By suppressing expression of TRAF6 and IRAK1, miR-146a regulates NF-κB activation in T cells through a negative feedback loop and controls the resolution of T cell responses in mice.
Abstract
T cell responses in mammals must be tightly regulated to both provide effective immune protection and avoid inflammation-induced pathology. NF-κB activation is a key signaling event induced by T cell receptor (TCR) stimulation. Dysregulation of NF-κB is associated with T cell–mediated inflammatory diseases and malignancies, highlighting the importance of negative feedback control of TCR-induced NF-κB activity. In this study we show that in mice, T cells lacking miR-146a are hyperactive in both acute antigenic responses and chronic inflammatory autoimmune responses. TCR-driven NF-κB activation up-regulates the expression of miR-146a, which in turn down-regulates NF-κB activity, at least partly through repressing the NF-κB signaling transducers TRAF6 and IRAK1. Thus, our results identify miR-146a as an important new member of the negative feedback loop that controls TCR signaling to NF-κB. Our findings also add microRNA to the list of regulators that control the resolution of T cell responses.
T cells of the adaptive immune system in mammals play a central role in the fight against pathogen invasion. The initiation and resolution of the T cell responses must be tightly regulated to allow for effective immune protection while avoiding inflammation-induced pathology. T cell activation is triggered by antigen engagement of the TCR, the sole molecule which determines the specificity of a T cell (Hannum et al., 1984). TCR engagement induces a signaling cascade leading to the activation of three major transcription factors: NFAT, AP-1, and NF-κB (Isakov and Altman, 2002). Among them, NF-κB plays a particularly important role and is involved in regulating almost all important aspects of T cell activation, including proliferation, survival, and effector functions (Schulze-Luehrmann and Ghosh, 2006). Dysregulated NF-κB activation in T cells has been associated with the development of T cell–mediated inflammatory diseases and malignancies (Karin and Greten, 2005; Jost and Ruland, 2007), highlighting the importance of a tightly regulated negative feedback control of TCR-induced NF-κB activation. Considering the pivotal role of NF-κB in regulating T cell activation, it is no surprise that studies from the past two decades have identified multiple layers of modulation that contribute to this negative feedback control system falling into three major categories: (1) TCR recycling induced by phosphorylation of the CD3γ chain by PKCθ; (2) degradation of key signaling molecules, including the CARD11–Bcl-10–MALT1 (CBM) signalosome component CARD11, through phosphorylation by casein kinase 1α (CK-1α), and Bcl-10 through phosphorylation by IKKβ or through ubiquitination by NEDD4, Itch, and cIAP2; and (3) negative feedback loop, including the well-characterized NF-κB–induced expression of inhibitory genes such as IκBα, which binds to nuclear NF-κB complexes and inhibits their function by shuttling NF-κB back into cytoplasm, and A20, which is a deubiquitinase, and through removing the K63-linked polyubiquitin from the signalosome component MALT1, leading to the degradation of MALT1 and the termination of signal transmission to NF-κB (Schulze-Luehrmann and Ghosh, 2006; Ruland, 2011). This list is still under active expansion (Ghosh and Hayden, 2008; Ruland, 2011).
MicroRNAs (miRNAs), a family of small noncoding RNAs which regulate gene expression by base-pairing to the 3′ untranslated region of their target genes, have recently emerged as a novel class of posttranscriptional regulators of various biological processes ranging from development to function, especially in the immune system (Ambros, 2004; Bartel, 2004; Baltimore et al., 2008; Tsitsiou and Lindsay, 2009). Their capacity to coordinately regulate multiple target genes makes miRNAs particularly suitable for regulating complex signaling cascades like NF-κB. Several years ago in an effort to screen for miRNAs involved in innate immune cell response to microbial infection, we identified three miRNAs (miR-132, miR-155, and miR-146a) that are sharply induced upon LPS stimulation (Taganov et al., 2006). Among them, miR-146a attracted our attention for its NF-κB–dependent induction and its targeted degradation of NF-κB signaling transducers TRAF6 and IRAK1, the signature of an NF-κB negative feedback regulator (Taganov et al., 2006). To study the physiological role of miR-146a, we generated miR-146a–deficient (miR-146a−/−) mice and reported that they developed chronic hyperinflammatory and autoimmune disorder characterized by splenomegaly, lymphadenopathy, and multiorgan leukocyte infiltration (Boldin et al., 2011). The gradual accumulation of hyperproliferating myeloid cells, activated CD4 and CD8 T cells, and autoantibodies in miR-146a−/− mice indicates that miR-146a is a critical physiological brake to prevent the overactivation of the innate as well as the adaptive immune systems (Boldin et al., 2011). We showed that constitutive NF-κB activation is a major contributor to the development of myeloid hyperproliferation and malignancies in these mice, confirming miR-146a as a physiologically negative feedback regulator of NF-κB activation in the innate immune system in vivo (Boldin et al., 2011; Zhao et al., 2011). Study of the autoimmune symptoms in the miR-146a−/− mice led to the finding of a regulatory T cell (Treg cell) deficiency resulting in poorly controlled Th1 response, likely caused by the dysregulation of the IFN-γ signaling pathway (Lu et al., 2010). However, the molecular mechanisms underlying the physiological role of miR-146a as a negative regulator of the adaptive immune system, especially its role as an autonomous regulator to modulate T cell and B cell responses to antigen stimulation, remain largely unexplored.
This study focuses on the physiological role of miR-146a in regulating T cell response to antigen stimulation in mice and its possible involvement in TCR signaling to NF-κB during T cell activation. We found that in vivo, T cells lacking miR-146a are hyperresponsive to antigen stimulation and are prone to induce T cell–mediated hyperinflammatory disease. Using a loss-of-function approach, we found that in the absence of miR-146a, both CD4 and CD8 T cells exhibited hyperresponsiveness after TCR stimulation, indicated by higher proliferation, lengthened survival, exaggerated activation phenotype, and enhanced effector cytokine production. In contrast, overexpression of miR-146a produced the opposite effects. This change of T cell activation kinetics is correlated with altered NF-κB activity and changed expression patterns of a set of NF-κB responsive genes that are responsible for these activities. Study of miR-146a expression in CD4 and CD8 T cells revealed that NF-κB activation induced by TCR stimulation up-regulates the expression of miR-146a, which in turn down-regulates NF-κB activity, at least in part through repressing the NF-κB signaling transducer TRAF6 and IRAK1. Thus, our results identify miR-146a as a new and quite critical constituent of the negative feedback regulatory network modulating TCR signaling to NF-κB and add miRNA as a novel molecular player in the control of the resolution of T cell responses.
RESULTS
miR-146a–deficient T cells are hyperresponsive to acute antigen stimulation in vivo
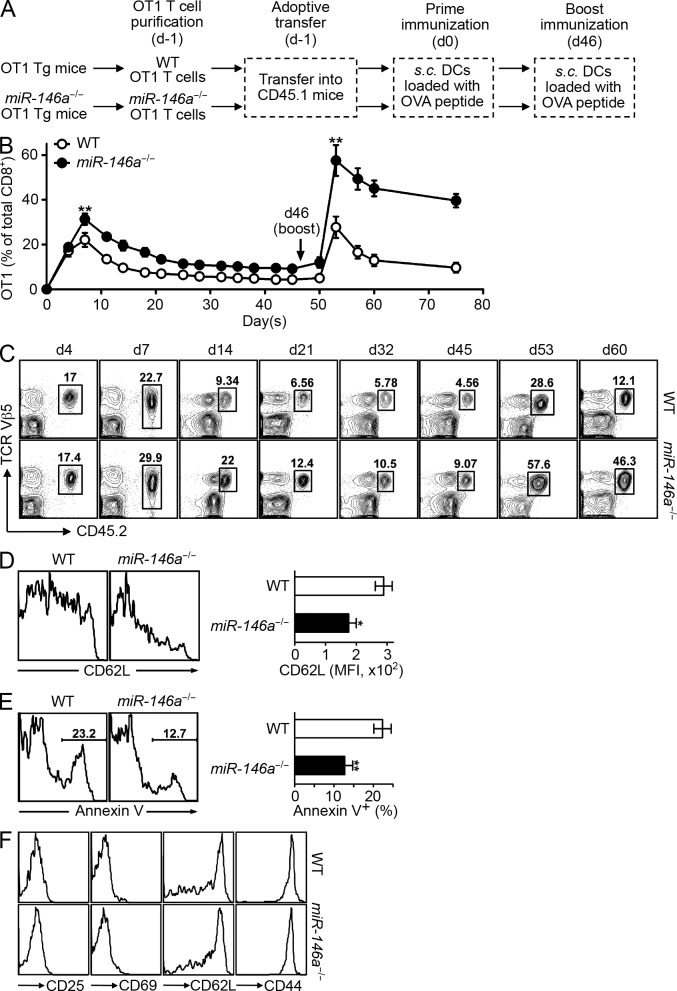
To study the physiological role of miR-146a in the regulation of T cell responses to antigen stimulation, we generated OVA-specific OT1 TCR transgenic (Tg) mice that were lacking miR-146a. Resting OT1 T cells of WT or miR-146−/− background were purified and adoptively transferred into CD45.1 congenic recipient mice, followed by antigenic stimulation with OVA peptide–pulsed DCs and periodic bleeding to monitor the transferred T cells for their response to antigen priming and boosting (Fig. 1 A). At the time of adoptive transfer, both WT and miR-146a−/− OT1 T cells displayed a similar naive T cell phenotype (CD25−CD69−CD62LhiCD44lo; not depicted). This experimental design allowed us to investigate the role of miR-146a as a T cell–autonomous regulator, modulating their response to antigen stimulation in vivo. We found that after antigen priming, compared with WT OT1 T cells, miR-146a−/− OT1 T cells were hyperresponsive, indicated by a higher magnitude of peak response (approximately day 7), delayed contraction after the peak response (approximately day 7 to day 21), and the persistence of a larger pool of antigen-experienced T cells after priming response (after day 21; Fig. 1, B and C). Compared with WT OT1 T cells, in addition to their higher peak response that implied higher proliferation, miR-146a−/− OT1 T cells also displayed a more activated phenotype at the peak response (shown as lower CD62L expression on day 7; Fig. 1 D) and exhibited increased survival during the contraction phase (shown as less Annexin V+ cells on day 14; Fig. 1 E). The antigen-experienced miR-146a−/− OT1 T cells that persisted after priming response were of typical memory cell phenotype (CD25−CD69−CD62Lhi/loCD44hi), similar to their WT counterpart (Fig. 1 F) although in higher numbers, suggesting a possible influence on the recall response. Indeed, upon antigen boosting at day 46, we observed a hyperresponse of the miR-146a−/− memory OT1 T cells compared with the WT memory OT1 T cells, which was similar in kinetics with that of the primary response (shown as higher magnitude of the peak response, delayed contraction, and higher number of antigen-experienced T cells persisting after the response) but was distinctly more exaggerated (Fig. 1, B and C). It is likely caused by the combined effect of more memory T cells initiating the response and the hyperresponsiveness of these cells, suggesting a dual role for miR-146a in the regulation of the T cell response upon secondary antigen stimulation.
