exclusively removing the Smad3-binding site from the CNS1 we were then able to narrow down T reg cell reduction in GALT to Smad3 signaling, dissecting it from roles of other transcription factors. Here, we provide direct evidence that binding of Smad3 to the foxp3 CNS1 is dispensable for T reg cell development with the exception of the gut.
MATERIALS AND METHODS
Gene targeting and mice.
The foxp3CNS1mut gene targeting vector consisted of a 5′ long homologous arm (−1468 to +3289 bp), a loxP2272-flanked neomycin resistance gene in opposite orientation to the locus, and a 3′ short homologous arm (+3290 to +4985 bp; − and + refer to upstream or downstream of transcriptional start, respectively; Ensembl ENSMUST00000115740). The Smad-binding site in the enhancer was deleted by site-directed mutagenesis (Sawano and Miyawaki, 2000), confirmed by PCR and sequencing, and foxp3CNS1mut embryonic stem (ES) cells were generated by gene targeting in v6.5 ES cells (129/B6 F1 hybrid). Chimeric mice were bred to germline Cre recombinase deleter mice (B6.FVB-Tg(EIIa-cre)C5379Lmgd/J). The mutant allele was then crossed to B6 for at least two generations. All animal care and procedures were executed according to National Institutes of Health guidelines and in accordance with the guidelines of the Animal Care and Use Committee of the Dana Farber Cancer Institute.
ChIP analysis.
CD4+CD25− splenocytes were enriched using the EasySep mouse CD4+ T cell enrichment kit (STEMCELL Technologies) and CD25 Microbead kit (Miltenyi Biotec). The cells were stimulated for 1 h in the presence of 5 ng/ml TGF-β, 5 µg/ml anti-CD3 (precoated), and 1 µg/ml anti-CD28. Cell lysates were immunoprecipitated with anti-Smad3 (Abcam) or control IgG using the ChIP-IT Express kit (Active Motif). Immunoprecipitated DNA was analyzed by PCR (enhancer-1 forward, 5′-CCCATGTTGGCTTCCAGTCTCCTTTATGG-3′; enhancer-1 reverse, 5′-AGGTACAGAGAGGTTAAGAGCCTGGGT-3′, product size 156 bp).
In vitro conversion.
Naive T cells (Sytox blue [Invitrogen]-negative CD4+CD25−CD44−/loCD62Lhi) were sorted from spleen and LN to >98% purity and an aliquot was used for intracellular staining for foxp3. 96-well culture plates were coated with 5 µg/ml anti-CD3 in PBS for 75 min at 37°C. 50,000 naive T cells were plated per well in 200 µl medium (RMPI 1640, 10% FCS, penicillin/streptomycin, gentamicin, 1× GlutaMAX, 1× nonessential amino acids, 1× sodium pyruvate, β-mercaptoethanol, 100 U/ml mIL-2, and 0–2 ng TGF-β/ml) for 2.5 d. Culture medium components were purchased from Gibco, rmIL-2 from PeproTech, and rhTGFβ1 from R&D Systems.
Isolation of PPs, IELs, and LPLs.
The small intestines were cleaned from fat and connective tissue and flushed with CMF (HBSS/10 mM Hepes/2% FCS), and 5–9 PPs were excised from a small intestine and incubated for two rounds of rotation (20 min at 37°C) in freshly prepared HBSS/10 mM Hepes/20% FCS/0.5 mM DTT/1 mM EDTA with strong vortexing after each round to remove epithelial cells and IEL. Single cell suspensions were prepared using frosted glass slides in FACS staining buffer (PBS/5% FCS). The intestines were opened longitudinally and cut into 0.5–1-cm pieces. To remove mucus, pieces were vortexed in large volume of RPMI 1640 for several rounds. IELs were recovered by incubation as described for PP, and suspension cells were roughly filtered through a metal filter and applied to gradient centrifugation as follows: cells of one intestine were resuspended in 24 ml of ice-cold 40% Percoll, pH 7.2, distributed over three FCS-precoated 15-ml tubes, underlaid with 5 ml of ice-cold 100% Percoll, pH 7.2, and centrifuged for 20 min at 600 g and 4°C with no brake. After centrifugation, the top layer was discarded and the interphase containing IEL was recovered. Intestinal pieces after IEL/epithelial cells removal were digested in HBSS/10 mM Hepes/80 CDU/ml Liberase-TM (Roche)/25 µg/ml DNase I (Roche) in three rounds of rotation (30 min at 37°C) with strong vortexing or pipetting after each digestion cycle. After complete digestion of the tissue, suspension cells were filtered as for IEL and applied to gradient centrifugation as for IEL in one to two 15-ml tubes per organ.