Figure 1.
miR-146a–deficient T cells are hyperresponsive to acute antigen stimulation in vivo. (A) Schematic representation of the experimental design to study the influence of miR-146a deficiency on OVA-specific OT1 T cells in response to antigen stimulation in vivo. (B–F) Flow cytometry analysis of the WT and miR-146a−/− OT1 T cells (gated as CD8+Vβ5+CD45.2+) in the PB of recipient CD45.1 congenic mice receiving 5 × 106 WT or miR-146a−/− OT1 Tg T cells (day −1) followed by prime (day 0) and boost (day 46) immunizations. (B) Percentage of WT and miR-146a−/− OT1 T cells of total CD8+ T cells at the indicated time points. (C) Representative contour plots showing the quantitation of WT and miR-146a−/− OT1 T cells (pregated on CD8+) at the indicated time points. (D) Surface expression of activation marker CD62L on WT and miR-146a−/− OT1 T cells on day 7 (7 d after prime immunization). Representative histogram plots and measurements of mean fluorescence intensity (MFI) are shown. (E) Apoptosis of WT and miR-146a−/− OT1 T cells on day 14 (14 d after prime immunization) measured by Annexin V staining. Representative histogram plots and measurements of the percentage of Annexin V+ OT1 T cells are shown. (F) Phenotype of the WT and miR-146a−/− OT1 T cells on day 45 (45 d after prime immunization). Representative histogram plots are shown. Data are presented as mean ± SEM (n = 8) and are representative of three independent experiments. *, P < 0.05; **, P < 0.01 (miR-146a−/− OT1 T cells in comparison with the corresponding control WT OT1 T cells; for time course experiment, statistics of the peak response are shown).
In summary, these results define a physiological role for miR-146a as an autonomous negative regulator of acute antigen-specific T cell response in vivo, for both primary and recall responses. The results also suggest that miR-146a may play an especially profound role modulating the T cell response after repetitive antigen stimulation, through regulating both the pool size of the memory T cells resulting from the previous effector T cell response and the responsiveness of these memory T cells. This possibility prompted us to investigate the physiological role of miR-146a as a T cell–autonomous factor regulating T cell–associated autoimmunity in vivo, in which the responding T cells are exposed to the constant presence of autoantigen.
miR-146a–deficient T cells are hyperactive in chronic inflammatory response in vivo and promote the development of T cell–associated autoimmunity
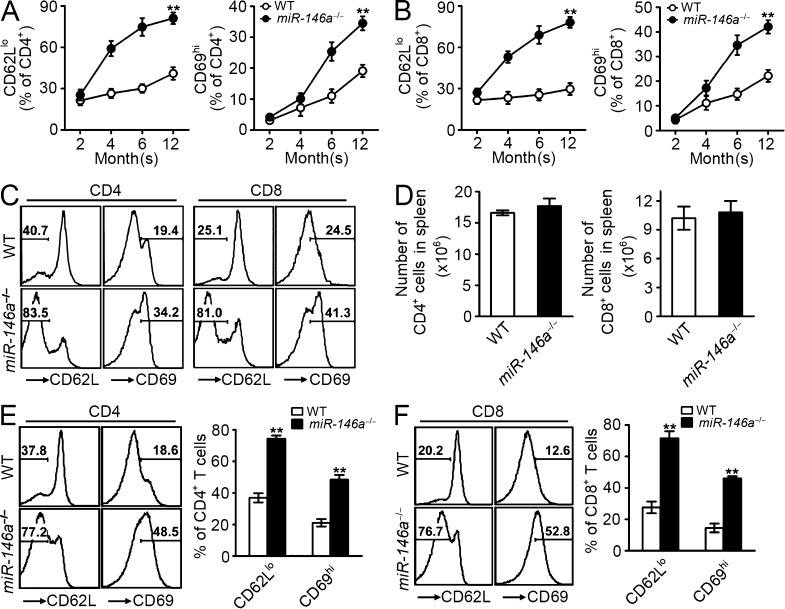
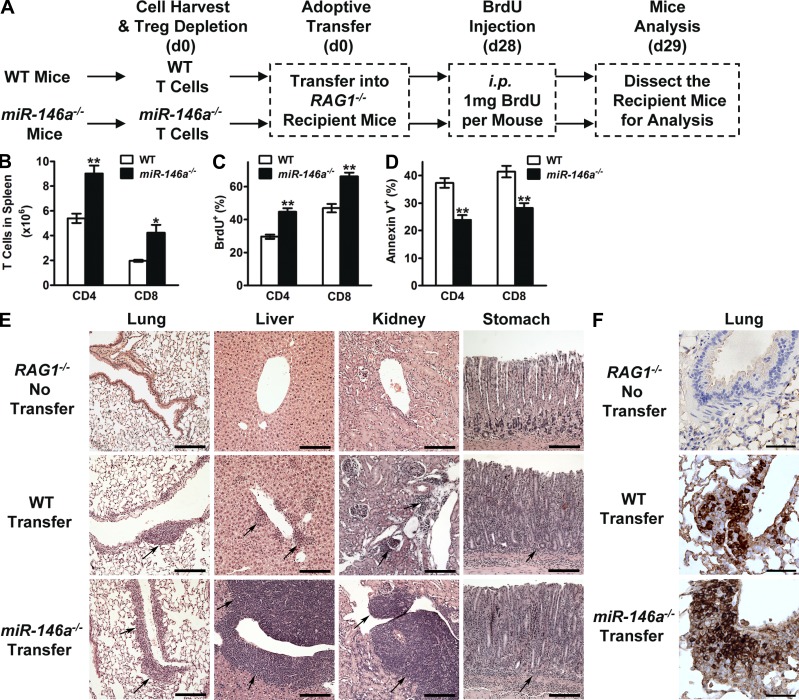
We have previously reported the development of T cell–related chronic hyperinflammatory autoimmune symptoms in miR-146a−/− mice starting from 6–8 mo, characterized by splenomegaly, lymphadenopathy, and multiorgan inflammation (Lu et al., 2010; Boldin et al., 2011; Zhao et al., 2011). There is a gradual accumulation of both CD4+ and CD8+ T cells of activated phenotype (CD62Llo and CD69hi) in the peripheral blood (PB) of miR-146a−/− mice that starts as early as 2 mo, long before their autoimmune symptoms became apparent (Fig. 2, A and B). The accumulation of activated T cells showed a sharp increase between 4 and 6 mo (Fig. 2, A and B), coincident with the appearance of autoimmune symptoms and the premature death of miR-146a−/− mice (Boldin et al., 2011). Examination of miR-146a−/− mice surviving to 1 yr revealed that by that time, the majority of their peripheral CD4 and CD8 T cells in their circulation (∼80% CD62Llo and ∼40% CD69hi; Fig. 2, A–C) and in the spleen (SP; ∼80% CD62Llo and ∼50% CD69hi; Fig. 2, D–F) were of the activated phenotype, associated with severe multiorgan inflammation and tissue destruction (Boldin et al., 2011). However, the interpretation of the role of miR-146a in this T cell hyperinflammatory phenotype is complicated by the multiple abnormalities in miR-146a−/− mice that may contribute to this phenomenon, including the impaired Treg cell function (Lu et al., 2010) and the systemic inflammation induced by myeloid hyperproliferation and malignancies (Zhao et al., 2011). We therefore designed an experiment in which we transferred into RAG1−/− recipient mice Treg cell–depleted WT and miR-146a−/− peripheral T cells, which were then allowed to manifest a chronic hyperinflammatory autoimmune response in recipient mice in the absence of Treg cell control (Fig. 3 A; Itoh et al., 1999). This adoptive transfer system allowed us to confine the miR-146a deficiency to the responding T cells and thus to directly assess the role of miR-146a as an autonomous factor in regulating chronic T cell inflammatory response in vivo. Compared with WT T cells, both miR-146a−/− CD4+ and CD8+ T cells were hyperresponsive, indicated by a higher number of T cells accumulating in SP (Fig. 3 B), more extensive proliferation (Fig. 3 C), lower apoptosis (Fig. 3 D), and a more activated phenotype (lower CD62L and higher CD69; not depicted). Analysis of tissues including lung, liver, kidney, and stomach of recipient mice receiving miR-146a−/− T cell transfer compared with those receiving WT T cell transfer also showed a more significant mononuclear cell infiltration into these tissues accompanying more severe tissue destruction, evidenced as the destruction of bronchiole and alveoli in lung, hepatocytes in liver, glomeruli and tubules in kidney, and the gastric mucosa in stomach (Fig. 3 E). Infiltrates primarily consisted of CD3+ T cells, confirming a T cell–associated autoimmune syndrome (Fig. 3 F). Thus, our results suggest that in addition to regulating acute T cell response to antigen stimulation, miR-146a is also an autonomous negative regulator of T cells in chronic inflammatory response in vivo and contributes to controlling the development of T cell–associated autoimmunity. These findings prompted us to study the expression of miR-146a in T cells, its regulation of the various aspects of T cell activation, and the molecular mechanisms underlying its regulatory function.
Figure 2.
Spontaneous accumulation of activated T cells in miR-146a−/− mice as they age. (A–C) Flow cytometry analysis to quantify the CD4 (gated as CD4+) and CD8 (gated as CD8+) T cells of activated phenotype (gated as CD62Llo or CD69hi) in PB of WT or miR-146a−/− mice. Measurements of the percentage of activated CD4 and CD8 T cells at the indicated age (A and B) and representative histogram plots at 1 yr old (C) are shown. (D) Flow cytometry analysis to quantify the total CD4 (gated as CD4+) and CD8 (gated as CD8+) T cells in the SP of 1-yr-old WT or miR-146a−/− mice. (E and F) Flow cytometry analysis to quantify the CD4 (E; gated as CD4+) and CD8 (F; gated as CD8+) T cells of activated phenotype (gated as CD62Llo or CD69hi) in the SP of 1-yr-old WT or miR-146a−/− mice. Measurements of the percentage of activated CD4 and CD8 T cells and representative histogram plots are shown. Data are presented as mean ± SEM (n = 8) and are representative of three independent experiments. **, P < 0.01 (miR-146a−/− T cells in comparison with the corresponding control WT T cells; for time course experiment, statistics of the end time point are shown).
Figure 3.
miR-146a–deficient T cells are hyperactive in chronic inflammatory response in vivo and promote the development of T cell–associated autoimmunity. (A) Schematic representation of the experimental design to study the role of miR-146a as an autonomous factor to regulate T cell response in a chronic T cell inflammatory autoimmune mouse disease model. (B) Flow cytometry analysis to quantify the number of WT and miR-146a−/− CD4 or CD8 T cells (gated as CD4+ and CD8+, respectively) in the SP of recipient mice on day 29 after adoptive transfer. (C) BrdU incorporation analysis of the in vivo proliferation of WT and miR-146a−/− CD4 and CD8 T cells (gated as CD4+BrdU+ and CD8+BrdU+, respectively) in the SP of recipient mice on day 29 after adoptive transfer. BrdU was injected into recipient mice 16 h before the analysis. (D) Annexin V staining analysis of the in vivo apoptosis of WT and miR-146a−/− CD4 and CD8 T cells (gated as CD4+Annexin V+ and CD8+Annexin V+, respectively) in the SP of recipient mice on day 29 after adoptive transfer. (E and F) Immunohistology analysis of tissue sections from recipient mice on day 29 after adoptive transfer. Tissue sections from RAG1−/− mice that did not receive cell transfer were included as control. (E) Hematoxylin and eosin staining. Arrows point to mononuclear cell infiltrates. (F) Anti–mouse CD3 staining. CD3 is shown in brown. Bars: (E, lung, liver, and kidney) 200 µm; (E, stomach) 400 µm; (F) 40 µm. Data are presented as mean ± SEM (n = 4) and are representative of three independent experiments. *, P < 0.05; **, P < 0.01 (miR-146a−/− T cells in comparison with the corresponding WT T cells).
miR-146a induction in mouse CD4 and CD8 T cells depends on NF-κB
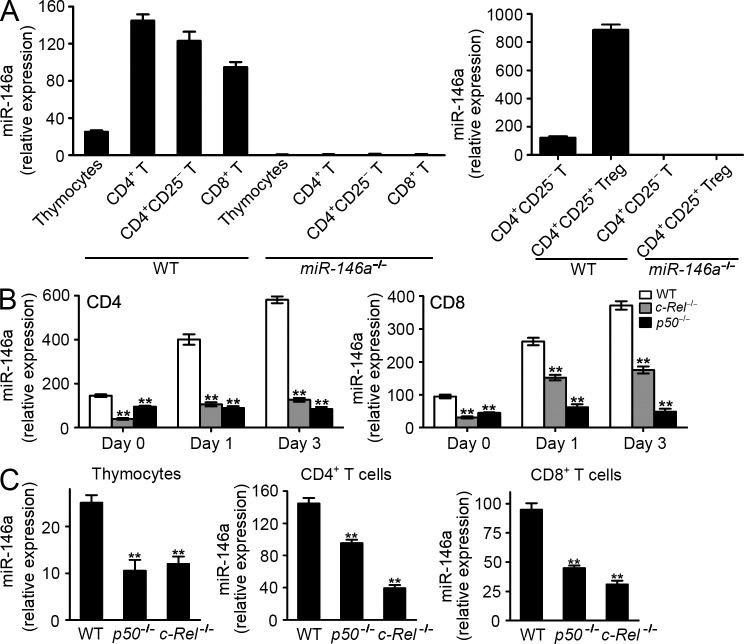
We and others have reported the expression of miR-146a in mouse peripheral CD4+ T cells, particularly in CD4+CD25+ Treg cells (Monticelli et al., 2005; Lu et al., 2010; Boldin et al., 2011). Further analysis of other T cell compartments revealed that miR-146a is also highly expressed in mouse CD8+ T cells, as well as non-Treg CD4+ T cells (CD4+CD25−), albeit at a lower level in CD4+CD25− T cells than in CD4+CD25+ Treg cells and at a slightly lower level in CD8+ T cells than in CD4+CD25− T cells (Fig. 4 A). Of note, both peripheral CD4+ and CD8+ T cells express significantly higher levels of miR-146a compared with that expressed in thymocytes, suggesting a significant role for miR-146a in mature T cell function (Fig. 4 A). Expression of miR-146a is further up-regulated in both CD4+ and CD8+ T cells upon TCR stimulation using anti-CD3/CD28, suggesting that miR-146a is directly involved in T cell activation (Fig. 4 B). To study the requirement of NF-κB for miR-146a expression in mouse primary T cells, we analyzed mice defective for the NF-κB subunit p50 (p50−/−) or c-Rel (c-Rel−/−). In the absence of either p50 or c-Rel, miR-146a expression was significantly down-regulated in both thymocytes and peripheral CD4+ and CD8+ T cells (Fig. 4 C), indicating that NF-κB is required for the basal expression of miR-146a in both progenitor and mature T cells. Moreover, after TCR stimulation, up-regulation of miR-146a was greatly reduced in CD4+ and CD8+ T cells lacking c-Rel and was almost abolished in T cells lacking p50 (Fig. 4 B), indicating that TCR-induced miR-146a expression is also dependent on NF-κB. Quantitatively, c-Rel seems to play a more significant role than p50 for regulating the basal expression of miR-146a, whereas p50 seems to be more important than c-Rel for regulating the TCR-induced expression of miR-146a. These results suggest that different constituents of NF-κB may be responsible for regulating the expression of miR-146a and its related functions in T cells at various developmental and activation stages. In this study, we focused on studying the expression of miR-146a in mature CD4+ and CD8+ T cells, its interaction with the NF-κB pathway upon TCR stimulation, and its regulation of T cell activation.
Figure 4.
miR-146a is expressed in T cells and is up-regulated upon TCR stimulation through NF-κB–dependent induction. (A) TaqMan quantitative PCR (Q-PCR) analysis of miR-146a expression in thymocytes and in total CD4+ T cells, CD4+CD25+ Treg cells, CD4+ T cells deprived of Treg cells (CD4+CD25−), and CD8+ T cells purified from the SP of 6-wk-old B6 WT mice. Cells harvested from the age-matched miR-146a−/− mice were included as negative control to show the specificity of the assay. (B) TaqMan Q-PCR analysis of miR-146a expression in WT CD4+ and CD8+ T cells or CD4+ and CD8+ T cells lacking NF-κB component p50 (p50−/−) or c-Rel (c-Rel−/−) purified from SP cells stimulated with anti-CD3/CD28 for the indicated time. (C) TaqMan Q-PCR analysis of miR-146a expression in thymocytes and peripheral CD4+ and CD8+ T cells purified from the SP of WT mice or mice deficient of NF-κB component p50 (p50−/−) or c-Rel (c-Rel−/−). Data are presented as mean ± SEM (n = 3) and are representative of three independent experiments. **, P < 0.01 (miR-146a−/− cells in comparison with the corresponding WT cells).
miR-146a modulates TCR-induced activation of NF-κB in CD4+ and CD8+ T cells
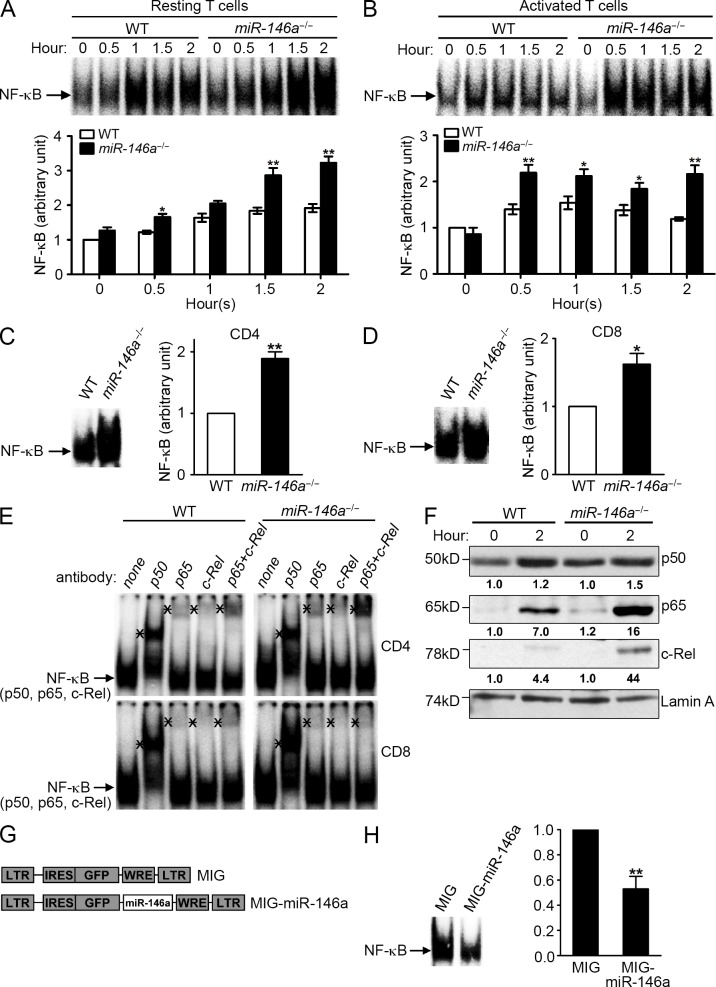
Work from us and others has implicated miR-146a as a negative feedback regulator of NF-κB activation in innate immune cells (Taganov et al., 2006; Williams et al., 2008; Hou et al., 2009; Boldin et al., 2011; Zhao et al., 2011). This prompted us to investigate the role of miR-146a in regulating NF-κB activity in T cells, given that NF-κB activation is a critical signaling event induced by TCR stimulation (Schulze-Luehrmann and Ghosh, 2006). Analysis of SP and LN cells containing peripheral T cells that were stimulated with anti-CD3/CD28 revealed the induction of higher (∼1.5–2-fold) levels of NF-κB activity in miR-146a−/− cells compared with WT cells as measured in an electrophoretic mobility shift assay (EMSA; Fig. 5 A), indicating that miR-146a negatively regulates the TCR-induced NF-κB activation in resting T cells. Analysis of purified CD4+ and CD8+ T cells confirmed the up-regulation of NF-κB activity in both types of T cells in the absence of miR-146a (Fig. 5, C and D). In addition to resting T cells, we also found an ∼1.5–2-fold higher induction of NF-κB in preactivated miR-146a−/− T cells that were restimulated with anti-CD3/CD28 compared with their WT counterpart (Fig. 5 B), indicating that miR-146a also negatively regulates the NF-κB activity induced in activated T cells by repetitive TCR stimulation.
Figure 5.
miR-146a modulates NF-κB activity in T cells induced by TCR stimulation. (A and B) EMSA of 10 µg of nuclear extracts from freshly isolated WT and miR-146a−/− SP and LN cells stimulated with anti-CD3 + anti-CD28 (1 µg/ml each) for the indicated time (A) or from WT and miR-146a−/− SP/LN cells prestimulated with anti-CD3 + anti-CD28 (1 µg/ml each) for 3 d and then rested and restimulated with plate-bound anti-CD3 (10 µg/ml) and anti-CD28 (1 µg/ml) for the indicated time (B). Representative EMSA images and the measurements of NF-κB in arbitrary units (mean ± SEM; WT 0 h = 1) from three independent experiments are shown. (C and D) EMSA of 10 µg of nuclear extracts from purified WT or miR-146a−/− CD4 (C) and CD8 (D) T cells stimulated with plate-bound anti-CD3 (10 µg/ml) and anti-CD28 (1 µg/ml) for 2 h. Representative EMSA images and the measurements of NF-κB in arbitrary units (mean ± SEM; WT = 1) from three independent experiments are shown. (E) Composition of NF-κB complexes induced by TCR stimulation in WT and miR-146a−/− CD4 and CD8 T cells assessed by EMSA with antibody supershift. 5 µg of specific antibodies to each NF-κB component was preincubated with 20 µg of nuclear extracts before addition of the NF-κB probe. Nuclear extracts were prepared from purified WT and miR-146a−/− CD4 or CD8 T cells stimulated with plate-bound anti-CD3 (10 µg/ml) and anti-CD28 (1 µg/ml) for 20 h. The mobilities of the inducible κB-binding activities are indicated by arrows. The supershifts of κB-binding mobilities induced by antibody inhibition are marked with “*.” Data are representative of three independent experiments. (F) Western blot analysis of 30 µg of nuclear extracts from WT and miR-146a−/− SP/LN cells stimulated with anti-CD3 + anti-CD28 (1 µg/ml each) for the indicated time for the expression of p50, p65, and c-Rel. Numbers under Western blots denote relative amounts normalized to Lamin A expression for each sample. Data are representative of three independent experiments. (G) Schematic representation of MSCV retroviral vector expressing GFP reporter gene (MIG) and MIG vector expressing murine miR-146a (MIG–miR-146a). IRES, internal ribosome entry site; LTR, long term repeats; MSCV, murine stem cell virus; WRE, woodchuck responsive element. (H) EMSA of 20 µg of nuclear extracts from WT SP/LN cells that were prestimulated with anti-CD3 + anti-CD28 (1 µg/ml each) for 3 d and transduced with either the control retroviral vector (MIG) or retroviral vector expressing miR-146a (MIG–miR-146a) and then were rested and restimulated with plate-bound anti-CD3 (10 µg/ml) and anti-CD28 (1 µg/ml) for 2 h. Representative EMSA image and the measurements of NF-κB in arbitrary units (mean ± SEM; WT cells transduced with MIG = 1) from three independent experiments are shown. *, P < 0.05; **, P < 0.01 (miR-146a−/− samples in comparison with the corresponding WT controls [A–D]; MIG–miR-146a–transduced sample in comparison with the corresponding MIG-transduced control [H]).
Multiple subunits of NF-κB are involved in TCR-induced NF-κB activation (Schulze-Luehrmann and Ghosh, 2006); we therefore analyzed the composition of NF-κB complexes induced by TCR stimulation in WT and miR-146a−/− T cells. As shown in Fig. 5 E, despite the difference in overall NF-κB activity, similar NF-κB complexes containing p50, p65, and c-Rel were induced in both types of T cells, suggesting that miR-146a regulates the activation of TCR-induced NF-κB without significantly altering the involvement of the various NF-κB subunits.
miR-146a regulation of TCR-driven NF-κB activation was also verified by increased nuclear translocation of multiple NF-κB subunits (p50, p65, and c-Rel) in miR-146a−/− T cells compared with WT T cells (Fig. 5 F). The steady-state expression of these NF-κB subunits in T cells is not affected by miR-146a deficiency (not depicted), indicating that miR-146a mainly regulates the activation status of these NF-κB subunits. Notably, no significant difference of TCR-driven activation of NFAT and AP-1 was observed between miR-146a−/− and WT T cells (not depicted), suggesting that NF-κB activation is the key TCR-induced signaling event modulated by miR-146a.