Isolation of CD4+ T cells from intestines of newborns.
The entire intestine of day 4 or 10 newborns was minced and digested with 100 U/ml Collagenase (Gibco) in three to four rounds of 30 min each at 37°C. Recovered suspension cells were filtered, unspecific binding sites were blocked with anti-CD16/32, and cells were than stained with biotinylated anti-CD4 antibody. CD4+ cells were enriched by MACS using magnetic streptavidin beads (Miltenyi Biotec) and LS columns (Miltenyi Biotec).
Induction of colitis.
Colitis was induced by three cycles of 2% DSS (MP Biomedicals) in drinking water for 1 wk and normal drinking water for 2 wk. Transfer colitis was induced by adoptive transfer of naive CD4+CD25− T cell in Rag−/− hosts. In brief, CD4+ T cells were purified by negative selection from spleen mononuclear cells of foxp3CNS1mut or foxp3WT mice, respectively, using MACS system (Miltenyi Biotec). The resulting CD4+ T cells were further enriched for CD24+ cells by immunomagnetic beads into CD24+ cells and transferred into RAG-1–deficient mice. Colitis activity was monitored by weight curves, endoscopy, and histological analysis, as specified in the next sections. Mice were maintained in isolated cages under specific pathogen-free conditions. No evidence of graft versus host disease, such as skin inflammation and histopathological evidence of small bowel inflammation, was observed in the reconstituted animals.
Mini-endoscopic analysis.
Colitis development was monitored with a high-resolution video endoscopic system (Karl Storz). Scoring of DSS colitis severity was performed according to MEICS score with five parameters (translucency, granularity, vascularity, fibrin production, and stool).
Flow cytometry, antibodies, and EdU.
For cell surface staining, cells were blocked with anti-CD16/32 (2.4G2; BD) and subsequently stained with antibodies in PBS/5% FCS for 45 min on ice. Cell sorting and analyses were performed on FACSAria. For intracellular staining, cells were first stained for surface molecules, subsequently fixed and permeabilized in the presence of blocking antibody (eBioscience, 00–5521), and then stained for 30 min with anti-Foxp3 (FJK-16s; eBioscience) and anti-Helios (22F6; BioLegend). For analysis of EdU incorporation, the supplier’s manual was followed (Molecular Probes). In brief, newborn mice were injected with 500 µg and adult mice i.p. with 1 mg EdU 16 h before analysis. Recovered cells were stained for surface and intracellular molecules as described in previous sections, washed twice with foxp3 staining kit permeabilization buffer and once with EdU staining kit permeabilization buffer, and subsequently resuspended in EdU reaction solution. EdU reaction was allowed to run for 35 min, and cells were washed three times in EdU staining kit permeabilization buffer and resuspended in PBS/5% FCS. Further antibodies used were CD4 (GK1.5), CD25 (PC61), CD44 (IM7), CD62L (MEL-14), and CD45 (30-F11; all from BioLegend).
Histology.
Whole mice were fixed in Bouin’s solution for 72 h and subsequently kept in 70% ethanol solution until organs were recovered. Organs were embedded in paraffin, and hematoxylin and eosin staining for general morphology was performed.
Statistics.