To verify the role of miR-146a as a negative regulator of NF-κB activation, we constructed a retroviral vector, MIG–miR-146a, that expresses miR-146a (Fig. 5 G). As expected, in contrast to miR-146a deficiency, overexpression of miR-146a in WT T cells through MIG–miR-146a transduction significantly down-regulated the NF-κB activity induced by TCR stimulation (Fig. 5 H). These experimental results establish a role for miR-146a as a negative feedback regulator modulating NF-κB activation induced by TCR stimulation in mouse CD4+ and CD8+ T cells, prompting us to investigate its influence on T cell activation that are under NF-κB control.
miR-146a regulates the various aspects of mouse T cell activation induced by TCR stimulation in an NF-κB–dependent manner
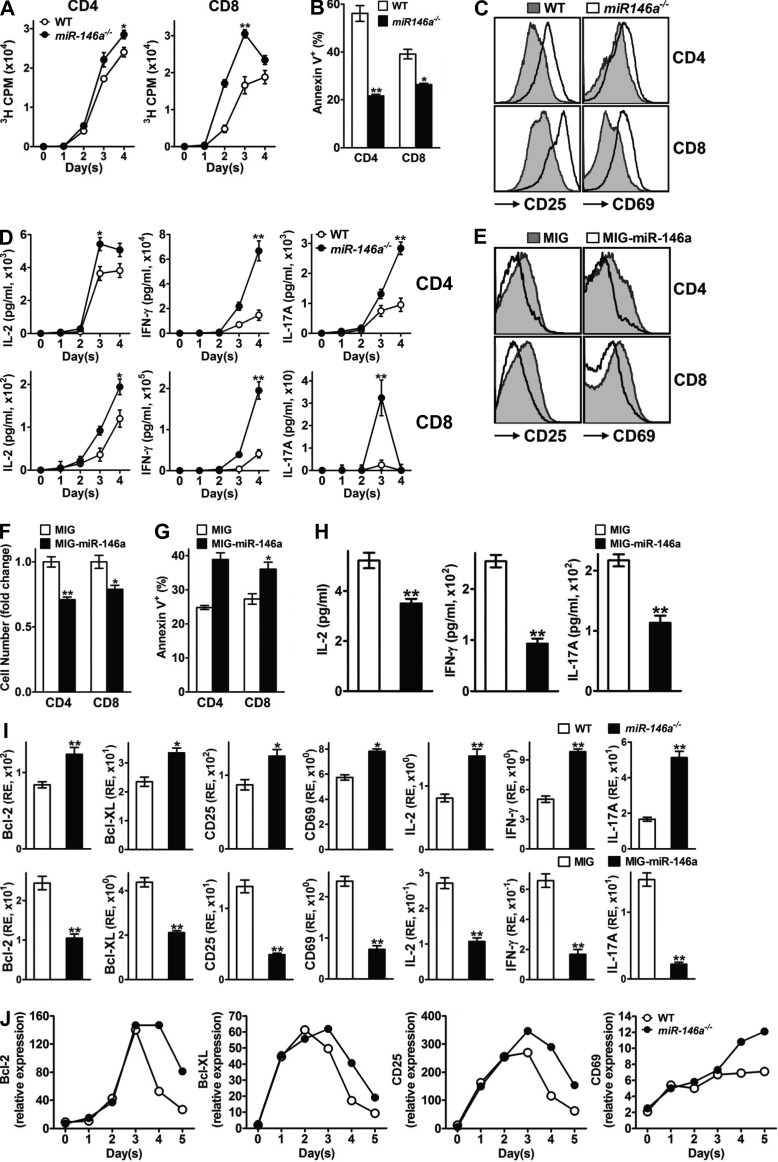
TCR-induced NF-κB activation regulates the expression of a large set of NF-κB responsive genes involved in almost all aspects of T cell activation, among which are those well characterized to be responsible for proliferation (IL-2, Cyclin-D1, Cyclin-D2, c-Myb, and c-Myc), survival (IL-2, Bcl-2, Bcl-XL, and c-FLIP), the effector phenotype (CD25 and CD69), and effector cytokine production (IL-2, IFN-γ, and IL-17A; Li and Verma, 2002; Vallabhapurapu and Karin, 2009). In light of the finding that miR-146a is a negative feedback regulator of TCR-induced NF-κB activation in mouse T cells, we asked whether miR-146a physiologically regulates these various aspects of mouse T cell activation and whether such regulation is mediated through NF-κB. To address these questions, first we conducted an extensive comparison of WT and miR-146a−/− T cells in response to anti-CD3/CD28 stimulation, focusing on those activities that are under NF-κB regulation. We found that compared with their WT counterparts, miR-146a−/− CD4+ and CD8+ T cells were overall hyperactive, manifested by higher proliferation (Fig. 6 A), diminished apoptosis (Fig. 6 B), an exaggerated effector phenotype with particularly high surface activation markers CD25 (IL-2Rα) and CD69 (Fig. 6 C), and increased production of effector cytokines including IL-2, IFN-γ, and IL-17A (Fig. 6 D). Of note, before anti-CD3/CD28 stimulation, both WT and miR-146a−/− T cells exhibited a similar naive T cell phenotype (CD25−CD69−CD62LhiCD44lo; not depicted), suggesting that the observed differences between WT and miR-146a−/− T cells ensued after TCR stimulation. On the contrary, overexpression of miR-146a in WT T cells through MIG–miR-146a–mediated transduction resulted in hypoactivation in response to anti-CD3/CD28 stimulation, shown as reduced proliferation (Fig. 6 F), enhanced apoptosis (Fig. 6 G), a dampened effector phenotype that was particularly lower in surface activation markers CD25 and CD69 (Fig. 6 E), and reduced production of effector cytokines (Fig. 6 H). Thus, these results indicate that miR-146a plays a physiological role of negatively modulating the various aspects of mouse T cell activation that are under NF-κB regulation.
Figure 6.
miR-146a regulates the various aspects of T cell activation induced by TCR stimulation. (A) [3H]Thymidine incorporation analysis of proliferation of purified WT or miR-146a−/− CD4 and CD8 T cells stimulated with plate-bound anti-CD3 (10 µg/ml) + anti-CD28 (1 µg/ml) for 4 d. (B) Annexin V staining analysis for apoptosis of WT or miR-146a−/− CD4 and CD8 T cells (gated as CD4+ and CD8+ from the SP/LN cell culture, respectively) stimulated with soluble anti-CD3 + anti-CD28 (1 µg/ml each) for 4 d. (C) Flow cytometry analysis of surface activation marker expression on WT or miR-146a−/− CD4 and CD8 T cells (gated as CD4+ and CD8+ from the SP/LN cell culture, respectively) stimulated with soluble anti-CD3 + anti-CD28 (1 µg/ml each) for 3 d. (D) ELISA analysis of effector cytokine production of purified WT or miR-146a−/− CD4 and CD8 T cells stimulated with plate-bound anti-CD3 (10 µg/ml) + anti-CD28 (1 µg/ml) for 4 d. (E) Flow cytometry analysis of surface activation marker expression on WT CD4 and CD8 T cells transduced with MIG–miR-146a or control MIG retroviral vectors and stimulated with soluble anti-CD3 + anti-CD28 (1 µg/ml each) for 3 d (gated as CD4+GFP+ and CD8+GFP+ from the SP/LN cell culture, respectively). (F) Relative cell number fold change of WT CD4 and CD8 T cells transduced with MIG–miR-146a or control MIG retroviral vectors and stimulated with soluble anti-CD3 + anti-CD28 (1 µg/ml each) for 3 d (gated as CD4+ and CD8+ from the SP/LN cell culture, respectively; counts of T cells transduced with MIG = 1). (G) Annexin V staining of WT CD4 and CD8 T cells transduced with MIG–miR-146a or control MIG retroviral vectors and stimulated with soluble anti-CD3 + anti-CD28 (1 µg/ml each) for 4 d (gated as CD4+GFP+ and CD8+GFP+ from the SP/LN cell culture, respectively). (H) ELISA analysis of effector cytokine production of WT SP/LN cells transduced with MIG–miR-146a or control MIG retroviral vectors and stimulated with soluble anti-CD3 + anti-CD28 (1 µg/ml each) for 4 d. (I) TaqMan Q-PCR analysis of the expression of NF-κB responsive genes in WT and miR-146a−/− SP/LN cells stimulated with soluble anti-CD3 + anti-CD28 (1 µg/ml each) for 3 d (top) or in WT SP/LN cells transduced with either MIG-miR-146a or control MIG retroviral vector and stimulated with soluble anti-CD3 + anti-CD28 (1 µg/ml each) for 3 d (bottom). RE, relative expression. (J) TaqMan Q-PCR analysis of the time course expression of NF-κB responsive genes in WT and miR-146a−/− SP/LN cells stimulated with soluble anti-CD3 + anti-CD28 (1 µg/ml each) for 5 d. Data are presented as mean of duplicate culture ± SEM (A, B, D, and F–I) and are representative of three independent experiments. *, P < 0.05; **, P < 0.01 (miR-146a−/− samples in comparison with the corresponding WT controls [A, B, D, and I, top]; MIG–miR-146a–transduced samples in comparison with the corresponding MIG-transduced controls [F–H and I, bottom]).
Next we analyzed the induction of the NF-κB responsive genes that are involved in T cell activation under the influence of miR-146a. After stimulation with anti-CD3/CD28, cells lacking miR-146a exhibited a significantly augmented induction of a set of NF-κB responsive genes, including Bcl-2, Bcl-XL, CD25, CD69, IL-2, IFN-γ, and IL-17A; conversely, overexpression of miR-146a in mouse T cells through MIG–miR-146a transduction resulted in a significantly decreased induction of these genes (Fig. 6 I). Analysis of the induction of these genes in purified CD4+ and CD8+ T cells confirmed that these genes were subjected to miR-146a regulation in both types of T cells (not depicted). Thus, miR-146a modulates the induction of genes that are under NF-κB regulation after TCR stimulation. Moreover, time course analysis revealed that miR-146a deficiency only altered the late expression of these NF-κB target genes without affecting their early expression (Fig. 6 J), strongly supporting the role of miR-146a as a negative feedback regulator of NF-κB activation in T cells. However, induction of certain other genes that have also been found to be NF-κB responsive was not affected in the absence or excess of miR-146a, including Cyclin-D1, Cyclin-D2, c-Myb, c-Myc, and c-FLIP (not depicted). This might be caused by an NF-κB sensitivity of these genes that is beyond the regulatory range of miR-146a or by the ability of other transcription factors to mask the effect.
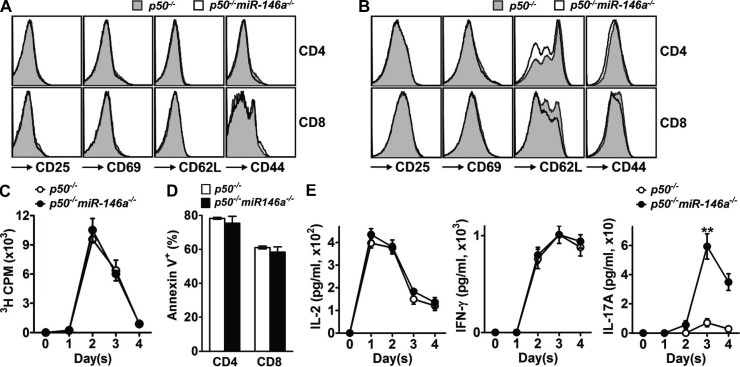
Finally, we analyzed the influence of miR-146a on T cell activation in the absence of p50, the key component of most NF-κB complexes induced by TCR stimulation in mouse T cells. No difference between p50−/− and p50−/−miR-146a−/− T cells upon TCR stimulation was evident studying surface activation marker expression (Fig. 7, A and B), proliferation (Fig. 7 C), survival (Fig. 7 D), and effector cytokine production (Fig. 7 E). This suggests that the regulatory function of miR-146a on T cell activation is mainly mediated through regulation of NF-κB, especially its p50 subunit. Notably, the production of IL-17A still showed certain difference between p50−/− and p50−/−miR-146a−/− T cells (Fig. 7 E), suggesting the involvement of p50-negative NF-κB complexes or other signaling pathways that are also under regulation of miR-146a for the induction of this cytokine.
Figure 7.
miR-146a regulates T cell activation in an NF-κB–dependent manner. (A and B) Flow cytometry analysis of surface activation marker expression on resting (A; gated as CD4+ and CD8+ from the SP cells freshly isolated from 6-wk-old mice) or activated (B; gated as CD4+ and CD8+ from the SP/LN cells stimulated with 1 µg/ml soluble anti-CD3 + 1 µg/ml soluble anti-CD28 for 3 d) p50−/− and p50−/−miR-146a−/− CD4 and CD8 T cells. (C) [3H]Thymidine incorporation analysis of proliferation of SP/LN cells harvested from p50−/− or p50−/−miR-146a−/− mice and stimulated with soluble anti-CD3 + anti-CD28 (1 µg/ml each) for 4 d. (D) Annexin V staining of p50−/− and p50−/−miR-146a−/− CD4 and CD8 T cells stimulated with soluble anti-CD3 + anti-CD28 (1 µg/ml each) for 4 d (gated as CD4+ and CD8+ from the SP/LN cell culture, respectively). (E) ELISA analysis of effector cytokine production of p50−/− and p50−/−miR-146a−/− SP/LN cells stimulated with soluble anti-CD3 and anti-CD28 (1 µg/ml each) for 4 d. Data are normalized to the input T cells in SP/LN cell culture. Data are presented as mean of duplicate culture ± SEM and are representative of three independent experiments. **, P < 0.01 (p50−/−miR-146a−/− samples in comparison with the corresponding p50−/− controls; for time course experiment, statistics of the peak responses are shown).
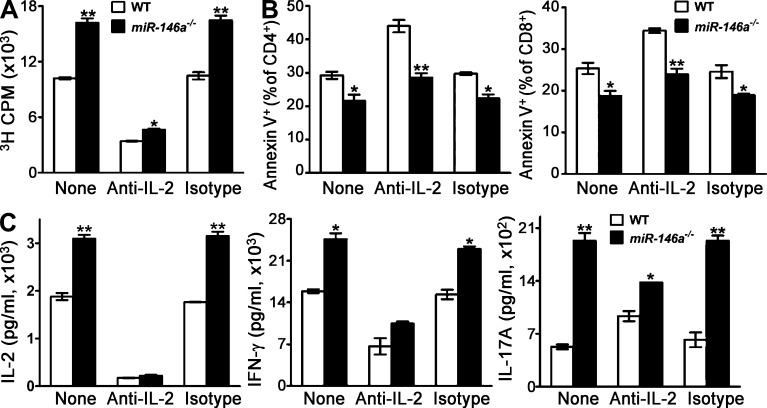
IL-2 is a major cytokine induced by NF-κB activation downstream of TCR stimulation and has been found to participate in many aspects of T cell activation (Malek, 2008). Because miR-146a was shown to regulate IL-2 production (Fig. 6, D and H), it brings up the possibility that miR-146a might regulate T cell activation mainly through its effect on IL-2. However, neutralizing of IL-2 in T cell culture only partially abrogated the difference between WT and miR-146a−/− T cells in response to TCR stimulation (Fig. 8, A–C), indicating that miR-146a regulation of T cell activation is not totally mediated through regulating IL-2 production, but rather likely through modulating the expression of multiple NF-κB responsive genes.
Figure 8.
miR-146a regulation of T cell activation is not totally mediated through regulating IL-2 production. (A–C) WT and miR-146a−/− SP/LN cells containing equal number of T cells were stimulated with soluble anti-CD3 + anti-CD28 (1 µg/ml each) for 3 d in the presence or absence of 10 µg/ml anti–IL-2 neutralizing antibody or isotype control antibody. (A) [3H]Thymidine incorporation analysis of the proliferation of WT and miR-146a−/− cells on day 3 after anti-CD3/CD28 stimulation. (B) Annexin V staining of WT and miR-146a−/− CD4 and CD8 cells (gated as CD4+ and CD8+ from the SP/LN cell culture, respectively) on day 3 after anti-CD3/CD28 stimulation. (C) ELISA analysis of effector cytokine production of WT and miR-146a−/− cells on day 2 (for IL-2) or day 3 (for IFN-γ and IL-17A) after anti-CD3/CD28 stimulation. The time points are chosen to represent the peak production of the corresponding cytokines during the 3-d time course experiment. Data are presented as mean of duplicate culture ± SEM and are representative of three independent experiments. *, P < 0.05; **, P < 0.01 (miR-146a−/− cells in comparison with the corresponding WT control cells).
Overall, these data suggest that miR-146a regulates the various aspects of T cell activation upon TCR stimulation in an NF-κB–dependent manner, likely through modulating the expression of a set of NF-κB responsive genes involved in these activities. This finding encouraged us to further investigate the molecular mechanisms underlying the miR-146a modulation of TCR signaling to NF-κB.
TRAF6 and IRAK1 are bona fide targets of miR-146a in mouse CD4+ and CD8+ T cells and are possible mediators for its regulation of TCR-induced NF-κB activity
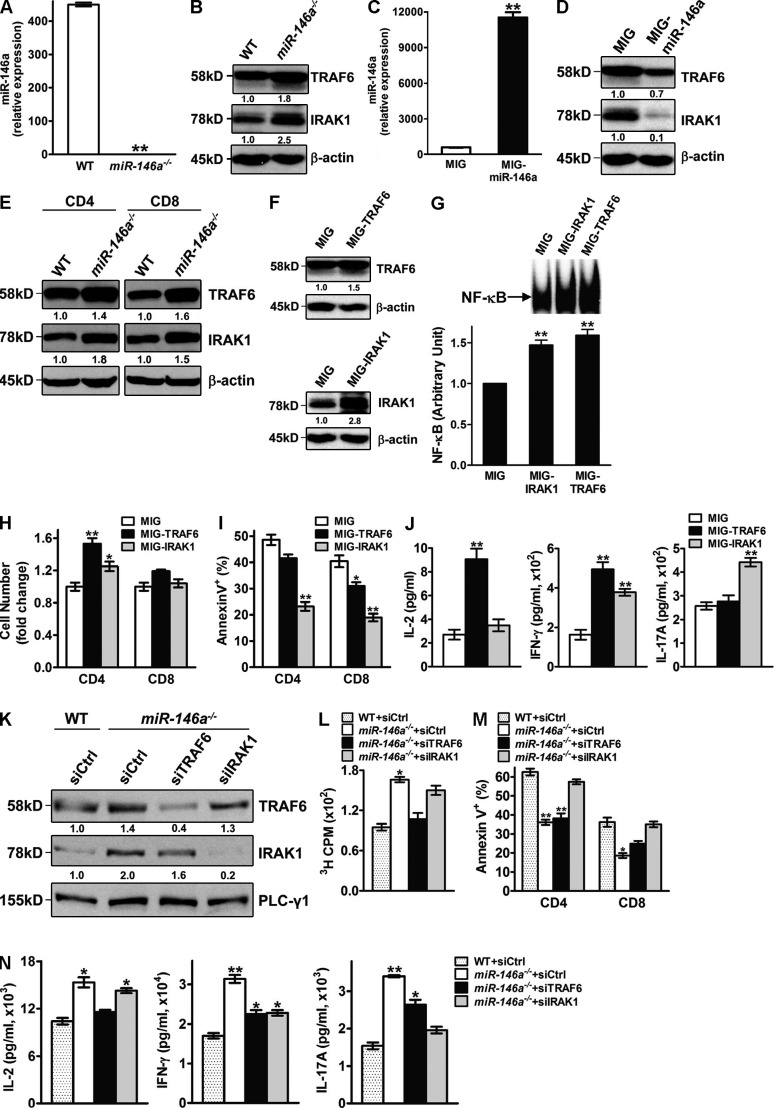
miRNAs function through posttranscriptional repression of their target genes (Ambros, 2004; Bartel, 2004). Among the many possible targets of miR-146a predicted by computational algorisms, we were particularly interested in TRAF6 and IRAK1, two adaptor molecules involved in the NF-κB activation pathway (Oeckinghaus et al., 2011) which are targets of miR-146a inhibition in epithelial cells, innate immune cells, and B cells (Taganov et al., 2006; Nahid et al., 2009; Chassin et al., 2010; Boldin et al., 2011). Mouse SP/LN cells stimulated with anti-CD3/CD28 for 3 d displayed an ∼1.8-fold increase of TRAF6 and an ∼2.5-fold increase of IRAK1 at the protein expression level in miR-146a−/− cells compared with WT cells (Fig. 9, A and B). On the contrary, overexpression of miR-146a in WT SP/LN cells stimulated with anti-CD3/aCD28 reduced the protein expression of TRAF6 for ∼1.4-fold and for IRAK1 up to 10-fold (Fig. 9, C and D). Analysis of purified CD4+ and CD8+ T cells confirmed an up-regulation of TRAF6 and IRAK1 proteins in both types of T cells in the absence of miR-146a (Fig. 9 E). Collectively, these results demonstrate TRAF6 and IRAK1 to be bona fide targets of miR-146a in primary mouse CD4+ and CD8+ T cells. To verify the connection between the up-regulated expression of TRAF6 and IRAK1 and the elevated activation of NF-κB in activated T cells lacking miR-146a, we constructed retroviral vectors expressing TRAF6 (MIG-TRAF6) and IRAK1 (MIG-IRAK1). Transduction of activated WT T cells with MIG-TRAF6 or MIG-IRAK1 resulted in an overexpression of IRAK1 or TRAF6 (Fig. 9 F) to a level similar to that observed in activated miR-146a−/− T cells (Fig. 9 B). Importantly, this up-regulation of TRAF6 and IRAK1 expression correlated with an increased induction of NF-κB activity after TCR stimulation (Fig. 9 G), which was similar to that observed in miR-146a−/− cells (Fig. 5 B), suggesting that TRAF6 and IRAK1 are possible mediators of the miR-146a regulation of NF-κB activation in primary mouse T cells. Consistent with their increased NF-κB activity, WT T cells overexpressing TRAF6 or IRAK1 showed enhanced proliferation, survival, and effector cytokine production in response to TCR stimulation (Fig. 9, H–J). In a complementary experiment, reduction of TRAF6 or IRAK1 expression in miR-146a−/− T cells through small interfering RNA (siRNA) targeting partially corrected their hyperactivation phenotype after TCR stimulation, further supporting the roles of these two signaling proteins in mediating miR-146a regulation of T cell activation (Fig. 9, K–N). Interestingly, changes of T cell activation induced by manipulating the expression of TRAF6 or IRAK1 are not identical: TRAF6 seems to impact more on proliferation (Fig. 9, H and L) and the production of IL-2 (Fig. 9, J and N), whereas IRAK1 seems to impact more on survival (Fig. 9, I and M) and the production of IL-17A (Fig. 9, J and N). These results suggest that miR-146a regulation of T cell activation is likely mediated through its combinatorial modulation of multiple targets including TRAF6 and IRAK1. Notably, in contrast to TRAF6 and IRAK1, analysis of other signaling components mediating TCR signaling to NF-κB including the CBM complex showed no obvious difference in their steady-state expression levels between miR-146a−/− and WT T cells (not depicted), suggesting that TRAF6 and IRAK1 are likely to be the major targets of miR-146a modulating TCR signaling to NF-κB.