Unpaired Student’s t test was used to determine the p-value in Prism (GraphPad Software).
Acknowledgments
The authors thank Dr. Carolin Daniel for help with the in vitro conversion assay and Drs. Adele Hill, Carolin Daniel, Michael Gleimer, Taras Kreslavsky, and Hans-Reimer Rodewald for helpful discussions and carefully reading the manuscript.
This paper was supported by the National Institutes of Health (NIH-5R37AI053102-09 to H. von Boehmer) and the German Research Foundation (DFG, Schl 1897/1-1 to S.M. Schlenner).
The authors declare no conflicting financial interests.
Footnotes
Abbreviations used:
- DSS
- dextran sulfate sodium
- GALT
- gut-associated lymphoid tissue
- IEL
- intraepithelial lymphocyte
- LPL
- lamina propria lymphocyte
- MLN
- mesenteric LN
- PP
- Peyer’s patch
References
- Akimova T., Beier U.H., Wang L., Levine M.H., Hancock W.W. 2011. Helios expression is a marker of T cell activation and proliferation. PLoS ONE. 6:e24226 10.1371/journal.pone.0024226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annacker O., Coombes J.L., Malmstrom V., Uhlig H.H., Bourne T., Johansson-Lindbom B., Agace W.W., Parker C.M., Powrie F. 2005. Essential role for CD103 in the T cell-mediated regulation of experimental colitis. J. Exp. Med. 202:1051–1061 10.1084/jem.20040662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes J.L., Siddiqui K.R., Arancibia-Cárcamo C.V., Hall J., Sun C.M., Belkaid Y., Powrie F. 2007. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-β and retinoic acid–dependent mechanism. J. Exp. Med. 204:1757–1764 10.1084/jem.20070590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.M., Rudensky A. 2006. The role of the transcription factor Foxp3 in the development of regulatory T cells. Immunol. Rev. 212:86–98 10.1111/j.0105-2896.2006.00426.x [DOI] [PubMed] [Google Scholar]
- Li M.O., Flavell R.A. 2008. TGF-beta: a master of all T cell trades. Cell. 134:392–404 10.1016/j.cell.2008.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.O., Sanjabi S., Flavell R.A. 2006. Transforming growth factor-beta controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity. 25:455–471 10.1016/j.immuni.2006.07.011 [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhang P., Li J., Kulkarni A.B., Perruche S., Chen W. 2008. A critical function for TGF-beta signaling in the development of natural CD4+CD25+Foxp3+ regulatory T cells. Nat. Immunol. 9:632–640 10.1038/ni.1607 [DOI] [PubMed] [Google Scholar]
- Mantel P.Y., Ouaked N., Rückert B., Karagiannidis C., Welz R., Blaser K., Schmidt-Weber C.B. 2006. Molecular mechanisms underlying FOXP3 induction in human T cells. J. Immunol. 176:3593–3602 [DOI] [PubMed] [Google Scholar]
- Marie J.C., Letterio J.J., Gavin M., Rudensky A.Y. 2005. TGF-β1 maintains suppressor function and Foxp3 expression in CD4+CD25+ regulatory T cells. J. Exp. Med. 201:1061–1067 10.1084/jem.20042276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie J.C., Liggitt D., Rudensky A.Y. 2006. Cellular mechanisms of fatal early-onset autoimmunity in mice with the T cell-specific targeting of transforming growth factor-beta receptor. Immunity. 25:441–454 10.1016/j.immuni.2006.07.012 [DOI] [PubMed] [Google Scholar]
- Moustakas A., Souchelnytskyi S., Heldin C.H. 2001. Smad regulation in TGF-beta signal transduction. J. Cell Sci. 114:4359–4369 [DOI] [PubMed] [Google Scholar]