Figure 9.
TRAF6 and IRAK1 are bona fide targets of miR-146a in mouse T cells and are possible mediators for its regulation of NF-κB activity. (A and B) TaqMan Q-PCR analysis of miR-146a expression (A; shown as mean of duplicates ± SEM) and Western blot analysis of TRAF6 and IRAK1 expression (B) in WT or miR-146a−/− SP/LN cells stimulated with soluble anti-CD3 + anti-CD28 (1 µg/ml each) for 3 d. Data are representative of three independent experiments. (C and D) TaqMan Q-PCR analysis of miR-146a expression (C; shown as mean of duplicates ± SEM) and Western blot analysis of TRAF6 and IRAK1 expression (D) in WT SP/LN cells transduced with MIG–miR-146a or control MIG retroviral vectors and stimulated with soluble anti-CD3 + anti-CD28 (1 µg/ml each) for 3 d. Data are representative of three independent experiments. (E) Western blot analysis of TRAF6 and IRAK1 expression in activated WT or miR-146a−/− CD4 and CD8 T cells. CD4 and CD8 T cells were purified through MACS sorting from WT or miR-146a−/− SP/LN cells stimulated with soluble anti-CD3 + anti-CD28 (1 µg/ml each) for 3 d. Data are representative of three independent experiments. (F–J) WT SP/LN cells were stimulated with soluble anti-CD3 + anti-CD28 (1 µg/ml each) and transduced with MIG-TRAF6, MIG-IRAK1, or control MIG retroviral vectors. By day 3, >90% of total cell culture was activated T cells (sum of CD4+ and CD8+ cells). Data are representative of at least three independent experiments. *, P < 0.05; **, P < 0.01 (MIG-TRAF6– or MIG-IRAK1–transduced samples in comparison with the corresponding MIG-transduced controls). (F) Western blot analysis of TRAF6 and IRAK1 expression in day 3 transduced cells. (G) EMSA analysis of NF-κB activity of 10 µg of nuclear extracts from day 3 transduced cells rested and restimulated with plate-bound anti-CD3 (10 µg/ml) + anti-CD28 (1 µg/ml) for 30 min. Representative EMSA image and the measurements of NF-κB in arbitrary units (mean ± SEM; cells transduced with MIG = 1) from three independent experiments are shown. (H) Relative CD4 and CD8 T cell number (gated as CD4+ and CD8+, respectively, cell count of MIG-transduced T cells = 1) on day 3. Data are presented as mean of duplicate culture ± SEM. (I) Annexin V staining analysis of apoptosis of transduced CD4 and CD8 T cells (gated as CD4+GFP+ and CD8+GFP+, respectively) on day 4. Data are presented as mean of duplicate culture ± SEM. (J) ELISA analysis of effector cytokine production from transduced cells on day 3. Data are presented as mean of duplicate culture ± SEM. (K–N) WT or miR-146a−/− SP/LN cells were stimulated with soluble anti-CD3 + anti-CD28 (1 µg/ml each) and transfected with nonsilencing control siRNA (siCtrl) or siRNA specific for mouse TRAF6 (siTRAF6) or IRAK1 (siIRAK1). Representative data from two independent experiments are shown. *, P < 0.05; **, P < 0.01 (miR-146a−/− cells transduced with various siRNAs in comparison with WT cells transduced with control siRNA). (K) Western blot analysis of TRAF6 and IRAK1 expression in the siRNA-transfected cells on day 3.5. (L) [3H]Thymidine incorporation analysis of proliferation of the siRNA-transfected cells on day 3. Data are presented as mean of duplicate culture ± SEM. (M) Annexin V staining analysis of apoptosis of the siRNA-transfected CD4 and CD8 T cells (gated as CD4+ and CD8+, respectively) on day 3.5. Data are presented as mean of duplicate culture ± SEM. (N) ELISA analysis of effector cytokine production from the siRNA-transfected cells on day 3.5. Data are presented as mean of duplicate culture ± SEM. Numbers under Western blots denote relative amounts normalized to β-actin or PLC-γ1 expression for each sample.
DISCUSSION
As the central player in the immune response fighting against pathogen invasions in mammals, the activation of T cells must be tightly regulated. Proper resolution of a T cell response is as essential as its initiation because of the possible severe pathological consequences of a hyperinflammatory response. NF-κB activation is a key signaling event downstream of TCR engagement and regulates almost all of the important aspects of T cell activation. Because of its pivotal role, NF-κB activation in T cells is regulated through a multilayered negative regulatory network (Schulze-Luehrmann and Ghosh, 2006). In this study, we identified miR-146a as an important new member of the negative feedback loop that modulates TCR signaling to NF-κB and demonstrated its physiological role in regulating both acute T cell response and chronic hyperinflammatory autoimmune T cell response in vivo. Based on these results, we propose a model of miR-146a as an important new constituent of the negative feedback regulatory network modulating TCR signaling to NF-κB. In this model, TCR stimulation activates NF-κB which induces the expression of miR-146a. miR-146a in turn down-regulates NF-κB activation through repressing NF-κB activators TRAF6 and IRAK1, providing a negative feedback control for the induction of NF-κB responsive genes that are involved in the various aspects of T cell activation. Therefore, our study adds miRNA, a key emerging posttranscriptional regulator of the immune system, as a novel molecular player controlling resolution, a critical physiological aspect of T cell responses.
Including miR-146a, three molecules have been identified that act in negative feedback loops modulating TCR signaling to NF-κB (the other two are IκBα and A20). It is interesting to ask why nature evolved so many pathways for this regulation. The answer is not redundancy because mice defective in any one of these three molecules develop autoimmune symptoms (Beg et al., 1995; Klement et al., 1996; Lee et al., 2000; Boldin et al., 2011). However, these three molecules enact their regulatory functions with distinct kinetics. IκBα and A20 belong to the Group I NF-κB early responsive genes whose expression is rapidly induced after NF-κB activation and peaks within 0.5 h and then are quickly down-regulated (Hoffmann et al., 2002; Werner et al., 2008; Hao and Baltimore, 2009). Newly made IκBα is particularly important for two perspectives: (1) it binds to nuclear NF-κB, shuttling it to the cytoplasm and terminating the response to a pulse of induction shorter than 30 min; and (2) if induction persists longer, it generates oscillations of active, nuclear NF-κB (Hoffmann et al., 2002). A20 provides a rheostat function, quickly reducing the secondary phases of NF-κB activity and possibly serving as a signaling cross talk mediator (Hoffmann et al., 2002; Werner et al., 2008). In contrast, miR-146a expression in TCR-stimulated T cells gradually increases through the time course of T cell activation (Fig. 4 B). These results put miR-146a into the group III NF-κB late responsive genes (Hao and Baltimore, 2009) and imply that it may have a gradually increased impact on NF-κB activity over the later course of induction. This notion is supported by our results of time course comparison of NF-κB activation in WT and miR-146a−/− T cells after TCR stimulation (not depicted). Similar to A20, miR-146a targets signaling molecules in the NF-κB pathway (e.g., TRAF6 and IRAK1) that are shared with other signaling pathways and therefore may have a similar rheostat function in addition to regulating NF-κB activation and may also serve as a mediator to modulate the cross talk between NF-κB pathways and other signaling pathways. For T cells, these three negative feedback regulators may form a temporal control to ensure proper resolution of T cell response after antigen stimulation: IκBα providing a potent but transient negative control at the initial phase of T cell activation, followed by A20 acting mainly at the early phase of T cell activation, and miR-146a providing a long-lasting negative control at the late phase. This working hypothesis is supported by our observation that miR-146a deficiency has a more profound impact on the late-phase (contraction phase and stabilization phase) antigen-specific T cell response in vivo (Fig. 1 B). Unfortunately, the in vivo T cell response data are lacking for mice defective in IκBα or A20, likely because of their early lethality (Beg et al., 1995; Klement et al., 1996; Lee et al., 2000). New mouse models that are conditionally knocked out of these two genes in the T cell compartment, alone or in combination with miR-146a deficiency, will provide valuable insights into the precise role of each of the NF-κB negative feedback regulators and their joint temporal control of the resolution of T cell response.
Our study demonstrated TRAF6 and IRAK1 to be bona fide targets of miR-146a in T cells that presumably mediate part or all of the regulation of TCR-induced NF-κB activity, extending our previous findings that these two signaling adaptor molecules are targets of miR-146a in monocytes and B cells (Taganov et al., 2006; Boldin et al., 2011). In TCR signaling to NF-κB, TRAF6 has been shown to mediate the signal transmission from the CBM signalosome to IKK through Lys-63–linked polyubiquitination of IKKγ (NEMO; Sun et al., 2004). Interaction of IRAK1 with the TCR to NF-κB signaling cascade remains to be demonstrated, but by analogy to its position in NF-κB signaling cascades downstream of TLR (Toll-like receptor) and the IL-1R families in innate immune cells, it may also be positioned between the CBM signalosome and IKK (Oeckinghaus et al., 2011). It is noteworthy that aside from the NF-κB pathway, TRAF6 and IRAK1 participate in other signaling pathways that are involved in T cell responses (King et al., 2006; Maitra et al., 2009; Pearce et al., 2009). For instance, TRAF6 regulates CD4+ T cell suppression by Treg cells through the PI3K (phosphatidylinositol 3-kinase)–Akt pathway (King et al., 2006), and it regulates CD8+ T cell memory development through the fatty acid metabolism pathway (Pearce et al., 2009). IRAK1 has been implicated in regulation of the differentiation of Th17 and Treg cells through modulating the relative activity of STAT3 and NFAT signaling pathways (Maitra et al., 2009). Therefore, targeting TRAF6 and IRAK1 allows miR-146a to modulate the cross talk between NF-κB and multiple other signaling pathways, supporting its rheostat role as proposed by our NF-κB negative feedback temporal control model. Moreover, besides TRAF6 and IRAK1, several other putative targets of miR-146a have been evaluated in T cells and have been implicated in miR-146a regulation of T cell function, including Stat1, which is involved in the IFN-γ receptor signaling pathway, and FADD (Fas-associated death domain), which is involved in the T cell apoptosis pathway (Curtale et al., 2010; Lu et al., 2010). Although our observation of a largely abrogated miR-146a regulatory function in T cells defective in the NF-κB subunit p50 strongly suggests that modulation of NF-κB activity through targeting TRAF6/IRAK1 is a predominant mechanism that miR-146a utilizes to regulate T cell activation, it does not exclude the participation of other miR-146a targets and their related signaling pathways. In fact, the multi-targeting nature of miRNAs enables them to simultaneously regulate multiple signaling events. The identification of further miR-146a targets in T cells, the dissection of miR-146a regulatory function into its modulation of individual target molecules, and the characterization of its overall modulation of specialized signaling network that lead to its control of T cell responses under various physiological and pathological conditions will be interesting topics for future study.
Our study indicates miR-146a to be a T cell–autonomous factor that not only regulates the resolution of acute T cell response but also regulates the resolution of chronic T cell hyperinflammatory response and controls the development of T cell–associated autoimmunity. Previously, we have shown that in miR-146a−/− mice, deficiency of miR-146a in other cell types may contribute to a hyperinflammatory autoimmune phenotype (Lu et al., 2010; Boldin et al., 2011; Zhao et al., 2011). First, the myeloid overproliferation and malignancies in miR-146a−/− mice generates an overall proinflammatory environment (Zhao et al., 2011); second, miR-146a−/− macrophages tend to produce an excess amount of proinflammatory cytokines (e.g., TNF and IL-6) upon stimulation (Boldin et al., 2011); third, miR-146a−/− Treg cells exhibit impaired capacity to suppress Th1 response (Lu et al., 2010). All three defects are well-established contributors to the development of autoimmune disease. Collectively, miR-146a provides a potent negative control for the chronic T cell hyperinflammatory response and the development of autoimmunity through multiple T cell–intrinsic and –extrinsic mechanisms. Notably, miR-146a dysregulation has been observed in several human autoimmune diseases, including rheumatoid arthritis and systemic lupus erythematosus (Nakasa et al., 2008; Stanczyk et al., 2008; Tang et al., 2009), implicating miR-146a in the development of these human diseases and suggesting it as a potential therapeutic target treating these diseases.
Consistent with our findings in mouse T cells, TCR stimulation has been implicated in the induction of miR-146a expression in human T cells, and the forced expression of miR-146a in the human Jurkat T cell line has been shown to affect IL-2 production and activation-induced cell death (Curtale et al., 2010), supporting the role of miR-146a as a feedback regulator in human T cell activation. The interaction of miR-146a with the NF-κB signaling pathway in human T cells and its physiological role in regulating T cell responses in humans during infection and during development of autoimmune diseases are interesting research directions to pursue.
miRNAs as a new class of posttranscriptional regulators are emerging as important players in the immune system. In T cells, a growing list of miRNAs have been implicated in regulating various stages of T cell activation, including miR-181a regulation of TCR sensitivity to antigen (Li et al., 2007) and miR-155 regulation of inflammatory T cell development (Rodriguez et al., 2007; Thai et al., 2007; O’Connell et al., 2010). Our study provides the first in vivo evidence of miR-146a being a critical physiological controller of the resolution of T cell responses, mediated through its targeting of a key signaling pathway downstream of TCR stimulation. These findings extend the regulatory function of miRNAs to cover this important stage of T cell activation and suggest miRNAs as attractive therapeutic targets for T cell–based vaccine development and for the treatment of autoimmune disorders and malignancies.
MATERIALS AND METHODS
Mice and materials.
C57BL/6J (B6), B6.SJL-PtprcaPepcb/BoyJ (CD45.1), and B6;129S7-Rag1tm1mom/J (RAG1−/−) mice were purchased from the Jackson Laboratory. OT1 Tg mice and p50−/− mice (provided by S. Hao, California Institute of Technology, Pasadena, CA) were bred at the California Institute of Technology. miR-146a−/− mice in C57BL/6J background were generated and maintained at the California Institute of Technology (Boldin et al., 2011). p50−/−miR-146a−/− mice were generated through breeding p50−/− and miR-146a−/− mice (Zhao et al., 2011). OT1 Tg mice deficient of miR-146a were generated through breeding OT1 Tg mice with miR-146a−/− mice. All mice were housed in the California Institute of Technology animal facility in accordance with institute regulations. 6–8-wk-old female mice were used for all experiments unless otherwise indicated. Animal experiments were approved by the Institutional Animal Care and Use Committees of the California Institute of Technology.
OVA257–269 peptide was purchased from GenScript. Polybrene was purchased from Millipore. MACS sorting reagents were purchased from Miltenyi Biotec.
Antibodies and flow cytometry.
Stimulatory anti–mouse CD3 (clone 145-2C11) and anti–mouse CD28 (clone 37.51) antibodies and neutralizing anti–mouse IL-2 (clone JES6-1A12) and isotype control (rat IgG2a, κ) antibodies were purchased from BioLegend. Fluorochrome-conjugated antibodies specific for mouse CD4, CD8, CD25 (IL-2Rα), CD69, CD62L, and CD44 were purchased from BioLegend and for mouse TCR Vβ5 and Annexin V were purchased from BD. Fc Block (anti–mouse CD16/32) was purchased from BioLegend. Cells were stained as previously described (Yang and Baltimore, 2005) and analyzed using either a FACSCalibur flow cytometer (BD) or a MACSQuant (Miltenyi Biotec). FlowJo software (Tree Star) was used to analyze the data.
miRNA quantitative RT-PCR.
Total RNA was isolated using TRIZOL reagent (Invitrogen). miR-146a expression was measured using TaqMan miRNA Assays (Applied Biosystems) according to the manufacturer’s instructions using a 7300 Real-time PCR thermocycler (Azco Biotech). snoRNA202 was used as internal control. The relative expression of miR-146a was calculated by the 2ΔΔCT method and was presented as the fold induction relative to 210 snoRNA202.
Messenger RNA (mRNA) quantitative RT-PCR.
Total RNA was isolated using TRIZOL reagent (Invitrogen). cDNA was prepared using a SuperScript III First-Strand Synthesis kit (Invitrogen). Gene expression was measured using TaqMan Gene Expression Assays (Applied Biosystems) according to the manufacturer’s instructions using a 7300 Real-time PCR thermocycler. Ube2d2 was used as an internal control because of its relative constant mRNA level during T cell activation (Hamalainen et al., 2001). The relative expression of the mRNA of the gene of interest was calculated by the 2ΔΔCT method and was presented as the fold induction relative to 20 Ube2d2.
EMSA.
EMSA was performed as previously described (Hoffmann et al., 2002). Nuclear protein extracts were prepared using Nuclear Protein Extraction kit (Sigma-Aldrich). Nuclear extracts containing equal amounts of protein were tested for binding to an NF-κB probe containing a consensus κB-binding site (5′-AGCTTGCTACAAGGGACTTTCCGCTGTCTACTTT-3′; NF-κB–binding sequence in bold). In antibody supershift assays to identify components of NF-κB complexes, 5 µg of purified antibodies specific to each NF-κB components (anti-p50 [Active Motif] and anti-p65(C-20) and anti–c-Rel(C) [Santa Cruz Biotechnology, Inc.]) was preincubated with nuclear extracts for 15 min on ice before addition of the NF-κB probe.
Western blot.
Western blot was performed as previously described (Boldin et al., 2011). Total protein was extracted using RIPA buffer (Thermo Fisher Scientific) supplemented with Protease Inhibitor Cocktail (Roche). Nuclear protein was extracted using the Nuclear Protein Extraction kit (Sigma-Aldrich). Protein extracts were resolved on 10% Bis-Tris NuPAGE gel. The following antibodies were used to blot for the protein of interest: anti–mouse IRAK1 (Cell Signaling Technology), anti–mouse TRAF6 (MBL), anti–NF-κB p50 (Abcam), and anti–NF-κB p65 and c-Rel (Santa Cruz Biotechnology, Inc.). β-Actin or PLC-γ1 (Santa Cruz Biotechnology, Inc.) was used as internal control for total protein extracts, and Lamin A (Santa Cruz Biotechnology, Inc.) was used as internal control for nuclear protein extracts. The data were analyzed using Fluorchem SP software (Quansys Biosciences).
Immunohistology.
Tissues collected from the experimental mice were fixed with 10% neutral-buffered formalin and embedded in paraffin for sectioning. Hematoxylin and eosin staining and anti–mouse CD3 (clone 2C-11; BioLegend) immunostaining were performed by Pacific Pathology Inc. using company standard procedures.
MIG–miR-146a, MIG-TRAF6, and MIG-IRAK1 retroviruses and T cell transduction.
The MIG–miR-146a, MIG-TRAF6, and MIG-IRAK1 constructs were generated by inserting into the MIG retroviral vector (Yang and Baltimore, 2005) the mouse miR-146a coding sequence or mouse TRAF6 or IRAK1 gene cDNAs. Retroviruses were made using HEK293.T cells as previously described (Yang and Baltimore, 2005). For T cell transduction, SP and LN cells were stimulated with soluble anti-CD3 and anti-CD28 (1 µg/ml each). On days 1 and 2 of the stimulation, cells were spin infected with retroviral-containing supernatant supplemented with 10 µg/ml polybrene for 90 min at 770 g at 30°C.
siRNA transfection of T cells.
SP/LN cells were cultured at 5 × 105 cells/well in a round-bottom 96-well plate in the presence of soluble anti-CD3 + anti-CD28 (1 µg/ml each). Cells were treated with 1 µM Accell SMARTpool siRNA specific for either mouse TRAF6 or IRAK1 or nonsilencing control in Accell delivery media according to the manufacturer’s instructions (Thermo Fisher Scientific). Cells were analyzed after 72-h culture.
In vivo acute antigen-specific T cell response assays.
Resting WT or miR-146a−/− OVA-specific OT1 T cells were purified from the SP and LN cells harvested from WT OT1 Tg mice or OT1 Tg mice deficient of the miR-146a gene through MACS sorting (positive selection for CD8+ cells). 2–5 × 106 purified WT or miR-146a−/− OT1 T cells were adoptively transferred into CD45.1 congenic recipient mice through i.v. injection (day −1). The next day (day 0), the recipient mice received a prime immunization with 5 × 106 CD45.1 mouse bone marrow DCs loaded with OVA257–269 peptide by s.c. injection as previously described (Yang et al., 2008). 46 d later (day 46), the recipient mice were given a boost immunization with 5 × 106 CD45.1 mouse bone marrow DCs loaded with OVA257–269 by s.c. injection. During the experiment, the recipient mice were periodically bled, and their PB cells were analyzed for the presence of WT or miR-146a−/− OT1 T cells (gated as CD8+Vβ5+CD45.2+) and the surface activation phenotype and apoptosis (through Annexin V staining) of these T cells using flow cytometry.
In vivo chronic inflammatory autoimmune T cell response assays.
WT and miR-146a−/− SP and LN cells containing 10 × 106 T cells depleted of CD4+CD25+ Treg cells through MACS sorting were adoptively transferred into RAG1−/− recipient mice separately via i.v. injection. On day 28 after adoptive transfer, recipient mice received i.p. injection of BrdU (1 mg/mouse). On day 29 (16 h after BrdU injection), SP cells were harvested from recipient mice and were analyzed for the presence of WT or miR-146a−/− CD4 and CD8 T cells (gated as CD4+ and CD8+, respectively) and the in vivo proliferation (via BrdU incorporation), apoptosis (via Annexin V staining), and surface activation marker expression of these T cells using flow cytometry. Tissues were also collected and fixed with 10% neutral-buffered formalin for immunohistology analysis of tissue pathology and T cell infiltration.
In vitro T cell activation assays.
For mixed SP/LN T cell stimulation, cells were cultured at 2 × 106 cells/well in a 24-well plate or at 2 × 105 cells/well in a round-bottom 96-well plate in the presence of soluble anti-CD3 + anti-CD28 (1 µg/ml each) with or without addition of anti–IL-2 neutralizing antibody or its isotype control antibody (10 µg/ml) for 4 d. At the indicated times, cells cultured in 24-well plate were collected and assayed by flow cytometry for surface activation marker expression and for apoptosis using an Annexin V Apoptosis Detection kit (BD) according to the manufacturer’s instructions. For cells cultured in a 96-well plate, at the indicated times, cell culture supernatants were collected and assayed for effector cytokine (IL-2, IFN-γ, and IL-17A) production by ELISA, and the cells were pulsed with [3H]thymidine to assay for proliferation.
For purified T cell stimulation, resting CD4 and CD8 T cells were purified from SP and LN cells through MACS sorting (positive selection for CD4+ and CD8+ cells, respectively) and were cultured at 105 cells/well in a 96-well round-bottom plate precoated with 10 µg/ml anti-CD3 + 1 µg/ml anti-CD28. At the indicated time points, cell culture supernatants were collected and assayed for effector cytokine (IL-2, IFN-γ, and IL-17A) production by ELISA, and the cells were pulsed with [3H]thymidine to assay for proliferation.
Statistic analysis.
Student’s two-tailed t test was used for paired comparisons. Data are presented as mean ± SEM, unless otherwise indicated. P < 0.05 was considered significant.
Acknowledgments
We thank the California Institute of Technology animal facility for providing animal support, Shengli Hao for providing p50−/− mice, Pin Wang, Shengli Hao, and Michael Bethune for critical reading of this manuscript, and all members of the Baltimore group for insightful discussions.
This work was supported by the National Institutes of Health P01 CA132681A and 5R01AI079243-02 grants.