- Nolting J., Daniel C., Reuter S., Stuelten C., Li P., Sucov H., Kim B.G., Letterio J.J., Kretschmer K., Kim H.J., von Boehmer H. 2009. Retinoic acid can enhance conversion of naive into regulatory T cells independently of secreted cytokines. J. Exp. Med. 206:2131–2139 10.1084/jem.20090639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polansky J.K., Kretschmer K., Freyer J., Floess S., Garbe A., Baron U., Olek S., Hamann A., von Boehmer H., Huehn J. 2008. DNA methylation controls Foxp3 gene expression. Eur. J. Immunol. 38:1654–1663 10.1002/eji.200838105 [DOI] [PubMed] [Google Scholar]
- Ruan Q., Kameswaran V., Tone Y., Li L., Liou H.C., Greene M.I., Tone M., Chen Y.H. 2009. Development of Foxp3(+) regulatory T cells is driven by the c-Rel enhanceosome. Immunity. 31:932–940 10.1016/j.immuni.2009.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samon J.B., Champhekar A., Minter L.M., Telfer J.C., Miele L., Fauq A., Das P., Golde T.E., Osborne B.A. 2008. Notch1 and TGFbeta1 cooperatively regulate Foxp3 expression and the maintenance of peripheral regulatory T cells. Blood. 112:1813–1821 10.1182/blood-2008-03-144980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samstein R.M., Josefowicz S.Z., Arvey A., Treuting P.M., Rudensky A.Y. 2012. Extrathymic generation of regulatory T cells in placental mammals mitigates maternal-fetal conflict. Cell. 150:29–38 10.1016/j.cell.2012.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawano A., Miyawaki A. 2000. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 28:E78 10.1093/nar/28.16.e78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serre K., Bénézech C., Desanti G., Bobat S., Toellner K.M., Bird R., Chan S., Kastner P., Cunningham A.F., Maclennan I.C., Mohr E. 2011. Helios is associated with CD4 T cells differentiating to T helper 2 and follicular helper T cells in vivo independently of Foxp3 expression. PLoS ONE. 6:e20731 10.1371/journal.pone.0020731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Wang Y.F., Jayaraman L., Yang H., Massagué J., Pavletich N.P. 1998. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-beta signaling. Cell. 94:585–594 10.1016/S0092-8674(00)81600-1 [DOI] [PubMed] [Google Scholar]
- Sun C.M., Hall J.A., Blank R.B., Bouladoux N., Oukka M., Mora J.R., Belkaid Y. 2007. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J. Exp. Med. 204:1775–1785 10.1084/jem.20070602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto T., Wakabayashi Y., Sekiya T., Inoue N., Morita R., Ichiyama K., Takahashi R., Asakawa M., Muto G., Mori T., et al. 2010. Smad2 and Smad3 are redundantly essential for the TGF-beta-mediated regulation of regulatory T plasticity and Th1 development. J. Immunol. 185:842–855 10.4049/jimmunol.0904100 [DOI] [PubMed] [Google Scholar]
- Thornton A.M., Korty P.E., Tran D.Q., Wohlfert E.A., Murray P.E., Belkaid Y., Shevach E.M. 2010. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J. Immunol. 184:3433–3441 10.4049/jimmunol.0904028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tone Y., Furuuchi K., Kojima Y., Tykocinski M.L., Greene M.I., Tone M. 2008. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 9:194–202 10.1038/ni1549 [DOI] [PubMed] [Google Scholar]
- Xu L., Kitani A., Strober W. 2010a. Molecular mechanisms regulating TGF-beta-induced Foxp3 expression. Mucosal Immunol. 3:230–238 10.1038/mi.2010.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Kitani A., Stuelten C., McGrady G., Fuss I., Strober W. 2010b. Positive and negative transcriptional regulation of the Foxp3 gene is mediated by access and binding of the Smad3 protein to enhancer I. Immunity. 33:313–325 10.1016/j.immuni.2010.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Josefowicz S., Chaudhry A., Peng X.P., Forbush K., Rudensky A.Y. 2010. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 463:808–812 10.1038/nature08750 [DOI] [PMC free article] [PubMed] [Google Scholar]