D. Baltimore is a director and chairman of the Scientific Advisory board of Regulus Therapeutics Inc., a biotech company developing miRNA-based drugs. Authors have no further conflicting interests.
Footnotes
Abbreviations used:
- CBM
- CARD11–Bcl-10–MALT1
- EMSA
- electrophoretic mobility shift assay
- miRNA
- microRNA
- mRNA
- messenger RNA
- PB
- peripheral blood
- Q-PCR
- quantitative PCR
- siRNA
- small interfering RNA
- SP
- spleen
- Tg
- transgenic
References
- Ambros V. 2004. The functions of animal microRNAs. Nature. 431:350–355 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- Baltimore D., Boldin M.P., O’Connell R.M., Rao D.S., Taganov K.D. 2008. MicroRNAs: new regulators of immune cell development and function. Nat. Immunol. 9:839–845 10.1038/ni.f.209 [DOI] [PubMed] [Google Scholar]
- Bartel D.P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116:281–297 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Beg A.A., Sha W.C., Bronson R.T., Baltimore D. 1995. Constitutive NF-kappa B activation, enhanced granulopoiesis, and neonatal lethality in I kappa B alpha-deficient mice. Genes Dev. 9:2736–2746 10.1101/gad.9.22.2736 [DOI] [PubMed] [Google Scholar]
- Boldin M.P., Taganov K.D., Rao D.S., Yang L., Zhao J.L., Kalwani M., Garcia-Flores Y., Luong M., Devrekanli A., Xu J., et al. 2011. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J. Exp. Med. 208:1189–1201 10.1084/jem.20101823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassin C., Kocur M., Pott J., Duerr C.U., Gütle D., Lotz M., Hornef M.W. 2010. miR-146a mediates protective innate immune tolerance in the neonate intestine. Cell Host Microbe. 8:358–368 10.1016/j.chom.2010.09.005 [DOI] [PubMed] [Google Scholar]
- Curtale G., Citarella F., Carissimi C., Goldoni M., Carucci N., Fulci V., Franceschini D., Meloni F., Barnaba V., Macino G. 2010. An emerging player in the adaptive immune response: microRNA-146a is a modulator of IL-2 expression and activation-induced cell death in T lymphocytes. Blood. 115:265–273 10.1182/blood-2009-06-225987 [DOI] [PubMed] [Google Scholar]
- Hamalainen H.K., Tubman J.C., Vikman S., Kyrölä T., Ylikoski E., Warrington J.A., Lahesmaa R. 2001. Identification and validation of endogenous reference genes for expression profiling of T helper cell differentiation by quantitative real-time RT-PCR. Anal. Biochem. 299:63–70 10.1006/abio.2001.5369 [DOI] [PubMed] [Google Scholar]
- Ghosh S., Hayden M.S. 2008. New regulators of NF-kappaB in inflammation. Nat. Rev. Immunol. 8:837–848 10.1038/nri2423 [DOI] [PubMed] [Google Scholar]
- Hannum C., Freed J.H., Tarr G., Kappler J., Marrack P. 1984. Biochemistry and distribution of the T cell receptor. Immunol. Rev. 81:161–176 10.1111/j.1600-065X.1984.tb01109.x [DOI] [PubMed] [Google Scholar]
- Hao S., Baltimore D. 2009. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat. Immunol. 10:281–288 10.1038/ni.1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A., Levchenko A., Scott M.L., Baltimore D. 2002. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 298:1241–1245 10.1126/science.1071914 [DOI] [PubMed] [Google Scholar]
- Hou J., Wang P., Lin L., Liu X., Ma F., An H., Wang Z., Cao X. 2009. MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J. Immunol. 183:2150–2158 10.4049/jimmunol.0900707 [DOI] [PubMed] [Google Scholar]
- Isakov N., Altman A. 2002. Protein kinase C(theta) in T cell activation. Annu. Rev. Immunol. 20:761–794 10.1146/annurev.immunol.20.100301.064807 [DOI] [PubMed] [Google Scholar]
- Itoh M., Takahashi T., Sakaguchi N., Kuniyasu Y., Shimizu J., Otsuka F., Sakaguchi S. 1999. Thymus and autoimmunity: production of CD25+CD4+ naturally anergic and suppressive T cells as a key function of the thymus in maintaining immunologic self-tolerance. J. Immunol. 162:5317–5326 [PubMed] [Google Scholar]
- Jost P.J., Ruland J. 2007. Aberrant NF-kappaB signaling in lymphoma: mechanisms, consequences, and therapeutic implications. Blood. 109:2700–2707 [DOI] [PubMed] [Google Scholar]
- Karin M., Greten F.R. 2005. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat. Rev. Immunol. 5:749–759 10.1038/nri1703 [DOI] [PubMed] [Google Scholar]
- King C.G., Kobayashi T., Cejas P.J., Kim T., Yoon K., Kim G.K., Chiffoleau E., Hickman S.P., Walsh P.T., Turka L.A., Choi Y. 2006. TRAF6 is a T cell-intrinsic negative regulator required for the maintenance of immune homeostasis. Nat. Med. 12:1088–1092 10.1038/nm1449 [DOI] [PubMed] [Google Scholar]
- Klement J.F., Rice N.R., Car B.D., Abbondanzo S.J., Powers G.D., Bhatt P.H., Chen C.H., Rosen C.A., Stewart C.L. 1996. IkappaBalpha deficiency results in a sustained NF-kappaB response and severe widespread dermatitis in mice. Mol. Cell. Biol. 16:2341–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.G., Boone D.L., Chai S., Libby S.L., Chien M., Lodolce J.P., Ma A. 2000. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 289:2350–2354 10.1126/science.289.5488.2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Verma I.M. 2002. NF-kappaB regulation in the immune system. Nat. Rev. Immunol. 2:725–734 10.1038/nri910 [DOI] [PubMed] [Google Scholar]
- Li Q.J., Chau J., Ebert P.J., Sylvester G., Min H., Liu G., Braich R., Manoharan M., Soutschek J., Skare P., et al. 2007. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 129:147–161 10.1016/j.cell.2007.03.008 [DOI] [PubMed] [Google Scholar]
- Lu L.F., Boldin M.P., Chaudhry A., Lin L.L., Taganov K.D., Hanada T., Yoshimura A., Baltimore D., Rudensky A.Y. 2010. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 142:914–929 10.1016/j.cell.2010.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra U., Davis S., Reilly C.M., Li L. 2009. Differential regulation of Foxp3 and IL-17 expression in CD4 T helper cells by IRAK-1. J. Immunol. 182:5763–5769 10.4049/jimmunol.0900124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek T.R. 2008. The biology of interleukin-2. Annu. Rev. Immunol. 26:453–479 10.1146/annurev.immunol.26.021607.090357 [DOI] [PubMed] [Google Scholar]
- Monticelli S., Ansel K.M., Xiao C., Socci N.D., Krichevsky A.M., Thai T.H., Rajewsky N., Marks D.S., Sander C., Rajewsky K., et al. 2005. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 6:R71 10.1186/gb-2005-6-8-r71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahid M.A., Pauley K.M., Satoh M., Chan E.K. 2009. miR-146a is critical for endotoxin-induced tolerance: Implication in innate immunity. J. Biol. Chem. 284:34590–34599 10.1074/jbc.M109.056317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakasa T., Miyaki S., Okubo A., Hashimoto M., Nishida K., Ochi M., Asahara H. 2008. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum. 58:1284–1292 10.1002/art.23429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell R.M., Kahn D., Gibson W.S., Round J.L., Scholz R.L., Chaudhuri A.A., Kahn M.E., Rao D.S., Baltimore D. 2010. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 33:607–619 10.1016/j.immuni.2010.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeckinghaus A., Hayden M.S., Ghosh S. 2011. Crosstalk in NF-κB signaling pathways. Nat. Immunol. 12:695–708 10.1038/ni.2065 [DOI] [PubMed] [Google Scholar]
- Pearce E.L., Walsh M.C., Cejas P.J., Harms G.M., Shen H., Wang L.S., Jones R.G., Choi Y. 2009. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 460:103–107 10.1038/nature08097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A., Vigorito E., Clare S., Warren M.V., Couttet P., Soond D.R., van Dongen S., Grocock R.J., Das P.P., Miska E.A., et al. 2007. Requirement of bic/microRNA-155 for normal immune function. Science. 316:608–611 10.1126/science.1139253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruland J. 2011. Return to homeostasis: downregulation of NF-κB responses. Nat. Immunol. 12:709–714 10.1038/ni.2055 [DOI] [PubMed] [Google Scholar]
- Schulze-Luehrmann J., Ghosh S. 2006. Antigen-receptor signaling to nuclear factor kappa B. Immunity. 25:701–715 10.1016/j.immuni.2006.10.010 [DOI] [PubMed] [Google Scholar]
- Stanczyk J., Pedrioli D.M., Brentano F., Sanchez-Pernaute O., Kolling C., Gay R.E., Detmar M., Gay S., Kyburz D. 2008. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 58:1001–1009 10.1002/art.23386 [DOI] [PubMed] [Google Scholar]
- Sun L., Deng L., Ea C.K., Xia Z.P., Chen Z.J. 2004. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol. Cell. 14:289–301 10.1016/S1097-2765(04)00236-9 [DOI] [PubMed] [Google Scholar]
- Taganov K.D., Boldin M.P., Chang K.J., Baltimore D. 2006. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA. 103:12481–12486 10.1073/pnas.0605298103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Luo X., Cui H., Ni X., Yuan M., Guo Y., Huang X., Zhou H., de Vries N., Tak P.P., et al. 2009. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 60:1065–1075 10.1002/art.24436 [DOI] [PubMed] [Google Scholar]
- Thai T.H., Calado D.P., Casola S., Ansel K.M., Xiao C., Xue Y., Murphy A., Frendewey D., Valenzuela D., Kutok J.L., et al. 2007. Regulation of the germinal center response by microRNA-155. Science. 316:604–608 10.1126/science.1141229 [DOI] [PubMed] [Google Scholar]
- Tsitsiou E., Lindsay M.A. 2009. microRNAs and the immune response. Curr. Opin. Pharmacol. 9:514–520 10.1016/j.coph.2009.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhapurapu S., Karin M. 2009. Regulation and function of NF-kappaB transcription factors in the immune system. Annu. Rev. Immunol. 27:693–733 10.1146/annurev.immunol.021908.132641 [DOI] [PubMed] [Google Scholar]
- Werner S.L., Kearns J.D., Zadorozhnaya V., Lynch C., O’Dea E., Boldin M.P., Ma A., Baltimore D., Hoffmann A. 2008. Encoding NF-kappaB temporal control in response to TNF: distinct roles for the negative regulators IkappaBalpha and A20. Genes Dev. 22:2093–2101 10.1101/gad.1680708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A.E., Perry M.M., Moschos S.A., Larner-Svensson H.M., Lindsay M.A. 2008. Role of miRNA-146a in the regulation of the innate immune response and cancer. Biochem. Soc. Trans. 36:1211–1215 10.1042/BST0361211 [DOI] [PubMed] [Google Scholar]
- Yang L., Baltimore D. 2005. Long-term in vivo provision of antigen-specific T cell immunity by programming hematopoietic stem cells. Proc. Natl. Acad. Sci. USA. 102:4518–4523 10.1073/pnas.0500600102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Yang H., Rideout K., Cho T., Joo K.I., Ziegler L., Elliot A., Walls A., Yu D., Baltimore D., Wang P. 2008. Engineered lentivector targeting of dendritic cells for in vivo immunization. Nat. Biotechnol. 26:326–334 10.1038/nbt1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.L., Rao D.S., Boldin M.P., Taganov K.D., O’Connell R.M., Baltimore D. 2011. NF-kappaB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc. Natl. Acad. Sci. USA. 108:9184–9189 10.1073/pnas.1105398108 [DOI] [PMC free article] [PubMed] [Google Scholar]